Receptor-Targeted Carbon Nanodot Delivery through Polymer Caging and Click Chemistry-Supported LRP1 Ligand Attachment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Chemicals
2.2. OAA Synthesis
2.3. DBCO–PEG–Ligand Conjugate Synthesis
2.4. RCD Synthesis
2.5. Characterization of RCD
2.6. RCD@OAA Preparation
2.7. RCD@1696–Ligands and RCD@1696–PEG Preparation and Characterization
2.8. Ellman’s Assay
2.9. TCEP Assay
2.10. Cell Viability Assay
2.11. Cellular Internalization
2.12. Ligand Competition Assay
2.13. Statistical Analysis
3. Results
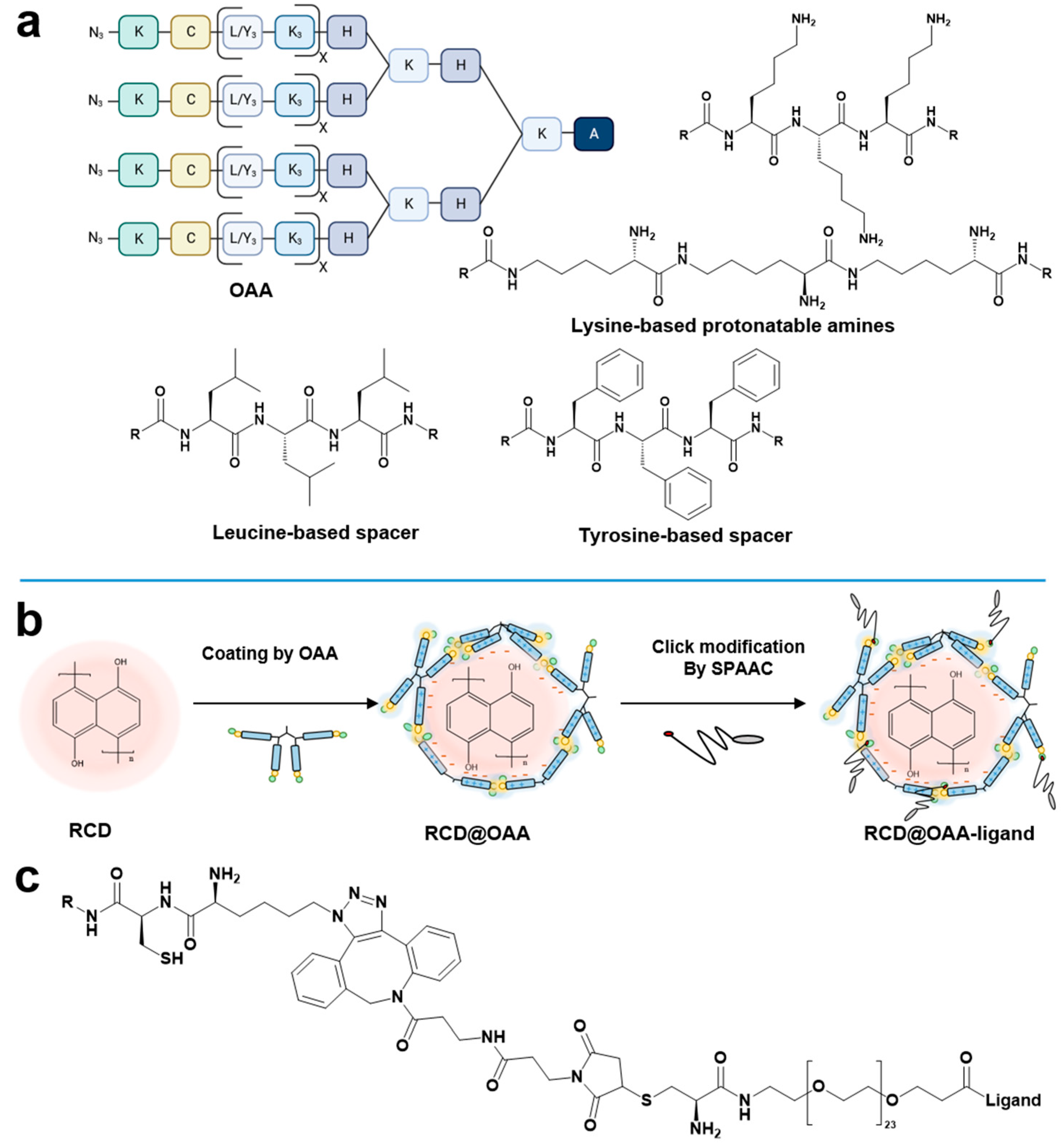
3.1. Design and Synthesis of Four-Armed OAA for Caging of RCD
- (i)
- Each arm of the OAAs contained two or three lysine tripeptides with corresponding numbers of positively charged amines, facilitating successful layer coating of the negatively charged hydroxynaphthalene-derived RCDs via the electrostatic interactions. Preferably, the lysine units were incorporated as ε-amino amidated lysine instead of the standard α-amino amidated lysine residues, increasing the contour length of each arm without changing the number of amino acids and positively charged amines, respectively.
- (ii)
- To enhance the stability of RCD@OAA, alternating with two lysine tripeptide blocks, two blocks of tyrosine or leucine tripeptides were included, serving either as aromatic or hydrophobic interaction domains stabilizing the coating of RCDs.
- (iii)
- Thiol groups of cysteines located close to the four N-terminal ends were included that should cage the coated RCDs by disulfide bridge formation.
- (iv)
- To enable controlled shielding and targeting of caged RCDs with ligand–PEG–DBCO via SPAAC, N-terminal azidolysines were included.
3.2. LRP1 Targeted DBCO–PEG–Ligand Peptide Conjugates
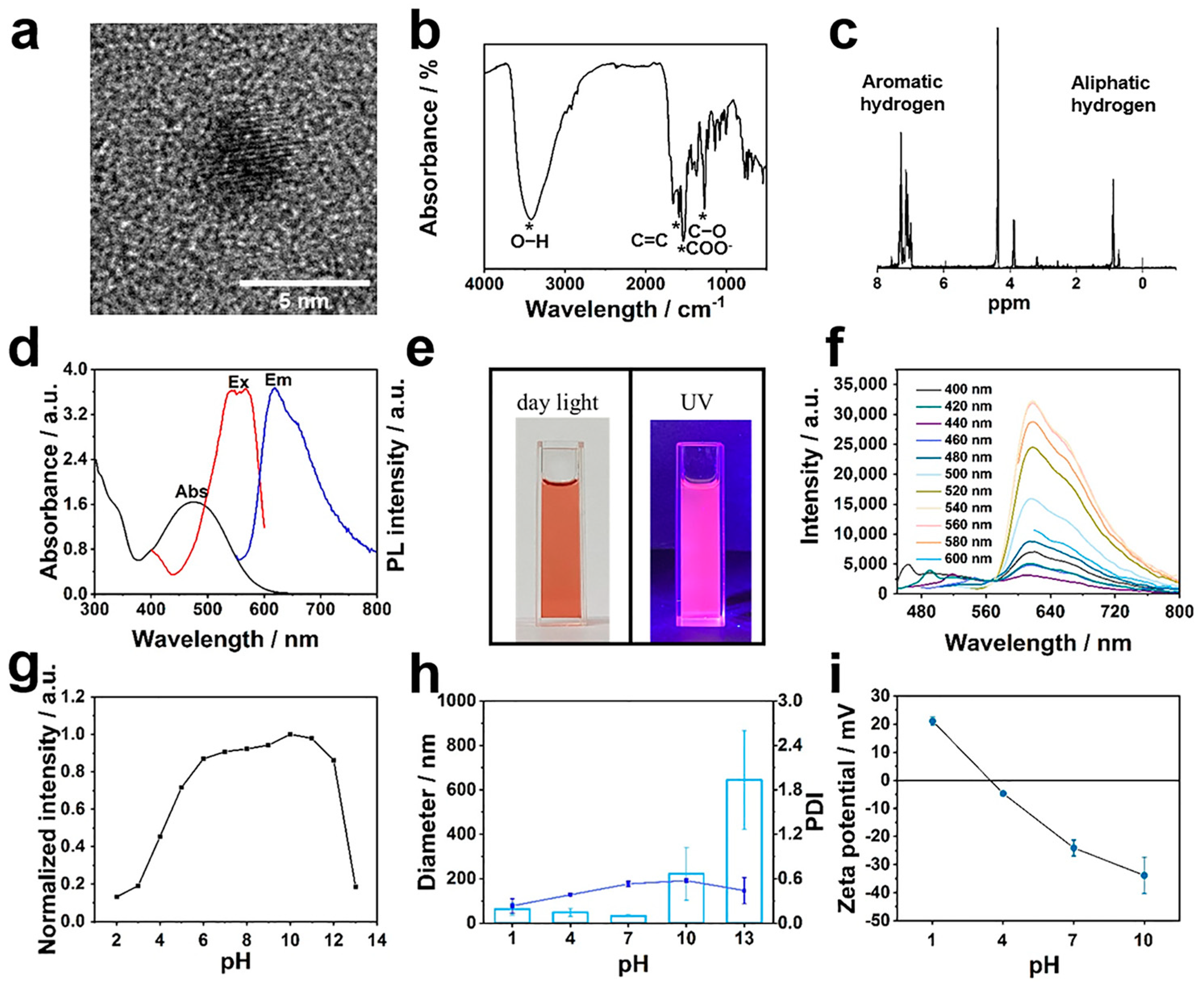
3.3. RCD Synthesis and Characterization
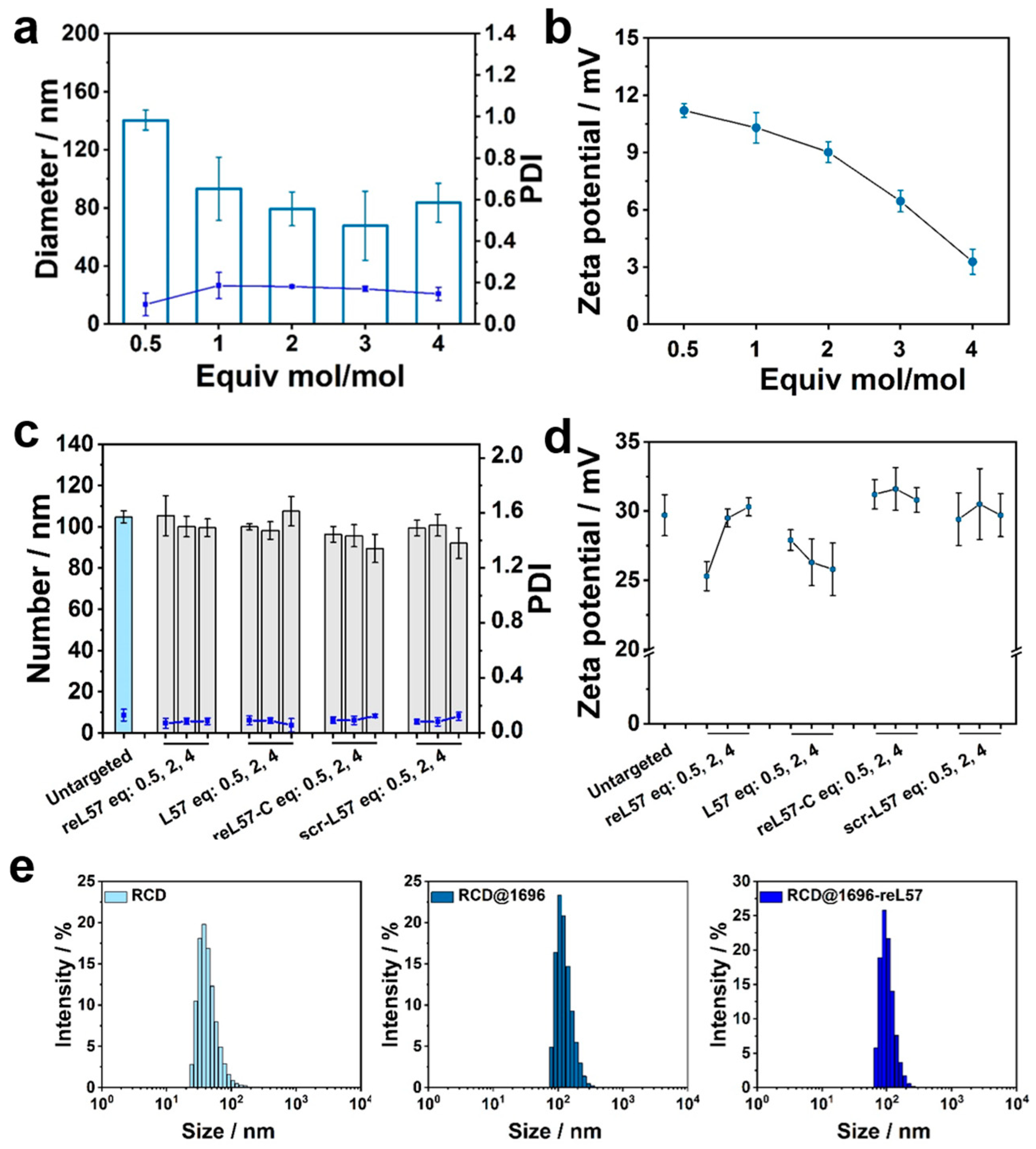
3.4. Screening of Four-Armed OAAs for RCD Coating
3.5. Click Modification of RCD@1696 with DBCO–PEG–Ligand Conjugates
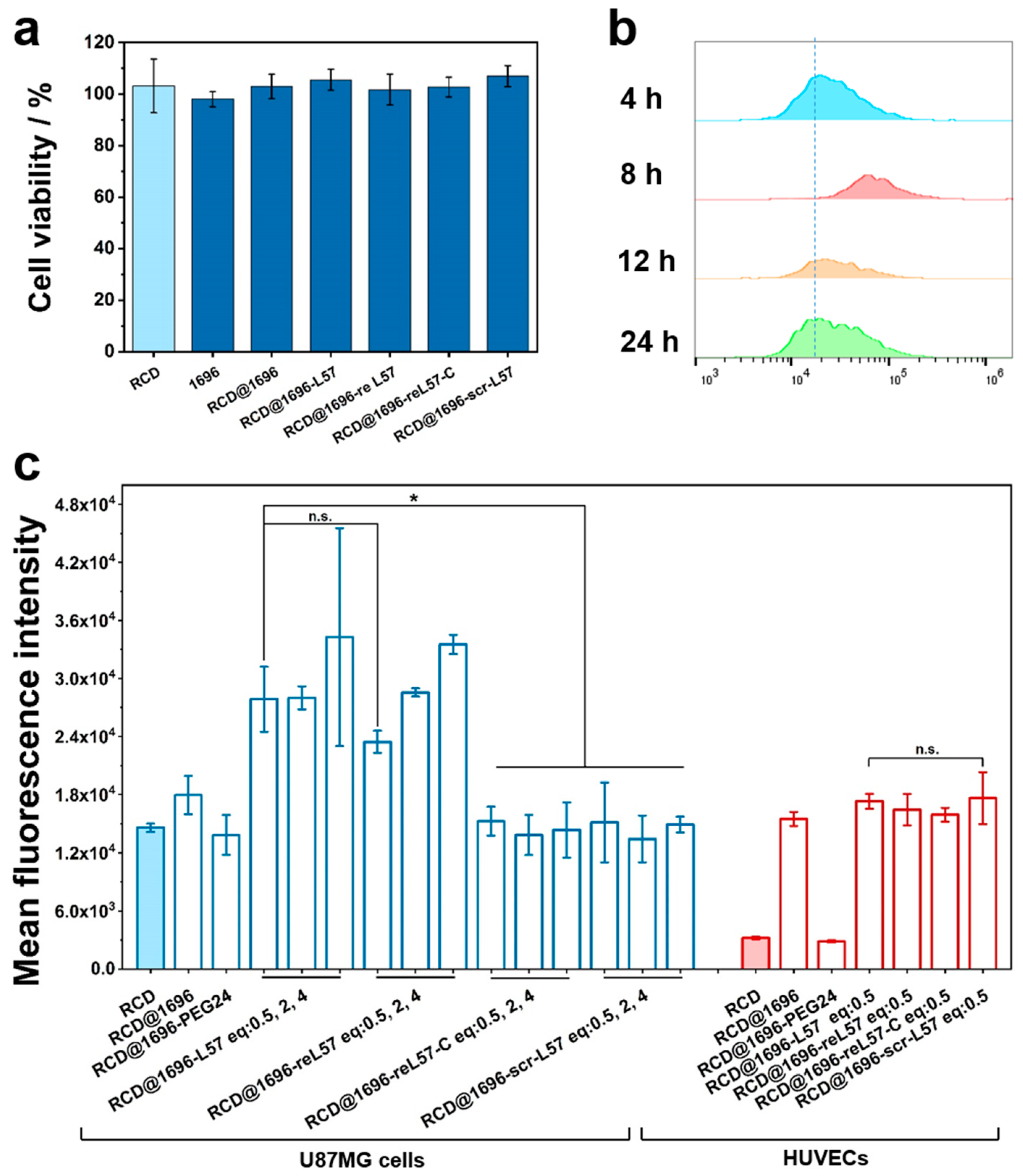
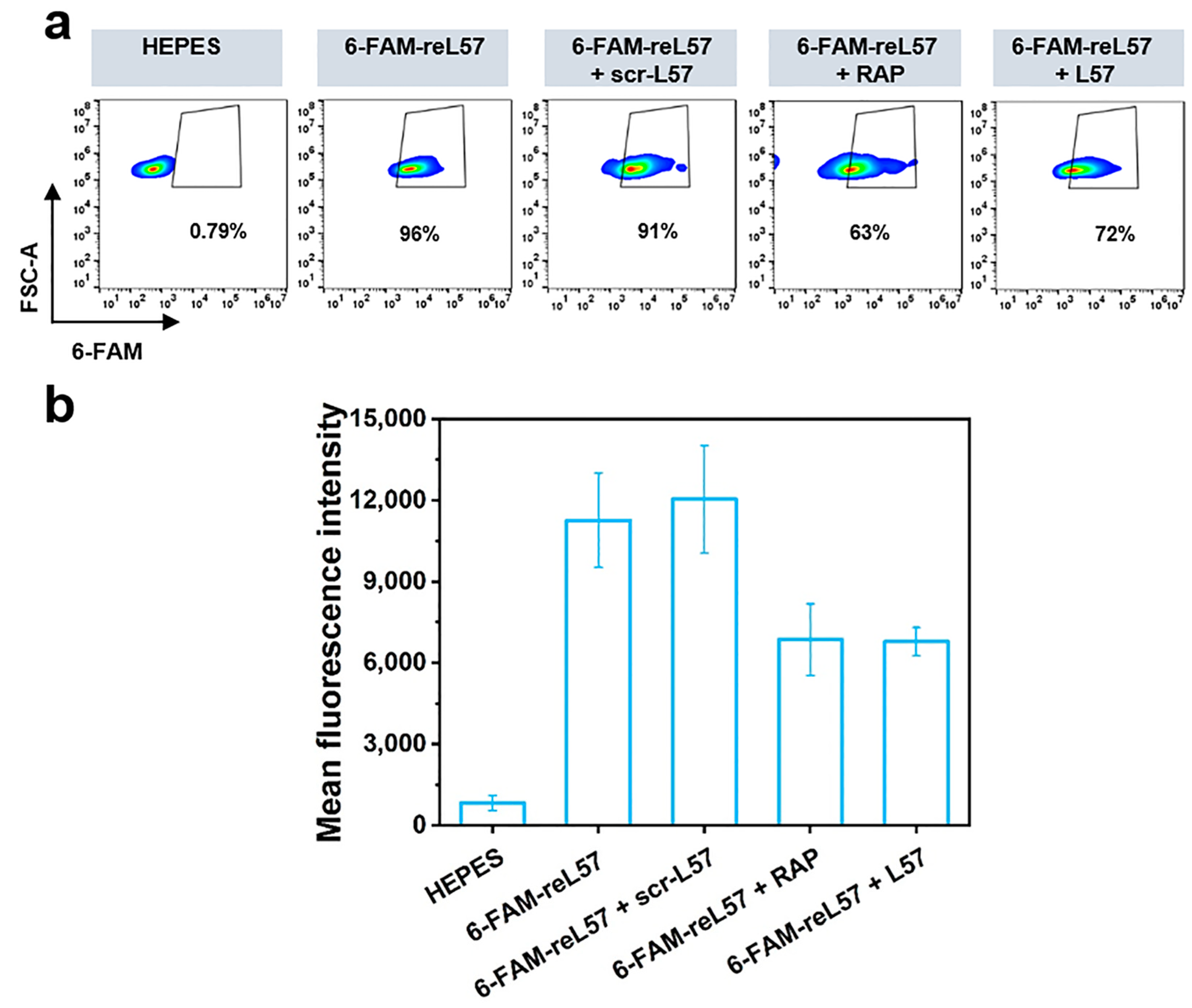
3.6. Evaluation of reL57 as Targeting Ligand for Receptor-Mediated RCD Delivery
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ostrom, Q.T.; Price, M.; Neff, C.; Cioffi, G.; Waite, K.A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2015–2019. Neuro. Oncol. 2022, 24, v1–v95. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Wu, Z.; Zeng, F.; Cai, H.; Wang, D.; Gu, L.; Zhu, H.; Lui, S.; Guo, G.; Song, B.; et al. Retro-Enantio Isomer of Angiopep-2 Assists Nanoprobes across the Blood-Brain Barrier for Targeted Magnetic Resonance/Fluorescence Imaging of Glioblastoma. Signal Transduct. Target. Ther. 2021, 6, 309. [Google Scholar] [CrossRef] [PubMed]
- Demchenko, A.P. Photobleaching of Organic Fluorophores: Quantitative Characterization, Mechanisms, Protection. Methods Appl. Fluoresc. 2020, 8, 022001. [Google Scholar] [CrossRef] [PubMed]
- Helmerich, D.A.; Beliu, G.; Matikonda, S.S.; Schnermann, M.J.; Sauer, M. Photoblueing of Organic Dyes Can Cause Artifacts in Super-Resolution Microscopy. Nat. Methods 2021, 18, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Zhu, S.; Feng, T.; Yang, M.; Yang, B. Evolution and Synthesis of Carbon Dots: From Carbon Dots to Carbonized Polymer Dots. Adv. Sci. 2019, 6, 1901316. [Google Scholar] [CrossRef]
- Kütahya, C.; Zhai, Y.; Li, S.; Liu, S.; Li, J.; Strehmel, V.; Chen, Z.; Strehmel, B. Distinct Sustainable Carbon Nanodots Enable Free Radical Photopolymerization, Photo-ATRP and Photo-CuAAC Chemistry. Angew. Chem. Int. Ed. 2021, 60, 10983–10991. [Google Scholar] [CrossRef]
- Rigodanza, F.; Burian, M.; Arcudi, F.; Đorđević, L.; Amenitsch, H.; Prato, M. Snapshots into Carbon Dots Formation through a Combined Spectroscopic Approach. Nat. Commun. 2021, 12, 2640. [Google Scholar] [CrossRef]
- Lesani, P.; Mohamad Hadi, A.H.; Lu, Z.; Palomba, S.; New, E.J.; Zreiqat, H. Design Principles and Biological Applications of Red-Emissive Two-Photon Carbon Dots. Commun. Mater. 2021, 2, 108. [Google Scholar] [CrossRef]
- Lu, S.; Sui, L.; Liu, J.; Zhu, S.; Chen, A.; Jin, M.; Yang, B. Near-Infrared Photoluminescent Polymer–Carbon Nanodots with Two-Photon Fluorescence. Adv. Mater. 2017, 29, 1603443. [Google Scholar] [CrossRef]
- Li, D.; Han, D.; Qu, S.-N.; Liu, L.; Jing, P.-T.; Zhou, D.; Ji, W.-Y.; Wang, X.-Y.; Zhang, T.-F.; Shen, D.-Z. Supra-(Carbon Nanodots) with a Strong Visible to Near-Infrared Absorption Band and Efficient Photothermal Conversion. Light Sci. Appl. 2016, 5, e16120. [Google Scholar] [CrossRef]
- Guo, X.-L.; Ding, Z.-Y.; Deng, S.-M.; Wen, C.-C.; Shen, X.-C.; Jiang, B.-P.; Liang, H. A Novel Strategy of Transition-Metal Doping to Engineer Absorption of Carbon Dots for Near-Infrared Photothermal/Photodynamic Therapies. Carbon 2018, 134, 519–530. [Google Scholar] [CrossRef]
- Zhang, T.; Cheng, Q.; Lei, J.H.; Wang, B.; Chang, Y.; Liu, Y.; Xing, G.; Deng, C.; Tang, Z.; Qu, S. Constructing Oxygen-Related Defects in Carbon Nanodots with Janus Optical Properties: Noninvasive NIR Fluorescent Imaging and Effective Photocatalytic Therapy. Adv. Mater. 2023, 35, 2302705. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yuan, F.; Li, X.; Li, Y.; Zhong, H.; Fan, L.; Yang, S. 53% Efficient Red Emissive Carbon Quantum Dots for High Color Rendering and Stable Warm White-Light-Emitting Diodes. Adv. Mater. 2017, 29, 1702910. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Yan, L.; Cao, M.; Huang, L.; Yang, K.; Wu, S.; Lan, M.; Niu, G.; Zhang, W. Near-Infrared Light-Triggered Lysosome-Targetable Carbon Dots for Photothermal Therapy of Cancer. ACS Appl. Mater. Interfaces 2021, 13, 53610–53617. [Google Scholar] [CrossRef]
- Han, Y.; Liu, H.; Fan, M.; Gao, S.; Fan, D.; Wang, Z.; Chang, J.; Zhang, J.; Ge, K. Near-Infrared-II Photothermal Ultra-Small Carbon Dots Promoting Anticancer Efficiency by Enhancing Tumor Penetration. J. Colloid Interface Sci. 2022, 616, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Arvanitis, C.D.; Ferraro, G.B.; Jain, R.K. The Blood–Brain Barrier and Blood–Tumour Barrier in Brain Tumours and Metastases. Nat. Rev. Cancer 2020, 20, 26–41. [Google Scholar] [CrossRef]
- Hajal, C.; Offeddu, G.S.; Shin, Y.; Zhang, S.; Morozova, O.; Hickman, D.; Knutson, C.G.; Kamm, R.D. Engineered Human Blood–Brain Barrier Microfluidic Model for Vascular Permeability Analyses. Nat. Protoc. 2022, 17, 95–128. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Phua, S.Z.F.; Bindra, A.K.; Zhao, Y. Degradability and Clearance of Inorganic Nanoparticles for Biomedical Applications. Adv. Mater. 2019, 31, 1805730. [Google Scholar] [CrossRef] [PubMed]
- Ji, D.-K.; Reina, G.; Guo, S.; Eredia, M.; Samorì, P.; Ménard-Moyon, C.; Bianco, A. Controlled Functionalization of Carbon Nanodots for Targeted Intracellular Production of Reactive Oxygen Species. Nanoscale Horiz. 2020, 5, 1240–1249. [Google Scholar] [CrossRef]
- Simonneau, C.; Duschmalé, M.; Gavrilov, A.; Brandenberg, N.; Hoehnel, S.; Ceroni, C.; Lassalle, E.; Kassianidou, E.; Knoetgen, H.; Niewoehner, J.; et al. Investigating Receptor-Mediated Antibody Transcytosis Using Blood–Brain Barrier Organoid Arrays. Fluids Barriers CNS 2021, 18, 43. [Google Scholar] [CrossRef]
- Tashima, T. Smart Strategies for Therapeutic Agent Delivery into Brain across the Blood–Brain Barrier Using Receptor-Mediated Transcytosis. Chem. Pharm. Bull. 2020, 68, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Wang, B.; Teng, Y.; Ho, W.; Hu, B.; Boakye-Yiadom, K.O.; Xu, X.; Zhang, X.Q. Rational Design of Engineered H-Ferritin Nanoparticles with Improved siRNA Delivery Efficacy across an In Vitro Model of the Mouse BBB. Nanoscale 2022, 6449–6464. [Google Scholar] [CrossRef]
- Shi, X.; Wang, Z.; Ren, W.; Chen, L.; Xu, C.; Li, M.; Fan, S.; Xu, Y.; Chen, M.; Zheng, F.; et al. LDL Receptor-Related Protein 1 (LRP1), a Novel Target for Opening the Blood-Labyrinth Barrier (BLB). Signal Transduct. Target. Ther. 2022, 7, 175. [Google Scholar] [CrossRef] [PubMed]
- Benitez-Amaro, A.; Pallara, C.; Nasarre, L.; Rivas-Urbina, A.; Benitez, S.; Vea, A.; Bornachea, O.; de Gonzalo-Calvo, D.; Serra-Mir, G.; Villegas, S.; et al. Molecular Basis for the Protective Effects of Low-Density Lipoprotein Receptor-Related Protein 1 (LRP1)-Derived Peptides against LDL Aggregation. Biochim. Biophys. Acta Biomembr. 2019, 1861, 1302–1316. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Hashimoto, T.; Yabuki, C.; Nagae, Y.; Tachikawa, M.; Strickland, D.K.; Liu, Q.; Bu, G.; Basak, J.M.; Holtzman, D.M.; et al. The Low Density Lipoprotein Receptor-Related Protein 1 Mediates Uptake of Amyloid β Peptides in an In Vitro Model of the Blood-Brain Barrier Cells. J. Biol. Chem. 2008, 283, 34554–34562. [Google Scholar] [CrossRef] [PubMed]
- Terstappen, G.C.; Meyer, A.H.; Bell, R.D.; Zhang, W. Strategies for Delivering Therapeutics across the Blood–Brain Barrier. Nat. Rev. Drug Discov. 2021, 20, 362–383. [Google Scholar] [CrossRef]
- Hartl, N.; Adams, F.; Merkel, O.M. From Adsorption to Covalent Bonding: Apolipoprotein E Functionalization of Polymeric Nanoparticles for Drug Delivery Across the Blood–Brain Barrier. Adv. Ther. 2021, 4, 2000092. [Google Scholar] [CrossRef]
- Kim, H.S.; Lee, S.J.; Lee, D.Y. Milk Protein-Shelled Gold Nanoparticles with Gastrointestinally Active Absorption for Aurotherapy to Brain Tumor. Bioact. Mater. 2022, 8, 35–48. [Google Scholar] [CrossRef]
- Habib, S.; Singh, M. Angiopep-2-Modified Nanoparticles for Brain-Directed Delivery of Therapeutics: A Review. Polymers 2022, 14, 712. [Google Scholar] [CrossRef]
- Sakamoto, K.; Shinohara, T.; Adachi, Y.; Asami, T.; Ohtaki, T. A Novel LRP1-Binding Peptide L57 That Crosses the Blood Brain Barrier. Biochem. Biophys. Rep. 2017, 12, 135–139. [Google Scholar] [CrossRef]
- Shi, Y.; Lammers, T.; Storm, G.; Hennink, W.E. Physico-Chemical Strategies to Enhance Stability and Drug Retention of Polymeric Micelles for Tumor-Targeted Drug Delivery. Macromol. Biosci. 2017, 17, 1600160. [Google Scholar] [CrossRef] [PubMed]
- Ghezzi, M.; Pescina, S.; Padula, C.; Santi, P.; Del Favero, E.; Cantù, L.; Nicoli, S. Polymeric Micelles in Drug Delivery: An Insight of the Techniques for Their Characterization and Assessment in Biorelevant Conditions. J. Control. Release 2021, 332, 312–336. [Google Scholar] [CrossRef] [PubMed]
- Novo, L.; Rizzo, L.Y.; Golombek, S.K.; Dakwar, G.R.; Lou, B.; Remaut, K.; Mastrobattista, E.; Van Nostrum, C.F.; Jahnen-Dechent, W.; Kiessling, F.; et al. Decationized Polyplexes as Stable and Safe Carrier Systems for Improved Biodistribution in Systemic Gene Therapy. J. Control. Release 2014, 195, 162–175. [Google Scholar] [CrossRef] [PubMed]
- Röder, R.; Helma, J.; Preiß, T.; Rädler, J.O.; Leonhardt, H.; Wagner, E. Intracellular Delivery of Nanobodies for Imaging of Target Proteins in Live Cells. Pharm. Res. 2017, 34, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Leng, Q.; Mixson, A.J. Modified Branched Peptides with a Histidine-Rich Tail Enhance in Vitro Gene Transfection. Nucleic Acids Res. 2005, 33, e40. [Google Scholar] [CrossRef] [PubMed]
- Salcher, E.E.; Kos, P.; Fröhlich, T.; Badgujar, N.; Scheible, M.; Wagner, E. Sequence-Defined Four-Arm Oligo(Ethanamino)Amides for pDNA and siRNA Delivery: Impact of Building Blocks on Efficacy. J. Control. Release 2012, 164, 380–386. [Google Scholar] [CrossRef]
- Lächelt, U.; Kos, P.; Mickler, F.M.; Herrmann, A.; Salcher, E.E.; Rödl, W.; Badgujar, N.; Bräuchle, C.; Wagner, E. Fine-Tuning of Proton Sponges by Precise Diaminoethanes and Histidines in pDNA Polyplexes. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 35–44. [Google Scholar] [CrossRef]
- Benli-Hoppe, T. Cationic Carrier Supported Peptide-Based Nanosystems for Tumor Targeting. Ph.D. Thesis, LMU Munich, Munich, Germany, 2023. [Google Scholar] [CrossRef]
- Prades, R.; Oller-Salvia, B.; Schwarzmaier, S.M.; Selva, J.; Moros, M.; Balbi, M.; Grazú, V.; de La Fuente, J.M.; Egea, G.; Plesnila, N.; et al. Applying the Retro-Enantio Approach to Obtain a Peptide Capable of Overcoming the Blood-Brain Barrier. Angew. Chem. Int. Ed. 2015, 54, 3967–3972. [Google Scholar] [CrossRef]
- Li, Y.; Lei, Y.; Wagner, E.; Xie, C.; Lu, W.; Zhu, J.; Shen, J.; Wang, J.; Liu, M. Potent Retro-Inverso D-Peptide for Simultaneous Targeting of Angiogenic Blood Vasculature and Tumor Cells. Bioconjug. Chem. 2013, 24, 133–143. [Google Scholar] [CrossRef]
- Wang, J.; Lei, Y.; Xie, C.; Lu, W.; Wagner, E.; Xie, Z.; Gao, J.; Zhang, X.; Yan, Z.; Liu, M. Retro-Inverso CendR Peptide-Mediated Polyethyleneimine for Intracranial Glioblastoma-Targeting Gene Therapy. Bioconjug. Chem. 2014, 25, 414–423. [Google Scholar] [CrossRef]
- Wei, X.; Zhan, C.; Chen, X.; Hou, J.; Xie, C.; Lu, W. Retro-Inverso Isomer of Angiopep-2: A Stable D-Peptide Ligand Inspires Brain-Targeted Drug Delivery. Mol. Pharm. 2014, 11, 3261–3268. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, Y.; Gao, Y.; Wang, P.; He, G.; Blum, N.T.; Lin, J.; Liu, Q.; Wang, X.; Huang, P. Six Birds with One Stone: Versatile Nanoporphyrin for Single-Laser-Triggered Synergistic Phototheranostics and Robust Immune Activation. Adv. Mater. 2020, 32, 2004481. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Li, X.; Liu, S.; Chen, J.; Li, M.; Yian Chew, S.; Leong, K.W.; Cheng, D. Codelivery of CRISPR-Cas9 and Chlorin e6 for Spatially Controlled Tumor-Specific Gene Editing with Synergistic Drug Effects. Sci. Adv. 2020, 6, eabb4005. [Google Scholar] [CrossRef] [PubMed]
- Pei, P.; Sun, C.; Tao, W.; Li, J.; Yang, X.; Wang, J. ROS-Sensitive Thioketal-Linked Polyphosphoester-Doxorubicin Conjugate for Precise Phototriggered Locoregional Chemotherapy. Biomaterials 2019, 188, 74–82. [Google Scholar] [CrossRef]
- Jiang, Y.; Yang, W.; Zhang, J.; Meng, F.; Zhong, Z. Protein Toxin Chaperoned by LRP-1-Targeted Virus-Mimicking Vesicles Induces High-Efficiency Glioblastoma Therapy In Vivo. Adv. Mater. 2018, 30, 1800316. [Google Scholar] [CrossRef]
- Li, L.; Xi, W.S.; Su, Q.; Li, Y.; Yan, G.H.; Liu, Y.; Wang, H.; Cao, A. Unexpected Size Effect: The Interplay between Different-Sized Nanoparticles in Their Cellular Uptake. Small 2019, 15, 1901687. [Google Scholar] [CrossRef]
- Donahue, N.D.; Acar, H.; Wilhelm, S. Concepts of Nanoparticle Cellular Uptake, Intracellular Trafficking, and Kinetics in Nanomedicine. Adv. Drug Deliv. Rev. 2019, 143, 68–96. [Google Scholar] [CrossRef]
- Xu, Y.; Li, C.; Lu, S.; Wang, Z.; Liu, S.; Yu, X.; Li, X.; Sun, Y. Construction of Emissive Ruthenium(II) Metallacycle over 1000 nm Wavelength for in Vivo Biomedical Applications. Nat. Commun. 2022, 13, 2009. [Google Scholar] [CrossRef]
- Fisher, C.; Beglova, N.; Blacklow, S.C. Structure of an LDLR-RAP Complex Reveals a General Mode for Ligand Recognition by Lipoprotein Receptors. Mol. Cell 2006, 22, 277–283. [Google Scholar] [CrossRef]
- Tao, L.; Tian, S.; Zhang, J.; Liu, Z.; Robinson-McCarthy, L.; Miyashita, S.I.; Breault, D.T.; Gerhard, R.; Oottamasathien, S.; Whelan, S.P.J.; et al. Sulfated Glycosaminoglycans and Low-Density Lipoprotein Receptor Contribute to Clostridium Difficile Toxin A Entry into Cells. Nat. Microbiol. 2019, 4, 1760–1769. [Google Scholar] [CrossRef]

| OAA ID Number | Sequence # (N → C) | Length of Arms * |
|---|---|---|
| 1658 | [[H2N-(N3)K-C-L3-K3-L3-K3-H]2-α,εK-H]2-α,εK-A-COOH | 5.40 nm |
| 1664 | [[H2N-(N3)K-C-Y3-K3-Y3-K3-H]2-α,εK-H]2-α,εK-A-COOH | 5.40 nm |
| 1696 | [[H2N-(N3)K-C-Y3-εK3-Y3-εK3-H]2-α,εK-H]2-α,εK-A-COOH | 8.42 nm |
| 1768 | [[H2N-(N3)K-C-K9-H]2-α,εK-H]2-α,εK-A-COOH | 4.32 nm |
| 1769 | [[H2N-(N3)K-C-εK9-H]2-α,εK-H]2-α,εK-A-COOH | 8.85 nm |
| Peptide Name | Sequence (N → C) |
|---|---|
| reL57 | DBCO–C–PEG24-hkglklisyfthkdfhkpwt-COOH |
| L57 | DBCO–C–PEG24-TWPKHFDKHTFYSILKLGKH-COOH |
| scr-L57 | DBCO–C–PEG24-KPFKHGTDLLKHFWYSHTKI-COOH |
| reL57-C | H2N–C–hkglklisyfthkdfhkpwt-COOH |
| PEG24 shielding domain | DBCO–PEG24-COOH |
| PEG5000 shielding domain | DBCO–PEG 5 kDa |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, F.; Benli-Hoppe, T.; Guo, W.; Seidl, J.; Wang, Y.; Huang, R.; Wagner, E. Receptor-Targeted Carbon Nanodot Delivery through Polymer Caging and Click Chemistry-Supported LRP1 Ligand Attachment. Polymers 2023, 15, 4039. https://doi.org/10.3390/polym15204039
Zhang F, Benli-Hoppe T, Guo W, Seidl J, Wang Y, Huang R, Wagner E. Receptor-Targeted Carbon Nanodot Delivery through Polymer Caging and Click Chemistry-Supported LRP1 Ligand Attachment. Polymers. 2023; 15(20):4039. https://doi.org/10.3390/polym15204039
Chicago/Turabian StyleZhang, Fengrong, Teoman Benli-Hoppe, Wei Guo, Johanna Seidl, Yi Wang, Rongqin Huang, and Ernst Wagner. 2023. "Receptor-Targeted Carbon Nanodot Delivery through Polymer Caging and Click Chemistry-Supported LRP1 Ligand Attachment" Polymers 15, no. 20: 4039. https://doi.org/10.3390/polym15204039





