The Synergistic Effect of GH13 and GH57 GBEs of Petrotoga mobilis Results in α-Glucan Molecules with a Higher Branch Density
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Commercial Enzymes
2.2. Enzyme Production and Purification
2.3. Enzyme Reactions and Analysis of Activity with Reducing End Assay
2.4. Chain Length Distribution with Anion Exchange Chromatography
2.5. Statistical Analysis
3. Results and Discussion
3.1. Activity of Glycogen Branching Enzymes on Linear Maltodextrin
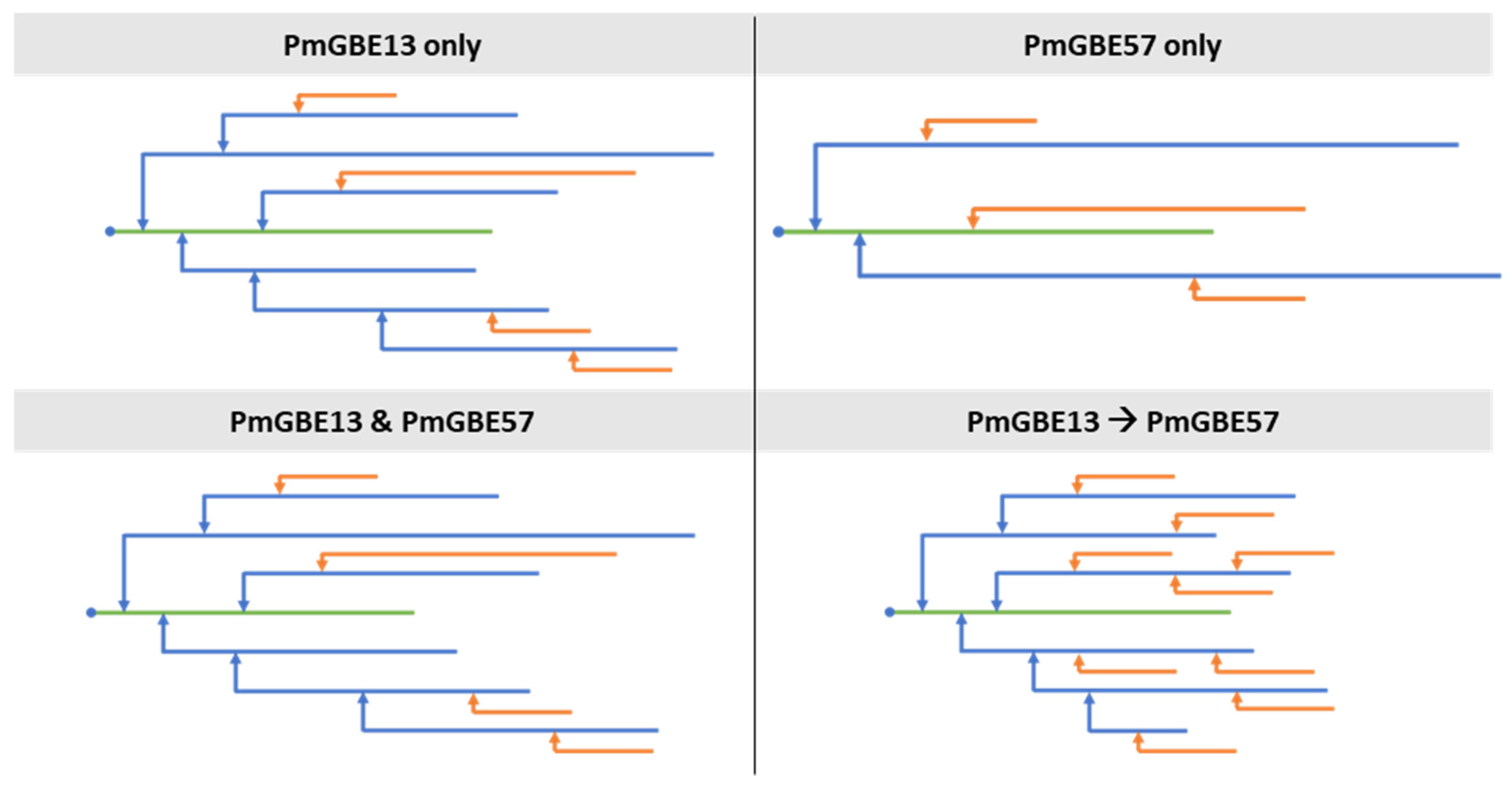
3.2. Chain Length Distribution of Modified Maltodextrin DP18 with a Combination of GH13 and GH57 Glycogen Branching Enzymes
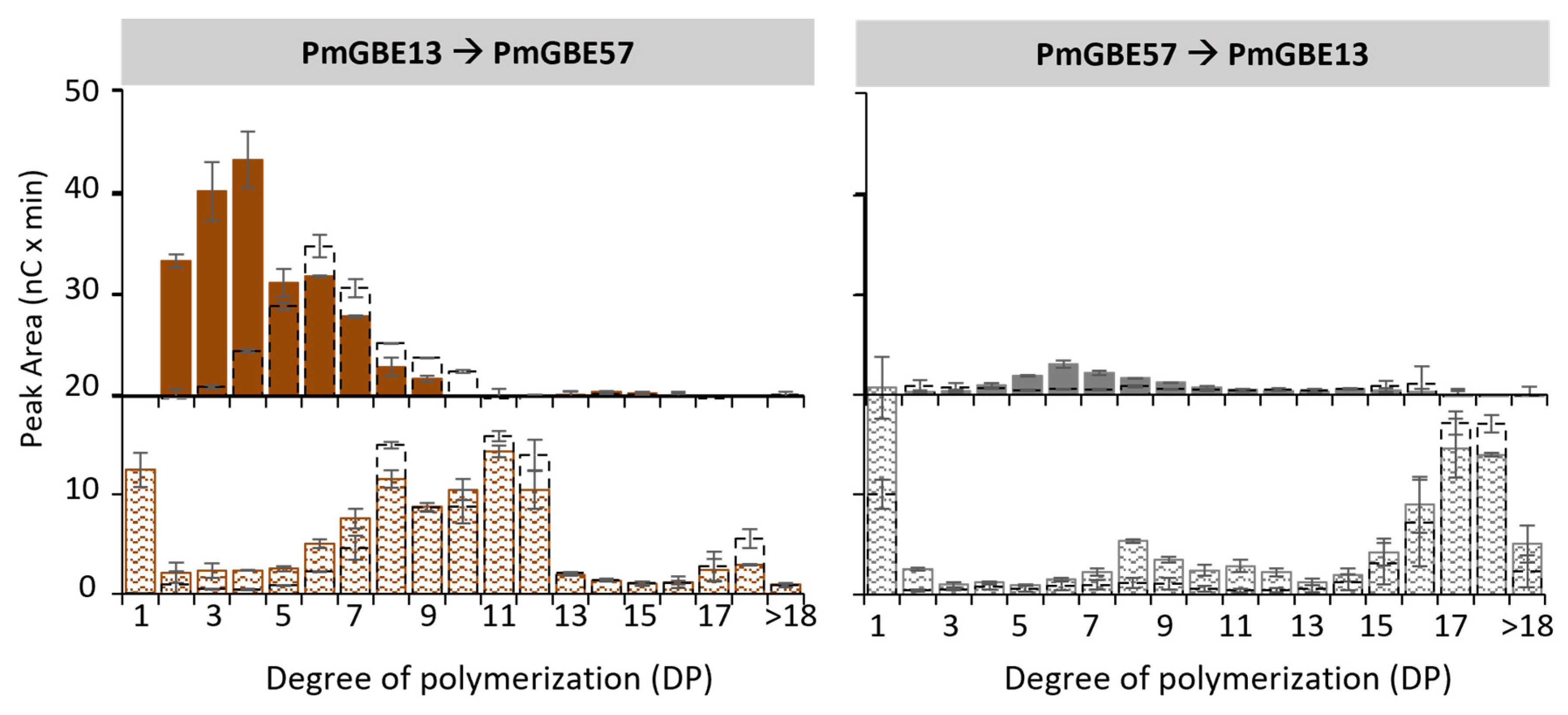
3.3. Two-Step Modification of Linear Maltodextrin DP18 with GH13 and GH57 Glycogen Branching Enzymes
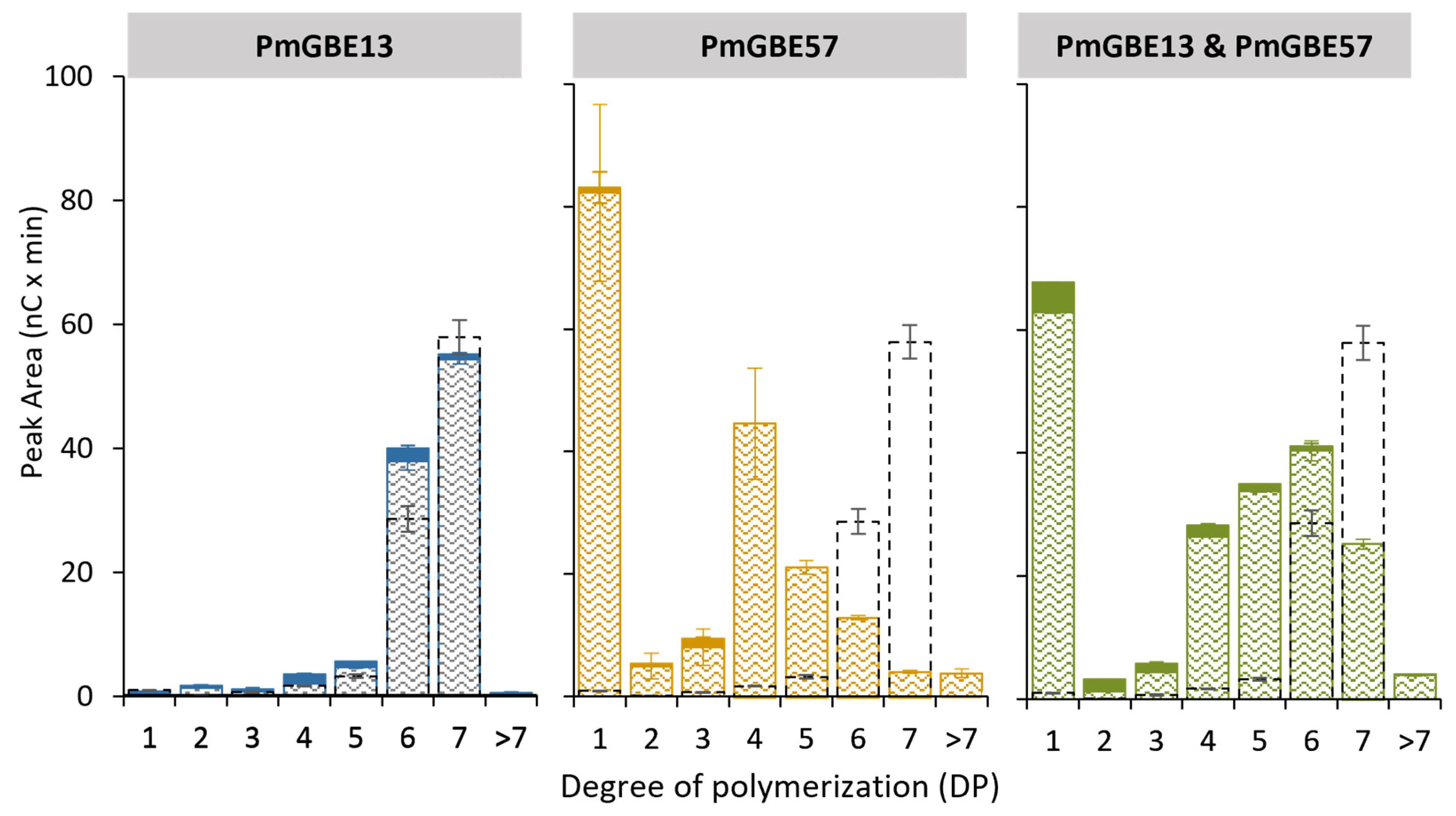
3.4. Modification of Maltodextrin DP7 with GH13 and GH57 Glycogen Branching Enzymes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Montero, M.; Eydallin, G.; Viale, A.M.; Almagro, G.; Muñoz, F.J.; Rahimpour, M.; Sesma, M.T.; Baroja-Fernández, E.; Pozueta-Romero, J. Escherichia Coli Glycogen Metabolism Is Controlled by the PhoP-PhoQ Regulatory System at Submillimolar Environmental Mg2+ Concentrations, and Is Highly Interconnected with a Wide Variety of Cellular Processes. Biochem. J. 2009, 424, 129–141. [Google Scholar] [CrossRef]
- Eydallin, G.; Viale, A.M.; Morán-Zorzano, M.T.; Muñoz, F.J.; Montero, M.; Baroja-Fernández, E.; Pozueta-Romero, J. Genome-Wide Screening of Genes Affecting Glycogen Metabolism in Escherichia Coli K-12. FEBS Lett. 2007, 581, 2947–2953. [Google Scholar] [CrossRef]
- Lillie, S.H.; Pringle, J.R. Reserve Carbohydrate Metabolism in Saccharomyces cerevisiae: Responses to Nutrient Limitation. J. Bacteriol. 1980, 143, 1384–1394. [Google Scholar] [CrossRef] [PubMed]
- Preiss, J. Bacterial Glycogen Synthesis and Its Regulation. Annu. Rev. Microbiol. 1984, 38, 419–458. [Google Scholar] [CrossRef] [PubMed]
- Preiss, J.; Romeo, T. Physiology, Biochemistry and Genetics of Bacterial Glycogen Synthesis. Adv. Microb. Physiol. 1990, 30, 183–238. [Google Scholar] [CrossRef]
- Murugesan, A.G.; Jeyasanthi, T.; Maheswari, S. Isolation and Characterization of Cypermethrin Utilizing Bacteria from Brinjal Cultivated Soil. Afr. J. Microbiol. Res. 2010, 4, 10–13. [Google Scholar]
- Patel, G.B.; Breuil, C. Accumulation of an Iodophilic Polysaccharide during Growth of Acetivibrio cellulolyticus on Cellobiose. Arch. Microbiol. 1981, 129, 265–267. [Google Scholar] [CrossRef]
- Boylen, C.W.; Ensign, J.C. Intracellular Substrates for Endogenous Metabolism during Long-Term Starvation of Rod and Spherical Cells of Arthrobacter crystallopoietes. J. Bacteriol. 1970, 103, 578–587. [Google Scholar] [CrossRef]
- Zevenhuizen, L.P.T.M. Formation and Function of the Glycogen-like Polysaccharide of Arthrobacter. Antonie Van Leeuwenhoek 1966, 32, 356–372. [Google Scholar] [CrossRef]
- Tetlow, I.J.; Emes, M.J. A Review of Starch-Branching Enzymes and Their Role in Amylopectin Biosynthesis. IUBMB Life 2014, 66, 546–558. [Google Scholar] [CrossRef]
- D’Hulst, C.; Mérida, Á. The Priming of Storage Glucan Synthesis from Bacteria to Plants: Current Knowledge and New Developments. New Phytol. 2010, 188, 13–21. [Google Scholar] [CrossRef]
- Wang, L.; Wise, M.J. Glycogen with Short Average Chain Length Enhances Bacterial Durability. Naturwissenschaften 2011, 98, 719–729. [Google Scholar] [CrossRef]
- Takata, H.; Takaha, T.; Okada, S.; Takagi, M.; Imanaka, T. Purification and Characterization of α-Glucan Phosphorylase from Bacillus stearothermophilus. J. Ferment. Bioeng. 1998, 85, 156–161. [Google Scholar] [CrossRef]
- Strange, R.E. Bacterial “Glycogen” and Survival. Nature 1968, 220, 606–607. [Google Scholar] [CrossRef]
- Zevenhuizen, L.P.T.M. Levels of Trehalose and Glycogen in Arthrobacter globiformis under Conditions of Nutrient Starvation and Osmotic Stress. Antonie Van Leeuwenhoek 1992, 61, 61–68. [Google Scholar] [CrossRef]
- Kim, B.H.; Gadd, G.M. Bacterial Physiology and Metabolism; Cambridge University Press: Cambridge, UK, 2008; ISBN 9780521846363. [Google Scholar]
- Binderup, K.; Mikkelsen, R.; Preiss, J. Truncation of the Amino Terminus of Branching Enzyme Changes Its Chain Transfer Pattern. Arch. Biochem. Biophys. 2002, 397, 279–285. [Google Scholar] [CrossRef]
- Abad, M.C.; Binderup, K.; Rios-Steiner, J.; Arni, R.K.; Preiss, J.; Geiger, J.H. The X-Ray Crystallographic Structure of Escherichia Coli Branching Enzyme. J. Biol. Chem. 2002, 277, 42164–42170. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, E.; Suzuki, R. Distribution of Glucan-Branching Enzymes among Prokaryotes. Cell. Mol. Life Sci. 2016, 73, 2643–2660. [Google Scholar] [CrossRef]
- Henrissat, B.; Bairoch, A. Updating the Sequence-Based Classification of Glycosyl Hydrolases. Biochem. J. 1996, 316, 695–696. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Leemhuis, H.; Janeček, Š.; Martinovičová, M.; Pijning, T.; van der Maarel, M.J.E.C. Identification of Thermotoga maritima MSB8 GH57 α-Amylase AmyC as a Glycogen-Branching Enzyme with High Hydrolytic Activity. Appl. Microbiol. Biotechnol. 2019, 103, 6141–6151. [Google Scholar] [CrossRef] [PubMed]
- Pal, K.; Kumar, S.; Sharma, S.; Garg, S.K.; Alam, M.S.; Xu, H.E.; Agrawal, P.; Swaminathan, K. Crystal Structure of Full-Length Mycobacterium tuberculosis H37Rv Glycogen Branching Enzyme. Insights of N-Terminal β-Sandwich in Substrate Specificity and Enzymatic Activity. J. Biol. Chem. 2010, 285, 20897–20903. [Google Scholar] [CrossRef]
- Garg, S.K.; Alam, M.S.; Kishan, K.V.R.; Agrawal, P. Expression and Characterization of α-(1,4)-Glucan Branching Enzyme Rv1326c of Mycobacterium tuberculosis H37Rv. Protein Expr. Purif. 2007, 51, 198–208. [Google Scholar] [CrossRef]
- Sambou, T.; Dinadayala, P.; Stadthagen, G.; Barilone, N.; Bordat, Y.; Constant, P.; Levillain, F.; Neyrolles, O.; Gicquel, B.; Lemassu, A.; et al. Capsular Glucan and Intracellular Glycogen of Mycobacterium tuberculosis: Biosynthesis and Impact on the Persistence in Mice. Mol. Microbiol. 2008, 70, 762–774. [Google Scholar] [CrossRef] [PubMed]
- Chandra, G.; Chater, K.F.; Bornemann, S. Unexpected and Widespread Connections between Bacterial Glycogen and Trehalose Metabolism. Microbiology 2011, 157, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- Mendes, V.; Maranha, A.; Alarico, S.; Empadinhas, N. Biosynthesis of Mycobacterial Methylglucose Lipopolysaccharides. Nat. Prod. Rep. 2012, 29, 834–844. [Google Scholar] [CrossRef] [PubMed]
- Xiang, G. The Mycobacterium tuberculosis H37Rv GH57-Type Glycogen Branching Enzyme Homologue Rv3031 Is Involved in Methylglucose Lipopolysaccharide Biosynthesis. Dual Biological Role of Glyczoside Hydrolase Family Fifty-Seven Glycogen Branching Enzymes. Ph.D. Thesis, University of Groningen, Groningen, The Netherlands, 2022. [Google Scholar]
- Sorndech, W.; Tongta, S.; Blennow, A. Slowly Digestible- and Non-Digestible α-Glucans: An Enzymatic Approach to Starch Modification and Nutritional Effects. Starch/Stärke 2018, 70, 1700145. [Google Scholar] [CrossRef]
- Gaenssle, A.L.O.; Bax, H.H.M.; van der Maarel, M.J.E.C.; Jurak, E. GH13 Glycogen Branching Enzymes Can Adapt the Substrate Chain Length towards Their Preferences via α-1,4-Transglycosylation. Enzyme Microb. Technol. 2021, 150, 109882. [Google Scholar] [CrossRef]
- Gaenssle, A.L.O.; van der Maarel, M.J.E.C.; Jurak, E. Reliability Factor for Identification of Amylolytic Enzyme Activity in the Optimized Starch-Iodine Assay. Anal. Biochem. 2020, 597, 113696. [Google Scholar] [CrossRef]
- Ban, X.; Dhoble, A.S.; Li, C.; Gu, Z.; Hong, Y.; Cheng, L.; Holler, T.P.; Kaustubh, B.; Li, Z. Bacterial 1, 4-α-Glucan Branching Enzymes: Characteristics, Preparation and Commercial Applications. Crit. Rev. Biotechnol. 2020, 40, 380–396. [Google Scholar] [CrossRef]
- Roussel, X.; Lancelon-Pin, C.; Viksø-Nielsen, A.; Rolland-Sabaté, A.; Grimaud, F.; Potocki-Véronèse, G.; Buléon, A.; Putaux, J.-L.; D’Hulst, C. Characterization of Substrate and Product Specificity of the Purified Recombinant Glycogen Branching Enzyme of Rhodothermus obamensis. Biochim. Biophys. Acta 2013, 1830, 2167–2177. [Google Scholar] [CrossRef]
- Palomo, M.; Pijning, T.; Booiman, T.; Dobruchowska, J.M.; van der Vlist, J.; Kralj, S.; Planas, A.; Loos, K.; Kamerling, J.P.; Dijkstra, B.W.; et al. Thermus thermophilus Glycoside Hydrolase Family 57 Branching Enzyme. Crystal Structure, Mechanism of Action, and Products Formed. J. Biol. Chem. 2011, 286, 3520–3530. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Leemhuis, H.; van der Maarel, M.J.E.C. Characterization of the GH13 and GH57 Glycogen Branching Enzymes from Petrotoga mobilis SJ95 and Potential Role in Glycogen Biosynthesis. PLoS ONE 2019, 14, e0219844. [Google Scholar] [CrossRef] [PubMed]
- Bax, H.H.; van der Maarel, M.J.; Jurak, E. Alpha-1,4-Transglycosylation Activity of GH57 Glycogen Branching Enzymes Is Higher in the Absence of a Flexible Loop with a Conserved Tyrosine Residue. Polymers 2023, 15, 2777. [Google Scholar] [CrossRef] [PubMed]

| PmGBE13 | PmGBE57 | |
|---|---|---|
| Non-branching activity [mUNB/mg E] | 42.0 ± 1.6 | 5.7 ± 0.4 |
| Branching activity [mUB/mg E] | 154.2 ± 15.8 | 4.8 ± 0.4 |
| Ratio B:NB * | 3.7 ± 0.3 | 0.8 ± 0.0 |
| PmGBE13 Only | PmGBE57 Only | PmGBE13 and PmGBE57 | PmGBE13 → PmGBE57 | PmGBE57 → PmGBE13 | ||
|---|---|---|---|---|---|---|
| Linear chains | DP 2–5 (%) | 8.8 ± 4.0 a | 39.8 ± 3.6 d | 30.0 ± 3.5 c | 21.5 ± 0.1 b | 27.9 ± 6.0 c |
| DP 6–10 (%) | 52.1 ± 1.5 c | 11.8 ± 0.8 a | 40.4 ± 0.8 b | 42.7 ± 1.3 b | 15.5 ± 0.04 a | |
| DP 11–18 (%) | 38.3 ± 5.7 a | 43.9 ± 2.0 a | 28.4 ± 3.0 a | 34.8 ± 1.1 a | 51.5 ± 4.6 a | |
| DP > 18 (%) | 0.8 ± 0.2 a | 4.6 ± 0.8 b | 1.2 ± 0.3 a | 0.9 ± 0.2 a | 5.1 ± 1.4 b | |
| ACL (DP) | 9.6 ± 0.5 b | 9.7 ± 0.5 b | 8.1 ± 0.3 a | 8.6 ± 0.03 a | 11.1 ± 0.9 b | |
| Branched chains | DP 2–5 (%) | 23.3 ± 0.4 a | 30.4 ± 10.4 a | 29.9 ± 2.2 a | 73.8 ± 1.8 b | 32.4 ± 2.3 a |
| DP 6–10 (%) | 59.2 ± 3.2 b | 14.8 ± 1.4 a | 57.7 ± 1.5 b | 25.1 ± 1.7 a | 51.6 ± 5.3 b | |
| DP 11–18 (%) | 12.2 ± 3.3 b | 54.7 ± 8.8 c | 11.8 ± 3.2 b | 1.1 ± 0.1 a | 14.8 ± 2.1 b | |
| DP > 18 (%) | 0.5 ± 0.1 a | 0.2 ± 0.2 a | 0.6 ± 0.5 a | 0.003 ± 0.003 a | 1.1 ± 0.9 a | |
| ACL (DP) | 7.1 ± 0.1 b | 12.0 ± 0.1 c | 7.0 ± 0.3 b | 4.3 ± 0.1 a | 8.1 ± 1.2 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bax, H.H.M.; Gaenssle, A.L.; van der Maarel, M.J.E.C.; Jurak, E. The Synergistic Effect of GH13 and GH57 GBEs of Petrotoga mobilis Results in α-Glucan Molecules with a Higher Branch Density. Polymers 2023, 15, 4603. https://doi.org/10.3390/polym15234603
Bax HHM, Gaenssle AL, van der Maarel MJEC, Jurak E. The Synergistic Effect of GH13 and GH57 GBEs of Petrotoga mobilis Results in α-Glucan Molecules with a Higher Branch Density. Polymers. 2023; 15(23):4603. https://doi.org/10.3390/polym15234603
Chicago/Turabian StyleBax, Hilda Hubertha Maria, Aline Lucie Gaenssle, Marc Jos Elise Cornelis van der Maarel, and Edita Jurak. 2023. "The Synergistic Effect of GH13 and GH57 GBEs of Petrotoga mobilis Results in α-Glucan Molecules with a Higher Branch Density" Polymers 15, no. 23: 4603. https://doi.org/10.3390/polym15234603
APA StyleBax, H. H. M., Gaenssle, A. L., van der Maarel, M. J. E. C., & Jurak, E. (2023). The Synergistic Effect of GH13 and GH57 GBEs of Petrotoga mobilis Results in α-Glucan Molecules with a Higher Branch Density. Polymers, 15(23), 4603. https://doi.org/10.3390/polym15234603





