Synthesis of Mannosylated Polyethylenimine and Its Potential Application as Cell-Targeting Non-Viral Vector for Gene Therapy
Abstract
:1. Introduction
2. Experimental Section
2.1. Materials and Reagents
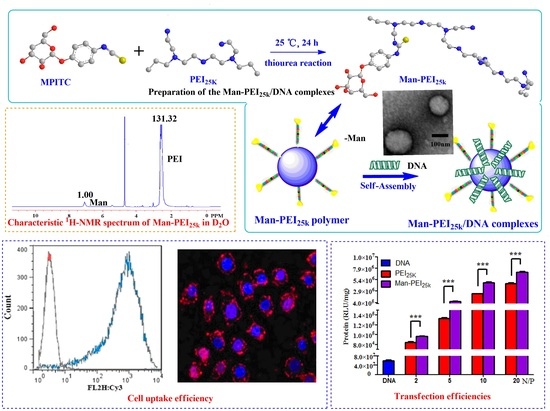
2.2. Synthesis of Man-PEI25k
2.3. Characterization of Copolymers
2.4. Characterization of the PEI/DNA Complexes
2.5. Gel Retardation Assay
2.6. Cytotoxicity Assay
2.7. Cell Uptake Study
2.8. In Vitro Transfection
2.9. Statistical Analysis
3. Results and Discussion
3.1. Synthesis and Characterization of Man-PEI25k



3.2. Characterization of Complexes
3.3. Gel Retardation Assays


3.4. Cytotoxicity Assay

3.5. Cell Uptake and Intracellular Distribution
3.6. Cell Transfection Assay


4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Höbel, S.; Aigner, A. Polyethylenimine (PEI)/siRNA-mediated gene knockdown in vitro and in vivo. Methods Mol. Biol. 2010, 623, 283–297. [Google Scholar] [PubMed]
- Swami, A.; Kurupati, R.K.; Pathak, A.; Singh, Y.; Kumar, P.; Gupta, K.C. A unique and highly efficient non-viral DNA/siRNA delivery system based on PEI-bisepoxide nanoparticles. Biochem. Biophys. Res. Commun. 2007, 362, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Kunath, K.; Merdan, T.; Hegener, O.; Häberlein, H.; Kissel, T. Integrin targeting using RGD-PEI conjugates for in vitro gene transfer. J. Gene Med. 2003, 5, 588–599. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, S.; Hashida, M. Targeted delivery systems of small interfering RNA by systemic administration. Drug Metab. Pharmacokinet. 2007, 22, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Zhu, J.L.; Zeng, X.; Jing, Y.; Zhang, X.Z.; Zhuo, R.X. Targeted gene delivery mediated by folate-polyethylenimine-block-poly(ethylene glycol) with receptor selectivity. Bioconjug. Chem. 2009, 20, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; He, M.L.; Chan, C.Y.; Chen, Y.C.; Li, X.P.; Li, Y.; Zheng, D.; Lin, M.C.; Kung, H.F.; Shuai, X.T.; et al. The use of folate-PEG-grafted-hybranched-PEI nonviral vector for the inhibition of glioma growth in the rat. Biomaterials 2009, 30, 4014–4020. [Google Scholar] [CrossRef] [PubMed]
- Diebold, S.S.; Kursa, M.; Wagner, E.; Cotton, M.; Zenke, M. Mannose polyethylenimine conjugates for targeted DNA delivery into dendritic cells. J. Bio. Chem. 1999, 274, 19087–19094. [Google Scholar] [CrossRef]
- Chen, Y.Z.; Ruan, G.X.; Yao, X.L.; Li, L.M.; Hu, Y.; Tabata, Y.; Gao, J.Q. Co-transfection gene delivery of dendritic cells induced effective lymph node targeting and anti-tumor vaccination. Pharm. Res. 2013, 30, 1502–1512. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Z.; Yao, X.L.; Ruan, G.X.; Zhao, Q.Q.; Tang, G.P.; Tabata, Y.; Gao, J.Q. Gene-carried chitosan-linked polyethylenimine induced high gene transfection efficiency on dendritic cells. Biotechnol. Appl. Biochem. 2012, 59, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.L.; Kim, Y.K.; Arote, R.; Jere, D.; Quan, J.S.; Yu, J.H.; Choi, Y.J.; Nah, J.W.; Cho, M.H.; Cho, C.S. Mannosylated chitosan-graft-polyethylenimine as a gene carrier for Raw 264.7 cell targeting. Int. J. Pharm. 2009, 375, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Chen, S.; Han, J.; Zhang, Z. Mannosylated biodegradable polyethyleneimine for targeted DNA delivery to dendritic cells. Int. J. Nanomed. 2012, 7, 2929–2942. [Google Scholar] [CrossRef]
- Zanta, M.A.; Boussif, O.; Adib, A.; Behr, J.P. In vitro gene delivery to hepatocytes with galactosylated polyethylenimine. Bioconjug. Chem. 1997, 8, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Diebold, S.S.; Plank, C.; Cotton, M.; Wagner, E.; Zenke, M. Mannose receptor-mediated gene delivery into antigen presenting dendritic cells. Somat. Cell Mol. Genet. 2002, 27, 65–74. [Google Scholar] [CrossRef]
- Hu, Y.; Xu, B.; Ji, Q.; Shou, D.; Sun, X.; Xu, J.; Gao, J.; Liang, W. A mannosylated cell-penetrating peptide-graft-polyethylenimine as a gene delivery vector. Biomaterials 2014, 35, 4236–4246. [Google Scholar] [CrossRef] [PubMed]
- Nimesh, S.; Thibault, M.M.; Lavertu, M.; Buschmann, M.D. Enhanced gene delivery mediated by low molecular weight chitosan/DNA complexes: Effect of pH and serum. Mol. Biotechnol. 2010, 46, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Ahn, H.H.; Lee, M.S.; Cho, M.H.; Shin, Y.N.; Lee, J.H.; Kim, K.S.; Kim, M.S.; Khang, G.; Hwang, K.C.; Lee, I.W.; et al. DNA/PEI nano-particles for gene delivery of rat bone marrow stem cells. Colloids Surf. Physicochem. Eng. Aspect 2008, 313–314, 116–120. [Google Scholar] [CrossRef]
- Ong, Z.Y.; Fukushima, K.; Coady, D.J.; Yang, Y.Y.; Ee, P.L.; Hedrick, J.L. Rational design of biodegradable cationic polycarbonates for gene delivery. J. Control. Release 2011, 152, 120–126. [Google Scholar] [PubMed]
- Liang, W.; Gong, H.; Yin, D.; Lu, S.; Fu, Q. High-molecular-weight polyethyleneimine conjuncted pluronic for gene transfer agents. Chem. Pharm. Bull. 2011, 59, 1094–1101. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, L.; Olaso, E.; Anasagasti, M.J.; Fuentes, A.M.; Vidal-Vanaclocha, F. Mannose receptor-mediated endothelial cell activation contributes to B16 melanoma cell adhesion and metastasis in liver. J. Cell. Physiol. 1998, 174, 322–330. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Y.; Xu, B.-H.; Xu, J.-J.; Shou, D.; Gao, J.-Q. Synthesis of Mannosylated Polyethylenimine and Its Potential Application as Cell-Targeting Non-Viral Vector for Gene Therapy. Polymers 2014, 6, 2573-2587. https://doi.org/10.3390/polym6102573
Hu Y, Xu B-H, Xu J-J, Shou D, Gao J-Q. Synthesis of Mannosylated Polyethylenimine and Its Potential Application as Cell-Targeting Non-Viral Vector for Gene Therapy. Polymers. 2014; 6(10):2573-2587. https://doi.org/10.3390/polym6102573
Chicago/Turabian StyleHu, Ying, Bei-Hua Xu, Jiao-Jiao Xu, Dan Shou, and Jian-Qing Gao. 2014. "Synthesis of Mannosylated Polyethylenimine and Its Potential Application as Cell-Targeting Non-Viral Vector for Gene Therapy" Polymers 6, no. 10: 2573-2587. https://doi.org/10.3390/polym6102573
APA StyleHu, Y., Xu, B.-H., Xu, J.-J., Shou, D., & Gao, J.-Q. (2014). Synthesis of Mannosylated Polyethylenimine and Its Potential Application as Cell-Targeting Non-Viral Vector for Gene Therapy. Polymers, 6(10), 2573-2587. https://doi.org/10.3390/polym6102573