Comparison of Biochemical, Anatomical, Morphological, and Physiological Responses to Salinity Stress in Wheat and Barley Genotypes Deferring in Salinity Tolerance
Abstract
1. Introduction
2. Material and Methods
2.1. Plant Material and Growth Conditions
2.2. Measurement of Growth Traits and Mineral Concentrations
2.3. Measurement of Photosynthesis Parameters, Chlorophyll Contents, and Chlorophyll Fluorescence
2.4. Lipid Peroxidation and Antioxidative Enzyme Activity Assay
2.5. Cell Ultrastructure
2.6. Statistical Analysis
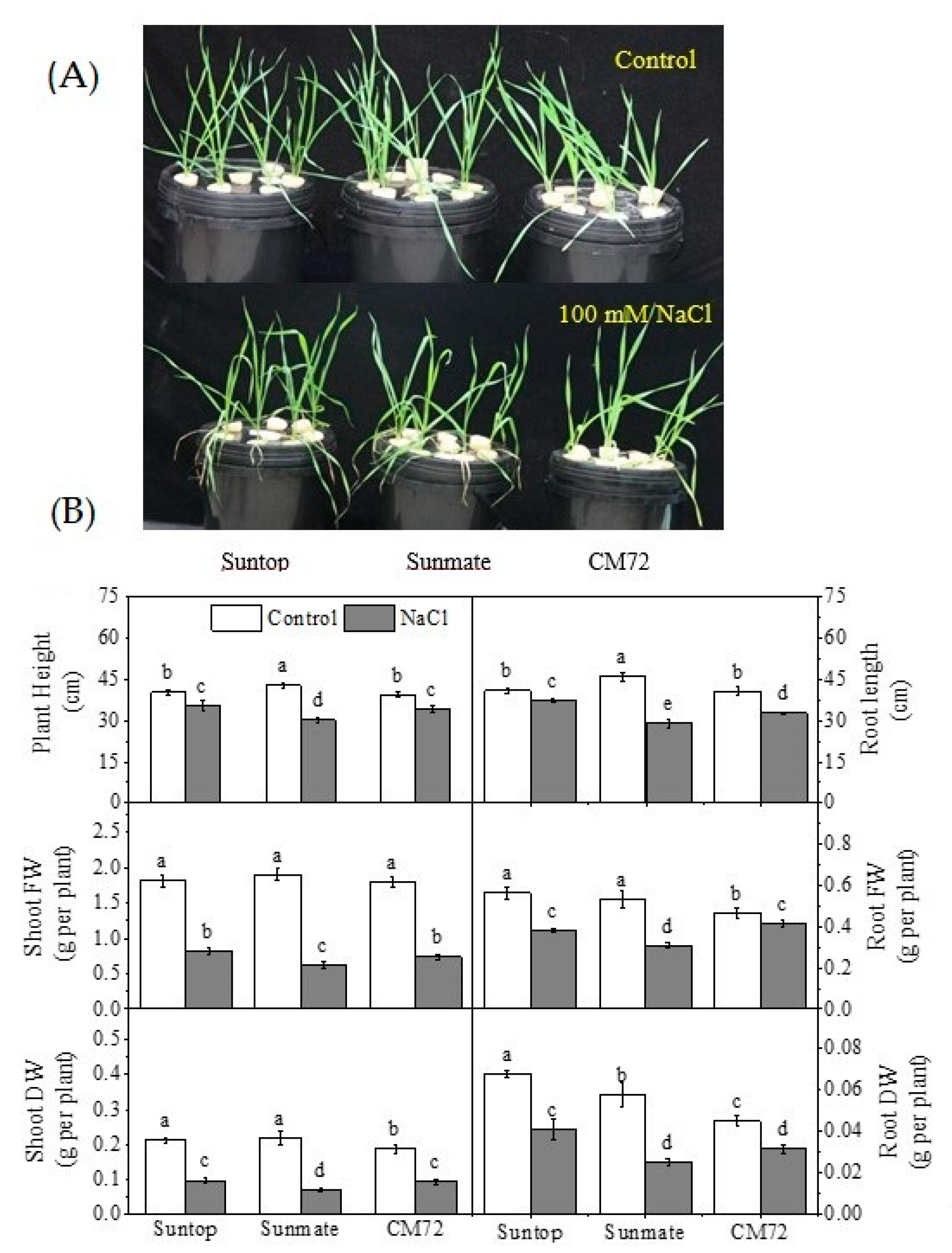
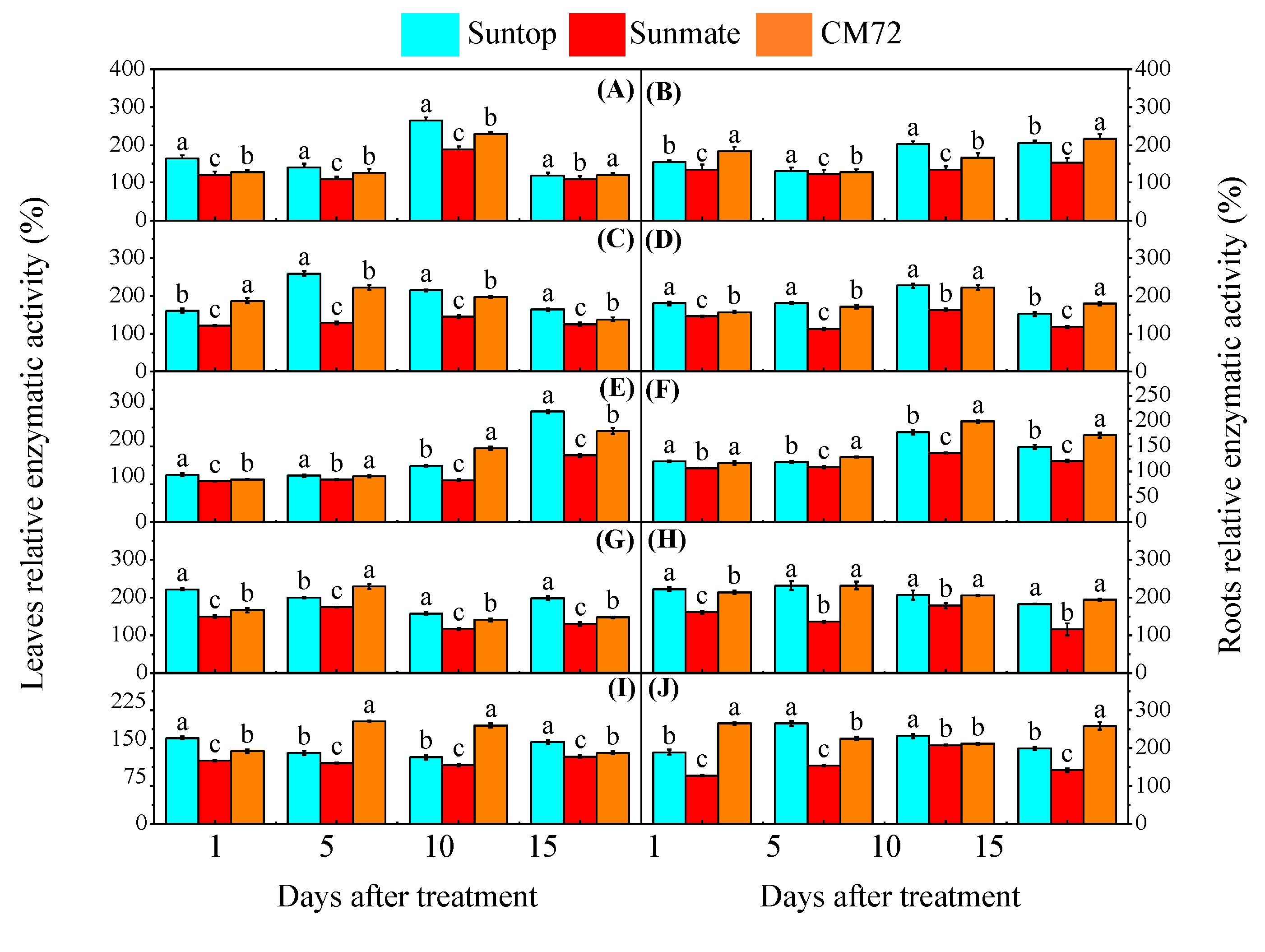
3. Results
3.1. Plant Growth Parameters
3.2. Chlorophyll and Photosynthetic Parameters
3.3. Shoots and Roots Na+, K+ Concentration, and Na+:K+ Ratio
3.4. Lipid Peroxidation Assay and Antioxidative Enzyme Activities
3.5. Leaf and Root Ultrastructure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ghassemi, F.; Jakeman, A.J.; Nix, H.A. Salinisation of Land and Water Resources: Human Causes, Extent, Management and Case Studies; CAB International Publishing: Wallingford, UK, 1995; p. 526. [Google Scholar]
- FAO. Global Network on Integrated Soil Management for Sustainable Use of Salt-Affected Soils. 2000. Available online: http://www.fao.org/ag/AGL/agll/spush/intro.htm (accessed on 10 May 2004).
- Zhang, H.X.; Hodson, J.N.; Williams, J.P.; Blumwald, E. Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc. Natl. Acad. Sci. USA 2001, 98, 12832–12836. [Google Scholar] [CrossRef] [PubMed]
- Flowers, T.J. Improving crop salt tolerance. J. Exp. Bot. 2004, 55, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Parida, A.K.; Das, A.B. Salt tolerance and salinity effects on plants: A review. Ecotoxicol. Environ. Saf. 2005, 60, 324–349. [Google Scholar] [CrossRef]
- FAO. The Special Challenge for Sub-Saharan Africa. How to Feed the World 2050; FAO: Rome, Italy, 2009.
- Ashraf, M.; Harris, P.J.C. Potential biochemical indicators of salinity tolerance in plants. Plant Sci. 2004, 166, 3–16. [Google Scholar] [CrossRef]
- Ferguson, L.; Grattan, S.R. How salinity damages citrus: osmotic effects and specific ion toxicities. Hort. Technol. 2005, 15, 95–99. [Google Scholar] [CrossRef]
- Munns, R.; James, R.A.; Lauchli, A. Approaches to increasing the salt tolerance of wheat and other cereals. J. Exp. Bot. 2006, 57, 1025–1043. [Google Scholar] [CrossRef]
- Apse, M.P.; Blumwald, E. Engineering salt tolerance in plants. Curr. Opin. Biotechol. 2002, 13, 146–150. [Google Scholar] [CrossRef]
- Machado, R.M.; Serralheiro, R.P. Soil salinity: effect on vegetable crop growth. Management practices to prevent and mitigate soil salinization. Horticulturae 2017, 3, 30. [Google Scholar] [CrossRef]
- Rengasamy, P. World salinization with emphasis on Australia. J. Exp. Bot. 2006, 57, 1017–1023. [Google Scholar] [CrossRef]
- Slabu, C.; Zörb, C.; Steffens, D.; Schubert, S. Is salt stress of faba bean (Vicia faba) caused by Na+ or Cl–toxicity? J. Plant Nutr. Soil Sci. 2009, 172, 644–651. [Google Scholar] [CrossRef]
- Tavakkoli, E.; Rengasamy, P.; McDonald, G.K. High concentrations of Na+ and Cl− ions in soil solution have simultaneous detrimental effects on growth of faba bean under salinity stress. J. Exp. Bot. 2010, 61, 4449–4459. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, J.M. The integration of activity in saline environments: Problems and perspectives. Funct. Plant Biol. 2013, 40, 759–774. [Google Scholar] [CrossRef]
- Schachtman, D.P.; Liu, W.H. Molecular pieces to the puzzle of the interaction between potassium and sodium uptake in plants. Trends Plant Sci. 1999, 4, 281–287. [Google Scholar] [CrossRef]
- Ball, M.C.; Chow, W.S.; Anderson, J.M. Salinity-induced potassium deficiency causes loss of functional photosystem II in leaves of the grey mangrove, Avicennia marina, through depletion of the atrazine-binding polypeptide. Funct. Plant Biol. 1987, 14, 351–361. [Google Scholar] [CrossRef]
- Botella, M.A.; Martinez, V.; Pardines, J.; Cerda, A. Salinity induced potassium deficiency in maize plants. J. Plant Physiol. 1997, 150, 200–205. [Google Scholar] [CrossRef]
- Pang, C.H.; Wang, B.S. Oxidative stress and salt tolerance in plants. In Progress in Botany; Lüttge, U., Beyschlag, W., Murata, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; Volume 69, pp. 231–245. [Google Scholar]
- Wong-Ekkabut, J.; Xu, Z.; Triampo, W.; Tang, I.M.; Tieleman, D.P.; Monticelli, L. Effect of lipid peroxidation on the properties of lipid bilayers: A molecular dynamics study. Biophys. J. 2007, 93, 4225–4236. [Google Scholar] [CrossRef]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012, 2012, 1–26. [Google Scholar] [CrossRef]
- Smirnoff, N. Plant resistance to environmental stress. Curr. Opin. Biotech. 1998, 9, 214–219. [Google Scholar] [CrossRef]
- Jena, N.R. DNA damage by reactive species: Mechanisms, mutation and repair. J. Biosci. 2012, 37, 503–517. [Google Scholar] [CrossRef]
- Noctor, G.; Foyer, C.H. Ascorbate and glutathione: Keeping active oxygen under control. Annu. Rev. Plant Biol. 1998, 49, 249–279. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Asada, K. Superoxide production in aprotic interior of chloroplast thylakoids. Arch. Biochem. Biophys. 1988, 267, 714–722. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Dietz, K.J.; Jacob, S.; Oelze, M.L.; Laxa, M.; Tognetti, V.; de Miranda, S.M.; Baier, M.; Finkemeier, I. The function of peroxiredoxins in plant organelle redox metabolism. J. Exp. Bot. 2006, 57, 1697–1709. [Google Scholar] [CrossRef]
- Türkan, I.; Demiral, T. Recent developments in understanding salinity tolerance. Environ. Exp. Bot. 2009, 67, 2–9. [Google Scholar] [CrossRef]
- Gupta, B.; Huang, B. Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Int. J. Genom. 2014, 2014, 1–18. [Google Scholar] [CrossRef]
- Pessarakli, M. Dry matter yield, nitrogen-15 absorption, and water uptake by green bean under sodium chloride stress. Crop Sci. 1991, 31, 1633–1640. [Google Scholar] [CrossRef]
- Wu, F.B.; Zhang, G.P.; Yu, J.S. Interaction of cadmium and four microelements for uptake and translocation in different barley genotypes. Comm. Soil Sci. Plant Analysis 2003, 34, 2003–2020. [Google Scholar] [CrossRef]
- Wu, F.B.; Zhang, G.P.; Dominy, P. Four barley genotypes respond differently to cadmium: Lipid peroxidation and activities of antioxidant capacity. Environ. Exp. Bot. 2003, 50, 67–78. [Google Scholar] [CrossRef]
- Chen, Z.; Zhou, M.; Newman, I.A.; Mendham, N.J.; Zhang, G.; Shabala, S. Potassium and sodium relations in salinized barley tissues as a basis of differential salt tolerance. Funct. Plant Biol. 2007, 34, 150–162. [Google Scholar] [CrossRef]
- Wu, F.B.; Lianghuan, W.; Fuhua, X. Chlorophyll meter to predict nitrogen sidedress requirements for short-season cotton (Gossypium hirsutum L.). Field Crop Res. 1998, 56, 309–314. [Google Scholar]
- Genty, B.; Briantais, J.M.; Baker, N.R. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta-Gen. Subj. 1989, 990, 87–92. [Google Scholar] [CrossRef]
- Leul, M.; Zhou, W.J. Alleviation of waterlogging damage in winter rape by uniconazole application: Effects on enzyme activity, lipid peroxidation, and membrane integrity. J. Plant Growth Regul. 1999, 18, 9–14. [Google Scholar] [CrossRef]
- Chen, F.; Wang, F.; Wu, F.B.; Mao, W.H.; Zhang, G.P.; Zhou, M.X. Modulation of exogenous glutathione in antioxidant defense system against Cd stress in the two barley genotypes differing in Cd tolerance. Plant Physiol. Biochem. 2010, 48, 663–672. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, J. Effect of abscisic acid on active oxygen species, antioxidative defence system and oxidative damage in leaves of maize seedlings. Plant Cell. Physiol. 2001, 42, 1265–1273. [Google Scholar] [CrossRef]
- Tang, Q.; Feng, M. Practical Statistics and Its DPS Statistics Software Package; China Agriculture Press: Beijing, China, 1997. [Google Scholar]
- Neill, S.J.; Desikan, R.; Clarke, A.; Hurst, R.D.; Hancock, J.T. Hydrogen peroxide and nitric oxide as signaling molecules in plants. J. Exp. Bot. 2002, 53, 1237–1247. [Google Scholar] [CrossRef]
- Netondo, G.W.; Onyango, J.C.; Beck, E. Sorghum and salinity. II. Gas exchange and chlorophyll fluorescence of sorghum under salt stress. Crop Sci. 2004, 44, 806–811. [Google Scholar]
- Taiz, L.; Zeiger, E. Plant Physiology; Sinauer Associates Inc.: Sunderland, MA, USA, 2002. [Google Scholar]
- Lawlor, D.W.; Cornic, G. Photosynthetic carbon assimilation and associated metabolism in relation to water deficits in higher plants. Plant Cell Environ. 2002, 25, 275–294. [Google Scholar] [CrossRef]
- Meloni, D.A.; Oliva, M.A.; Martinez, C.A.; Cambraia, J. Photosynthesis and activity of superoxide dismutase, peroxidase and glutathione reductase in cotton under salt stress. Environ. Exp. Bot. 2003, 49, 69–76. [Google Scholar] [CrossRef]
- Ibrahim, W.; Ahmed, I.M.; Chen, X.; Cao, F.; Zhu, S.; Wu, F. Genotypic differences in photosynthetic performance, antioxidant capacity, ultrastructure and nutrients in response to combined stress of salinity and Cd in cotton. BioMetals 2015, 28, 1063–1078. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.M.; Cao, F.; Zhang, M.; Chen, X.; Zhang, G.; Wu, F. Difference in yield and physiological features in response to drought and salinity combined stress during anthesis in Tibetan wild and cultivated barleys. PLoS ONE 2013, 8, e77869. [Google Scholar] [CrossRef] [PubMed]
- Yamane, K.; Kawasaki, M.; Taniguchi, M.; Miyake, H. Differential effect of NaCl and polyethylene glycol on the ultrastructure of chloroplasts in rice seedlings. J. Plant Physiol. 2003, 160, 573–575. [Google Scholar] [CrossRef] [PubMed]
- Naeem, M.S.; Warusawitharana, H.; Liu, H.; Liu, D.; Ahmad, R.; Waraich, E.A.; Xua, L.; Zhou, W.J. 5-aminolevulinic acid alleviates the salinity-induced changes in Brassica napus as revealed by the ultrastructural study of chloroplast. Plant Physiol. Biochem. 2012, 57, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Parida, A.K.; Das, A.B.; Mittra, B. Effects of NaCl stress on the structure, pigment complex composition, and photosynthetic activity of mangrove Bruguiera parviflora chloroplasts. Photosynthetica 2003, 41, 191–200. [Google Scholar] [CrossRef]
- Zhang, H.X.; Blumwald, E. Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat. Biotechnol. 2001, 19, 765–768. [Google Scholar] [CrossRef] [PubMed]
- Katsuhara, M.; Kawasaki, T. Salt stress induced nuclear and DNA degradation in meristematic cells of barley roots. Plant Cell Physiol. 1996, l37, 169–173. [Google Scholar] [CrossRef]
- Kader, M.A.; Lindberg, S. Uptake of sodium in protoplasts of salt-sensitive and salt-tolerant cultivars of rice, Oryza sativa L. determined by the fluorescent dye SBFI. J. Exp. Bot. 2005, 56, 3149–3158. [Google Scholar] [CrossRef]
- Genc, Y.; McDonald, G.K.; Tester, M. Reassessment of tissue Na+ concentration as a criterion for salinity tolerance in bread wheat. Plant Cell. Environ. 2007, 30, 1486–1498. [Google Scholar] [CrossRef]
- Reuter, D.; Robinson, J.B. Plant Analysis: An Interpretation Manual, 2nd ed.; CSIRO Publishing: Melbourne, Australia, 1997. [Google Scholar]
- Ozfidan-Konakci, C.; Yildiztugay, E.; Kucukoduk, M. Upregulation of antioxidant enzymes by exogenous gallic acid contributes to the amelioration in Oryza sativa roots exposed to salt and osmotic stress. Environ. Sci. Pollut. Res. Int. 2015, 22, 1487–1498. [Google Scholar] [CrossRef]
- Del, R.D.; Stewart, A.J.; Pellegrini, N. A review of recent studies on malondialdehyde as toxic molecule and biological marker of oxidative stress. Nutr. Metab. Cardiovasc Dis. 2005, 15, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.W.; Stals, E.; Panis, B.; Keulemans, J.; Swennen, R.L. High-throughput determination of malondialdehyde in plant tissues. Anal. Biochem. 2005, 347, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, P.; Hakeem, K.R.; Kumar, A.; Ashraf, M.; Akram, N.A. Salt-induced changes in photosynthetic activity and oxidative defense system of three cultivars of mustard (Brassica juncea L.). African J. Biotechnol. 2012, 11, 2694–2703. [Google Scholar]
- Azooz, M.M.; Youssef, A.M.; Ahmad, P. Evaluation of salicylic acid (SA) application on growth, osmotic solutes and antioxidant enzyme activities on broad bean seedlings grown under diluted seawater. Int. J. Plant Physiol. Biochem. 2011, 3, 253–264. [Google Scholar]
- Katsuhara, M.; Otsuka, T.; Ezaki, B. Salt stress-induced lipid peroxidation is reduced by glutathione S-transferase, but this reduction of lipid peroxides is not enough for a recovery of root growth in Arabidopsis. Plant Sci. 2005, 169, 369–373. [Google Scholar] [CrossRef]
- Bowler, C.; Montagu, M.V.; Inze, D. Superoxide dismutase and stress tolerance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1992, 43, 83–116. [Google Scholar] [CrossRef]
- Shalata, A.; Mittova, V.; Volokita, M.; Guy, M.; Tal, M. Response of the cultivated tomato and its wild salt-tolerant relative Lycopersicon pennellii to salt-dependent oxidative stress: The root antioxidative system. Physiol. Plant 2001, 112, 487–494. [Google Scholar] [CrossRef]
- Hernandez, J.A.; Jimenez, A.; Mullineaux, P.; Sevilla, F. Tolerance of pea (Pisum sativum L.) to long-term salt stress is associated with induction of antioxidant defences. Plant Cell Environ. 2000, 23, 853–862. [Google Scholar] [CrossRef]
- Feki, K.; Tounsi, S.; Brini, F. Comparison of an antioxidant system in tolerant and susceptible wheat seedlings in response to salt stress. Spanish J. Agric. Res. 2017, 15, e0805. [Google Scholar] [CrossRef]
- Koca, H.; Bor, M.; Özdemir, F.; Türkan, I. The effect of salt stress on lipid peroxidation, antioxidative enzymes and proline content of sesame cultivars. Environ. Exp. Bot. 2007, 60, 344–351. [Google Scholar] [CrossRef]
- Temel, A.; Gozukirmizi, N. Physiological and molecular changes in barley and wheat under salinity. Appl. Biochem. Biotechol. 2015, 175, 2950–2960. [Google Scholar] [CrossRef] [PubMed]
- Reddy, R.A.; Chaitanya, K.V.; Vivekanandan, M. Drought-induced responses of photosynthesis and antioxidant metabolism in higher plants. J. Plant Physiol. 2004, 161, 1189–1202. [Google Scholar] [CrossRef] [PubMed]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeeshan, M.; Lu, M.; Sehar, S.; Holford, P.; Wu, F. Comparison of Biochemical, Anatomical, Morphological, and Physiological Responses to Salinity Stress in Wheat and Barley Genotypes Deferring in Salinity Tolerance. Agronomy 2020, 10, 127. https://doi.org/10.3390/agronomy10010127
Zeeshan M, Lu M, Sehar S, Holford P, Wu F. Comparison of Biochemical, Anatomical, Morphological, and Physiological Responses to Salinity Stress in Wheat and Barley Genotypes Deferring in Salinity Tolerance. Agronomy. 2020; 10(1):127. https://doi.org/10.3390/agronomy10010127
Chicago/Turabian StyleZeeshan, Muhammad, Meiqin Lu, Shafaque Sehar, Paul Holford, and Feibo Wu. 2020. "Comparison of Biochemical, Anatomical, Morphological, and Physiological Responses to Salinity Stress in Wheat and Barley Genotypes Deferring in Salinity Tolerance" Agronomy 10, no. 1: 127. https://doi.org/10.3390/agronomy10010127
APA StyleZeeshan, M., Lu, M., Sehar, S., Holford, P., & Wu, F. (2020). Comparison of Biochemical, Anatomical, Morphological, and Physiological Responses to Salinity Stress in Wheat and Barley Genotypes Deferring in Salinity Tolerance. Agronomy, 10(1), 127. https://doi.org/10.3390/agronomy10010127






