Classification Binary Trees with SSR Allelic Sizes: Combining Regression Trees with Genetic Molecular Data in Order to Characterize Genetic Diversity between Cultivars of Olea europaea L.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. DNA Extraction and Microsatellite Analysis
2.3. Cluster Analysis by the Classification Binary Tree (CBT)
3. Results
3.1. Genetic Parameters from SSR Analysis
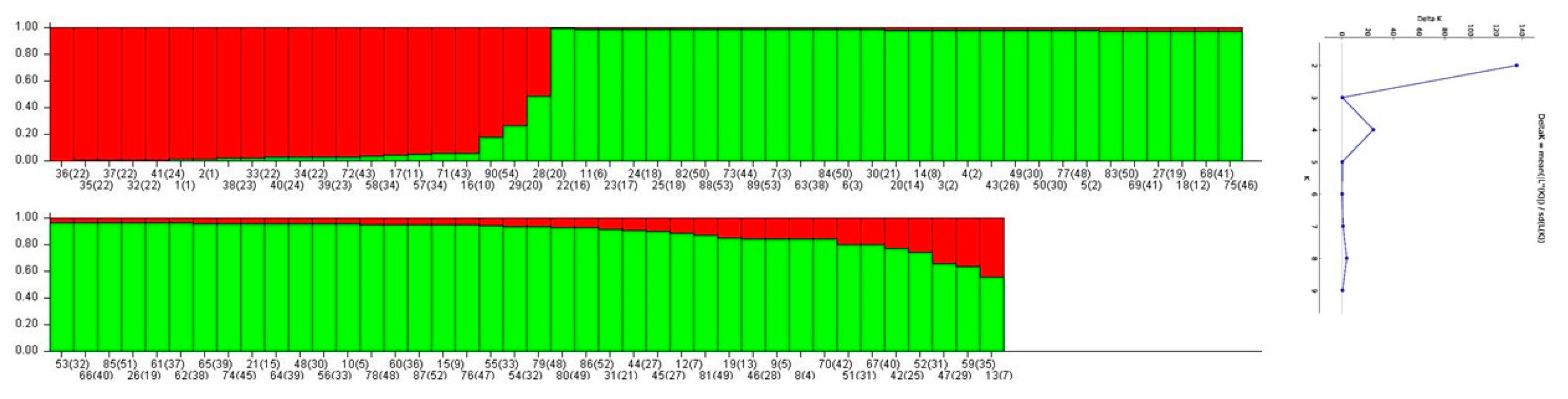
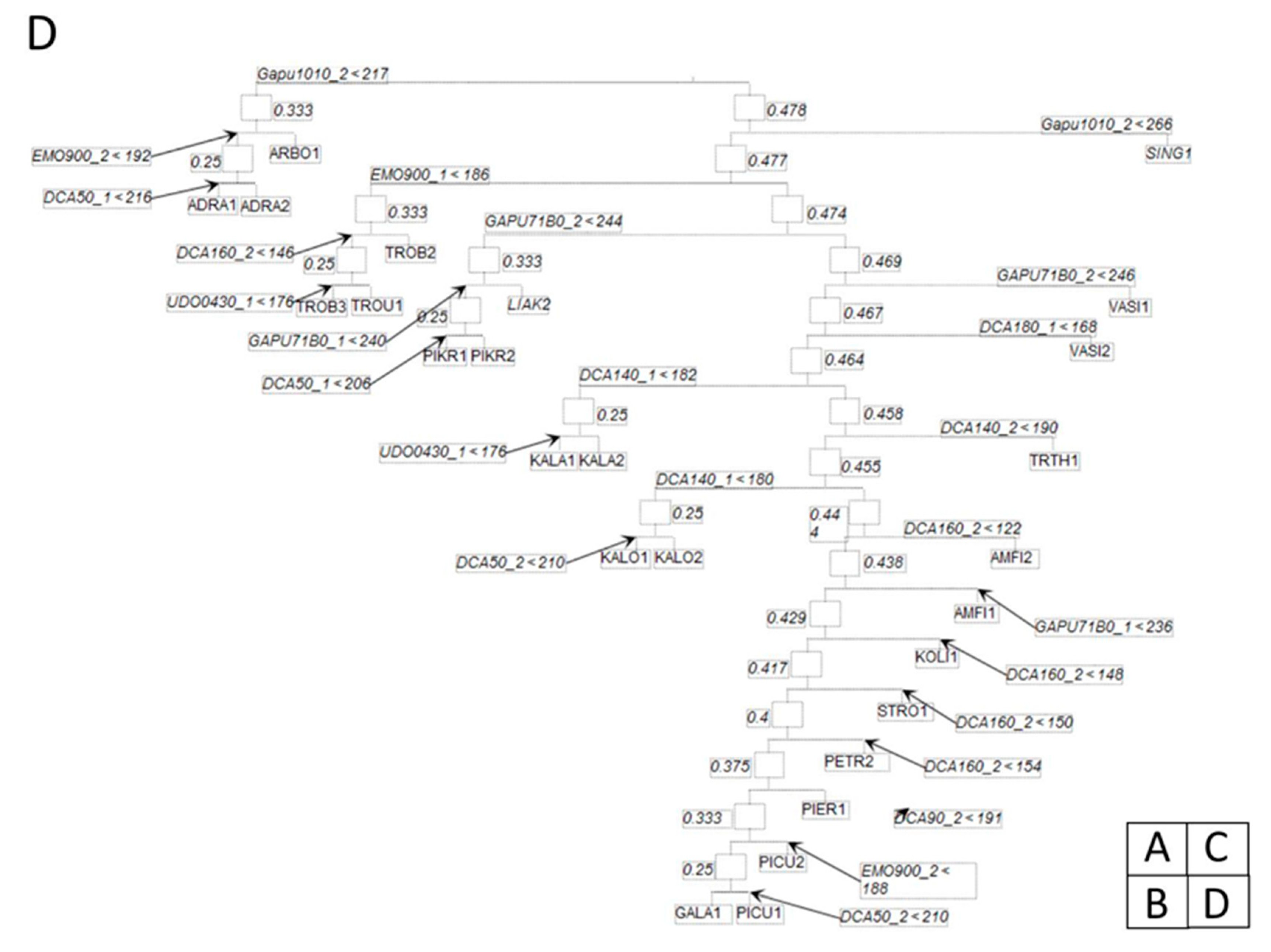
3.2. Classification Binary Trees
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Aybar, V.E.; de Melo, E.A.; Mourão, J.P.; Searles, P.S.; Matias, A.C.; del Río, C.; Reig, J.M.C.; Rousseaux, M.C. Evaluation of olive flowering at low latitude sites in Argentina using a chilling requirement model. Span. J. Agric. Res. 2015, 13, e09-001. [Google Scholar] [CrossRef] [Green Version]
- Ponti, L.; Gutierrez, A.P.; Ruti, P.M.; Dell’Aquila, A. Fine-scale ecological and economic assessment of climate change on olive in the Mediterranean basin reveals winners and losers. Proc. Natl. Acad. Sci. USA 2014, 111, 5598–5603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koubouris, G.; Kavroulakis, N.; Metzidakis, I.; Vasilakakis, M.; Sofo, A. Ultraviolet-B radiation or heat cause changes in photosynthesis, antioxidant enzyme activities and pollen performance in olive tree. Photosynthetica 2015, 53, 279–287. [Google Scholar] [CrossRef]
- Brito, C.; Dinis, L.-T.; Moutinho-Pereira, J.; Correia, C.M. Drought stress effects and olive tree acclimation under a changing climate. Plants 2019, 8, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avramidou, E.V.; Doulis, A.G.; Petrakis, P.V. Chemometrical and molecular methods in olive oil analysis: A review. J. Food Process. Preserv. 2018, 42, e13770. [Google Scholar] [CrossRef]
- Koubouris, G.; Avramidou, E.; Metzidakis, I.; Petrakis, P.; Sergentani, C.; Doulis, A. Phylogenetic and evolutionary applications of analyzing endocarp morphological characters by classification binary tree and leaves by SSR markers for the characterization of olive germplasm. Tree Genet. Genomes 2019, 15, 26. [Google Scholar] [CrossRef]
- Sebastiani, L.; Busconi, M. Recent developments in olive (Olea europaea L.) genetics and genomics: Applications in taxonomy, varietal identification, traceability and breeding. Plant Cell Rep. 2017, 36, 1345–1360. [Google Scholar] [CrossRef] [PubMed]
- Belaj, A.; De La Rosa, R.; Lorite, I.J.; Mariotti, R.; Cultrera, N.G.; Beuzón, C.R.; González-Plaza, J.J.; Muñoz-Mérida, A.; Trelles, O.; Baldoni, L. Usefulness of a new large set of high throughput EST-SNP markers as a tool for olive germplasm collection management. Front. Plant Sci. 2018, 9, 1320. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Long, C.; Pang, X.; Ning, D.; Wu, T.; Dong, M.; Han, X.; Guo, H. The newly developed genomic-SSR markers uncover the genetic characteristics and relationships of olive accessions. PeerJ 2020, 8, e8573. [Google Scholar] [CrossRef]
- Barranco, D.; Trujillo, I.; Rallo, P. Are Oblonga’ and Frantoio’ Olives the Same Cultivar? HortScience 2000, 35, 1323–1325. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.; Stone, C.J.; Olshen, R.A. Classification and Regression Trees; CRC Press: Boca Raton, FL, USA, 1984. [Google Scholar]
- Petrakis, P.V.; Agiomyrgianaki, A.; Christophoridou, S.; Spyros, A.; Dais, P. Geographical characterization of Greek virgin olive oils (Cv. Koroneiki) using 1H and 31P NMR fingerprinting with canonical discriminant analysis and classification binary trees. J. Agric. Food Chem. 2008, 56, 3200–3207. [Google Scholar] [CrossRef]
- Agiomyrgianaki, A.; Petrakis, P.V.; Dais, P. Detection of refined olive oil adulteration with refined hazelnut oil by employing NMR spectroscopy and multivariate statistical analysis. Talanta 2010, 80, 2165–2171. [Google Scholar] [CrossRef] [PubMed]
- Agiomyrgianaki, A.; Petrakis, P.V.; Dais, P. Influence of harvest year, cultivar and geographical origin on Greek extra virgin olive oils composition: A study by NMR spectroscopy and biometric analysis. Food Chem. 2012, 135, 2561–2568. [Google Scholar] [CrossRef] [PubMed]
- Belbin, L.; Faith, D.P.; Milligan, G.W. A comparison of two approaches to beta-flexible clustering. Multivar. Behav. Res. 1992, 27, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Strobl, C.; Boulesteix, A.-L.; Augustin, T. Unbiased split selection for classification trees based on the Gini index. Comput. Stat. Data Anal. 2007, 52, 483–501. [Google Scholar] [CrossRef] [Green Version]
- Rokach, L.; Maimon, O. Classification trees. In Data Mining and Knowledge Discovery Handbook; Springer: Berlin/Heidelberg, Germany, 2009; pp. 149–174. [Google Scholar]
- Steinberg, D.; Colla, P. CART: Classification and regression trees. Top Ten Algorithms Data Min. 2009, 9, 179. [Google Scholar]
- Wilkinson, L. Systat. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 256–257. [Google Scholar] [CrossRef]
- Nisbet, R.; Elder, J.; Miner, G. Handbook of Statistical Analysis and Data Mining Applications; Academic Press: Cambridge, MA, USA, 2009. [Google Scholar]
- Paradis, E. Analysis of Phylogenetics and Evolution with R.; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Sneath, P.; Sokal, R. Unweighted pair group method with arithmetic mean. In Numerical Taxonomy; Springer: Berlin/Heidelberg, Germany, 1973; pp. 230–234. [Google Scholar]
- Hart, G. The occurrence of multiple UPGMA phenograms. In Numerical Taxonomy; Springer: Berlin/Heidelberg, Germany, 1983; pp. 254–258. [Google Scholar]
- Loewenstein, Y.; Portugaly, E.; Fromer, M.; Linial, M. Efficient algorithms for accurate hierarchical clustering of huge datasets: Tackling the entire protein space. Bioinformatics 2008, 24, i41–i49. [Google Scholar] [CrossRef] [Green Version]
- Aksehirli-Pakyurek, M.; Koubouris, G.; Petrakis, P.; Hepaksoy, S.; Metzidakis, I.; Yalcinkaya, E.; Doulis, A. Cultivated and Wild Olives in Crete, Greece—Genetic Diversity and Relationships with Major Turkish Cultivars Revealed by SSR Markers. Plant Mol. Biol. Report. 2017, 35, 575–585. [Google Scholar] [CrossRef]
- Baldoni, L.; Cultrera, N.G.; Mariotti, R.; Ricciolini, C.; Arcioni, S.; Vendramin, G.G.; Buonamici, A.; Porceddu, A.; Sarri, V.; Ojeda, M.A. A consensus list of microsatellite markers for olive genotyping. Mol. Breed. 2009, 24, 213–231. [Google Scholar] [CrossRef]
- Marshall, T.C.; Slate, J.; Kruuk, L.E.B.; Pemberton, J.M. Statistical confidence for likelihood-based paternity inference in natural populations. Mol. Ecol. 1998, 7, 639–655. [Google Scholar] [CrossRef] [Green Version]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Marra, F.; Caruso, T.; Costa, F.; Di Vaio, C.; Mafrica, R.; Marchese, A. Genetic relationships, structure and parentage simulation among the olive tree (Olea europaea L. Subsp. Europaea) cultivated in Southern Italy revealed by SSR markers. Tree Genet. Genomes 2013, 9, 961–973. [Google Scholar] [CrossRef]
- Earl, D.A. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Lynch, M.; Ritland, K. Estimation of pairwise relatedness with molecular markers. Genetics 1999, 152, 1753–1766. [Google Scholar] [PubMed]
- Atkinson, E.J.; Therneau, T.M. An Introduction to Recursive Partitioning Using the RPART Routines; Mayo Foundation: Rochester, NY, USA, 2000. [Google Scholar]
- Therneau, T.M.; Atkinson, E.J. An Introduction to Recursive Partitioning Using the RPART Routines; Technical Report; Mayo Foundation: Rochester, NY, USA, 1997. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Mantia, L.; Lain, T.; Caruso, T.; Testolin, R. SSR-based DNA fingerprints reveal the genetic diversity of Sicilian olive (Olea europaea L.) germplasm. J. Hortic. Sci. Biotechnol. 2005, 80, 628–632. [Google Scholar] [CrossRef]
- Lopes, M.S.; Mendonça, D.; Sefc, K.M.; Gil, F.S.; da Câmara Machado, A. Genetic evidence of intra-cultivar variability within Iberian olive cultivars. HortScience 2004, 39, 1562–1565. [Google Scholar] [CrossRef] [Green Version]
- Belaj, A.; del Carmen Dominguez-García, M.; Atienza, S.G.; Urdíroz, N.M.; De la Rosa, R.; Satovic, Z.; Martín, A.; Kilian, A.; Trujillo, I.; Valpuesta, V. Developing a core collection of olive (Olea europaea L.) based on molecular markers (DArTs, SSRs, SNPs) and agronomic traits. Tree Genet. Genomes 2012, 8, 365–378. [Google Scholar] [CrossRef]
- Sion, S.; Taranto, F.; Montemurro, C.; Mangini, G.; Camposeo, S.; Falco, V.; Gallo, A.; Mita, G.; Saddoud Debbabi, O.; Ben Amar, F. Genetic Characterization of Apulian Olive Germplasm as Potential Source in New Breeding Programs. Plants 2019, 8, 268. [Google Scholar] [CrossRef] [Green Version]
- Albertini, E.; Torricelli, R.; Bitocchi, E.; Raggi, L.; Marconi, G.; Pollastri, L.; Di Minco, G.; Battistini, A.; Papa, R.; Veronesi, F. Structure of genetic diversity in Olea europaea L. cultivars from central Italy. Mol. Breed. 2011, 27, 533–547. [Google Scholar] [CrossRef]
- Díez, C.M.; Trujillo, I.; Barrio, E.; Belaj, A.; Barranco, D.; Rallo, L. Centennial olive trees as a reservoir of genetic diversity. Ann. Bot. 2011, 108, 797–807. [Google Scholar] [CrossRef] [Green Version]
- Khadari, B.; Breton, C.; Moutier, N.; Roger, J.; Besnard, G.; Bervillé, A.; Dosba, F. The use of molecular markers for germplasm management in a French olive collection. Theor. Appl. Genet. 2003, 106, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Abdessemed, S.; Muzzalupo, I.; Benbouza, H. Assessment of genetic diversity among Algerian olive (Olea europaea L.) cultivars using SSR marker. Sci. Hortic. 2015, 192, 10–20. [Google Scholar] [CrossRef]
- Omrani-Sabbaghi, A.; Shahriari, M.; Falahati-Anbaran, M.; Mohammadi, S.; Nankali, A.; Mardi, M.; Ghareyazie, B. Microsatellite markers based assessment of genetic diversity in Iranian olive (Olea europaea L.) collections. Sci. Hortic. 2007, 112, 439–447. [Google Scholar] [CrossRef]
- Ginko, E.; Dobeš, C.; Saukel, J. Suitability of root and rhizome anatomy for taxonomic classification and reconstruction of phylogenetic relationships in the tribes cardueae and cichorieae (asteraceae). Sci. Pharm. 2016, 84, 585. [Google Scholar] [CrossRef] [Green Version]
- Germino, M.J.; Barnard, D.M.; Davidson, B.E.; Arkle, R.S.; Pilliod, D.S.; Fisk, M.R.; Applestein, C. Thresholds and hotspots for shrub restoration following a heterogeneous megafire. Landsc. Ecol. 2018, 33, 1177–1194. [Google Scholar] [CrossRef]
- Mousavi, S.; Mariotti, R.; Regni, L.; Nasini, L.; Bufacchi, M.; Pandolfi, S.; Baldoni, L.; Proietti, P. The first molecular identification of an olive collection applying standard simple sequence repeats and novel expressed sequence tag markers. Front. Plant Sci. 2017, 8, 1283. [Google Scholar] [CrossRef]
- Beiki, A.H.; Saboor, S.; Ebrahimi, M. A new avenue for classification and prediction of olive cultivars using supervised and unsupervised algorithms. PLoS ONE 2012, 7, e44164. [Google Scholar] [CrossRef] [Green Version]
- Cultrera, N.G.; Sarri, V.; Lucentini, L.; Ceccarelli, M.; Alagna, F.; Mariotti, R.; Mousavi, S.; Ruiz, C.G.; Baldoni, L. High levels of variation within gene sequences of Olea europaea L. Front. Plant Sci. 2019, 9, 1932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruz, F.; Julca, I.; Gómez-Garrido, J.; Loska, D.; Marcet-Houben, M.; Cano, E.; Galán, B.; Frias, L.; Ribeca, P.; Derdak, S. Genome sequence of the olive tree, Olea europaea. Gigascience 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Garantonakis, N.; Varikou, K.; Birouraki, A. Parasitism of psytallia concolor (hymenoptera: Braconidae) on bactrocera oleae (diptera: Tephritidae) infesting different olive varieties. Phytoparasitica 2017, 45, 461–469. [Google Scholar] [CrossRef]


| Cultivar Full Name | Origin | Number of Independent Genotypes Per Cultivar | Cultivar Full Name | Origin | Number of Independent Genotypes Per Cultivar |
|---|---|---|---|---|---|
| Adramytini | Greece | 2 | Makris | Greece | 1 |
| Aggouromanakolia | Greece | 3 | Manzanilla | Spain | 3 |
| Amfissis | Greece | 2 | Mastoidis | Greece | 2 |
| Arbequina | Spain | 1 | Matolia | Greece | 2 |
| Arbosana | Spain | 2 | Megareitiki | Greece | 2 |
| Asprolia Alexandroupolis | Greece | 1 | Myrtolia | Greece | 2 |
| Asprolia Lefkados | Greece | 2 | Nevadillo Blanco | Spain | 1 |
| Chalkidikis | Greece | 1 | Nevadillo Negro | Spain | 1 |
| Chondrolia Chalkidikis | Greece | 1 | Oblonga | USA | 1 |
| Dafnelia | Greece | 1 | Petrolia | Greece | 2 |
| Dopia Zakynthou | Greece | 1 | Picual | Spain | 2 |
| Frantoio Rodou | Greece | 1 | Pierias | Greece | 2 |
| Frantoio | Italy | 1 | Pikrolia | Greece | 2 |
| Gaidourelia | Greece | 1 | Picholine Marocaine | France | 1 |
| Galatistas | Greece | 1 | Rahati | Greece | 2 |
| Gordal | Spain | 1 | San Agostino | Italy | 1 |
| Kalamon | Greece | 2 | San Francesco | Italy | 1 |
| Kalokairida | Greece | 2 | Sigoise | Algeria | 1 |
| Karydolia | Greece | 2 | Stroggylolia | Greece | 1 |
| Kolybada | Greece | 2 | Thiaki | Greece | 3 |
| Koroneiki | Greece | 6 | Tragolia | Greece | 2 |
| Kothreiki | Greece | 2 | Throubolia | Greece | 3 |
| Koutsourelia | Greece | 2 | Throuba Thassou | Greece | 1 |
| Leccino | Italy | 1 | Valanolia | Greece | 2 |
| Lefkolia Serron | Greece | 1 | Vasilikada | Greece | 2 |
| Lianolia Kerkyras | Greece | 2 | O. europaea subsp. cuspidata | Not cultivated | 1 |
| LianomanakoTyrou | Greece | 1 |
| Na | Ne | Ho | He | PI | PIC | I | F(null) | F | |
|---|---|---|---|---|---|---|---|---|---|
| DCA3 | 11 | 6.288 | 0.864 | 0.846 | 0.045 | 0.821 | 1.955 | −0.014 | −0.027 |
| DCA5 | 13 | 4.489 | 0.663 | 0.782 | 0.069 | 0.758 | 1.92 | 0.075 | 0.147 |
| DCA9 | 15 | 11.172 | 1.000 | 0.916 | 0.015 | 0.904 | 2.522 | −0.047 | −0.098 |
| DCA14 | 13 | 5.097 | 0.529 | 0.809 | 0.063 | 0.779 | 1.928 | 0.198 | 0.341 |
| DCA16 | 19 | 7.993 | 0.759 | 0.880 | 0.028 | 0.862 | 2.319 | 0.067 | 0.133 |
| DCA18 | 13 | 7.468 | 0.932 | 0.871 | 0.031 | 0.853 | 2.235 | −0.044 | −0.076 |
| GAPU101 | 12 | 2.037 | 0.043 | 0.513 | 0.026 | 0.489 | 1.244 | 0.846a | 0.916 |
| UDO043 | 8 | 3.956 | 0.819 | 0.849 | 0.087 | 0.724 | 1.706 | 0.224a | 0.354 |
| GAPU71B | 12 | 6.423 | 0.547 | 0.799 | 0.042 | 0.826 | 2.043 | 0.014 | 0.03 |
| EMO90 | 10 | 4.871 | 0.483 | 0.752 | 0.070 | 0.766 | 1.789 | 0.179 | 0.312 |
| mean | 12.6 | 5.979 | 0.663 | 0.801 | 0.778 | 1.966 | 0.203 | ||
| combined | 1.708 × 10−13 |
| A | B | ||
|---|---|---|---|
| Locus | Number of Splits | Alleles | Number of Splits |
| DCA5 | 14 | DCA16_2 | 10 |
| DCA16 | 14 | DCA5_1 | 8 |
| EMO90 | 10 | DCA9_2 | 8 |
| DCA18 | 8 | EMO9__2 | 7 |
| DCA9 | 8 | DCA14_2 | 6 |
| GAPU71B | 8 | DCA5_2 | 6 |
| GAPU101 | 6 | Gapu101_2 | 6 |
| DCA14 | 6 | DCA14_1 | 5 |
| UDO043 | 6 | UDO043_1 | 5 |
| DCA3 | 2 | DCA16_1 | 4 |
| DCA18_1 | 4 | ||
| DCA18_2 | 4 | ||
| GAPU71B_1 | 4 | ||
| GAPU71B_2 | 4 | ||
| EMO90_1 | 3 | ||
| DCA3_1 | 1 | ||
| DCA3_2 | 1 | ||
| UDO043_2 | 1 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avramidou, E.V.; Koubouris, G.C.; Petrakis, P.V.; Lambrou, K.K.; Metzidakis, I.T.; Doulis, A.G. Classification Binary Trees with SSR Allelic Sizes: Combining Regression Trees with Genetic Molecular Data in Order to Characterize Genetic Diversity between Cultivars of Olea europaea L. Agronomy 2020, 10, 1662. https://doi.org/10.3390/agronomy10111662
Avramidou EV, Koubouris GC, Petrakis PV, Lambrou KK, Metzidakis IT, Doulis AG. Classification Binary Trees with SSR Allelic Sizes: Combining Regression Trees with Genetic Molecular Data in Order to Characterize Genetic Diversity between Cultivars of Olea europaea L. Agronomy. 2020; 10(11):1662. https://doi.org/10.3390/agronomy10111662
Chicago/Turabian StyleAvramidou, Evangelia V., Georgios C. Koubouris, Panos V. Petrakis, Katerina K. Lambrou, Ioannis T. Metzidakis, and Andreas G. Doulis. 2020. "Classification Binary Trees with SSR Allelic Sizes: Combining Regression Trees with Genetic Molecular Data in Order to Characterize Genetic Diversity between Cultivars of Olea europaea L." Agronomy 10, no. 11: 1662. https://doi.org/10.3390/agronomy10111662







