Diversity and Effectivity of Indigenous Mesorhizobium Strains for Chickpea (Cicer arietinum L.) in Myanmar
Abstract
:1. Introduction
2. Materials and Methods
2.1. Analysis of Collected Soil Samples
2.2. Isolation of Indigenous Root Nodule Bacteria from Soil Samples of Major Chickpea (Cicer Arietinum) Growing Areas of Myanmar
2.3. DNA Extraction, PCR Analysis, and Phylogenetic Analysis
2.4. Nucleotide Sequence Accession Numbers
2.5. Myanmar Chickpea Cultivars and Mesorhizobium Strains
2.6. Screening the Effectiveness of Mesorhizobial Strains for Nitrogen Fixation on Yezin-4 chickpea Variety
2.7. Evaluation the Effectiveness of Selected Mesorhizboium Strains on Two Myanmar Chickpea Cultivars: Yezin-4 and Yezin-6
2.8. Statistical Analysis
3. Results
3.1. Diversity of Indigenous Mesorhizobium Strains for Myanmar Chickpea (Cicer arietinum L.) Cultivars
3.2. Screening of Effective Bacterial Strains by Yezin-6 for Nitrogen Fixation
3.3. Effectivity of Selected Mesorhizobium Strains on Yezin-4 and Yezin-6 of Two Myanmar Chickpea Varieties
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nedumaran, S.; Abinaya, P.; Jyosthnaa, P.; Shraavya, B.; Rao, P.; Bantilan, C. Grain Legumes Production, Consumption and Trade Trends in Developing Countries; Working Paper Series No 60. ICRISAT Research Program, Markets, Institutions and Policies. Patancheru 502 324; International Crops Research Institute for the Semi-Arid Tropics: Telangana, India, 2015; p. 64. [Google Scholar]
- Laranjoa, M.; Alexandrea, A.; Oliveiraa, S. Legume growth-promoting rhizobia: An overview on theMesorhizobium genus. Microbiol. Res. 2014, 169, 2–17. [Google Scholar] [CrossRef]
- Merga, B.; Haji, J. Economic importance of chickpea: production, value, and world trade. Cogent Food Agric. 2019, 5, 1615718. [Google Scholar]
- Korir, H.; Mungai, N.W.; Thuita, M.; Hamba, Y.; Masso, C. Co-inoculation effect of rhizobia and plant growth promoting rhizobacteria on common bean growth in a low phosphorus soil. Front. Plant Sci. 2017, 8, 141. [Google Scholar] [CrossRef] [Green Version]
- Boonkerd, N.; Singleton, P. Production of rhizobium biofertilizer. In Biotechnology of Biofertilizers; Kannaiyan, S., Ed.; Narosa Publishing House: New Delhi, India, 2002; pp. 122–128. [Google Scholar]
- Sessitsch, A.; Howieson, J.G.; Perret, X.; Antoun, H.; Martínez- Romero, E. Advances in rhizobium research. Crit. Rev. Plant Sci. 2002, 21, 323–378. [Google Scholar] [CrossRef]
- Than, H.; Han, T. Contribution of nitrogen fixation to crop production in Myanmar. Proc. Myanmar Agric. Sci. Res. Div. 1988, 131–144. [Google Scholar]
- Than, M.M.; Myint, H.; Lwin, T.; Myint, Y.Y. Comparison of Nodulation efficiency between native rhizobium strains and exotic TAL strains on green gram (Vigna radiata). In Proceedings of the 3rd Agricultural Conference, Yezin Agriculture University, Yezin, Myanmar, 5–6 June 2003; pp. 50–58. [Google Scholar]
- Brockwell, J.; Dudman, W.F.; Gibson, A.H.; Hely, F.W.; Erbinsion, A.C. An integrated programme for the improvement of legume inoculant strains. Trans. 9th Intl. Cong. Soil Sci. 1968, 2, 103–114. [Google Scholar]
- Date, R.A. Microbiological problems in the inoculation and nodulation of legumes. Plant Soil Fla. Proc. 1970, 34, 71–79. [Google Scholar] [CrossRef]
- Than, M.M. Evaluation and selection of root nodule bacteria (Mesorhizobium ciceri) and chickpea germplasm for high nitrogen fixation. Ph.D. Thesis, Yezin Agriculture University, Naypyidaw, Myanmar, 2010; p. 118. [Google Scholar]
- Ohyama, T.; Ito, M.; Kobayashi, K.; Araki, S.; Yasuyoshi, S.; Sasaki, O.; Yamazaki, T.; Soyama, K.; Tanemura, R.; Mizuno, Y.; et al. Analytical procedures of N, P, K contents in plant and manure materials using H2SO4–H2O2 Kjeldahl digestion method. Jpn. Bull. Facul. Agric. Niigata Univ. 1991, 43, 110–120. [Google Scholar]
- Cataldo, D.A.; Schrader, L.E.; Youngs, V.L. Analysis by digestion and colorimetric assay of total nitrogen in plant tissues high in nitrate. Crop Sci. 1974, 14, 854–856. [Google Scholar] [CrossRef]
- Murphy, J.; Riley, J. A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 1962, 27, 31–36. [Google Scholar] [CrossRef]
- Muramoto, J.; Goto, I.; Ninaki, M. Rapid analysis of exchangeable cations and cation exchange capacity (CEC) of soils by shaking extraction method. J. Soil Sci. Plant Nutr. 1992, 63, 210–215. [Google Scholar]
- Downs, R.; Hellmers, H. Water and nutrition. In Environment and the Experimental Control of Plant Growth; Academic Press: London, UK, 1975; pp. 95–116. [Google Scholar]
- Nakano, Y.; Yamakawa, T.; Ikeda, M.; Ishizuka, J. Nodulation of Rj-soybean varieties with Rhizobium fredii USDA193 under limited supply of nutrients. Soil Sci. Plant Nutr. 1997, 43, 929–932. [Google Scholar] [CrossRef] [Green Version]
- Vincent, J.M. A Manual for the Practical Study of Root Nodule Bacteria; Blackwell Scientific Publications, Ltd.: Oxford, UK, 1970. [Google Scholar]
- Somasegaran, P.; Hoben, H.J. Handbook for Rhizobia. Methods in Legume-Rhizobium Technology; Springer: New York, NY, USA, 1994; pp. 240–258. [Google Scholar]
- Kuykendall, L.D. Isolation and identification of genetically marked strains of nitrogen-fixing microsymbionts of soybeans. In Practical Symbiotic Nitrogen Fixation Methodology; Elkan, G.H., Ed.; Marcel Dekker: New York, NY, USA, 1987; pp. 205–220. [Google Scholar]
- Sarr, P.S.; Yamakawa, T.; Fujimoto, S.; Saeki, Y.; Thao, H.T.B.; Myint, A.K. Phylogenetic diversity and symbiotic effectiveness of root-nodulating bacteria associated with cowpea in the south-west area of japan. Microbes Environ. 2009, 24, 285–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Kuykendall, L.D. Transfer of R factors to and between genetically marked sublines of Rhizobium japonicum. Appl. Environ. Microbiol. 1979, 37, 862–866. [Google Scholar] [CrossRef] [Green Version]
- Tsurumaru, H.; Yamakawa, T.; Tanaka, M.; Sakai, M. Tn5 mutants of Bradyrhizobium japonicum Is-1 with altered compatibility with Rj2-soybean cultivars. Soil Sci. Plant Nutr. 2008, 54, 197–203. [Google Scholar] [CrossRef]
- Yamakawa, T.; Shirai, T.; Ishizuka, J. Effects of symbiosis with Rhizobium fredii on transport of fixed nitrogen in the xylem of soybean plant. Soil Sci. Plant Nutr. 2000, 46, 885–892. [Google Scholar] [CrossRef]
- Haider, J.; Hussam, A.K.M.A.; Ikeda, M.; Yamakawa, T.; Ishizuka, J. Effects ofnitrate application on growth, nodulation and nitrogen fixation of nitrate-tolerant mutant soybean. Soil Sci. Plant. Nutr. 1991, 37, 521–529. [Google Scholar] [CrossRef]
- Aouani, M.E.; Mhamdi, R.; Jebara, M.; Amarger, N. Characterization of rhizobia nodulating chickpea in Tunisia. Agronomie 2001, 21, 577–581. [Google Scholar] [CrossRef] [Green Version]
- Kuykendall, L.D.; Gaur, Y.D.; Dutta, S.K. Genetic diversity among Rhizobium strains from Cicer arietinum L. Lett. Appl. Microbiol. 1993, 17, 259–263. [Google Scholar] [CrossRef]
- Maâtallah, J.; Berraho, E.B.; Muñoz, S.; Sanjuan, J.; Lluch, C. Phenotypic and molecular characterization of chickpea rhizobia isolated from different areas of Morocco. J. Appl. Microbiol. 2002, 93, 531–540. [Google Scholar] [CrossRef] [Green Version]
- Nour, S.M.; Cleyet-Marel, J.C.; Beck, D.; Effosse, A.; Fernandez, M.P. Genotypic and phenotypic diversity of Rhizobium isolated from chickpea (Cicer arietinum L.). Can. J. Microbiol. 1994, 40, 345–354. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.M.; Zhu, W.F.; Bontemps, C.; Young, J.P.W.; Wei, G.H. Mesorhizobium alhagi sp. nov., isolated from wild Alhagi sparsifolia in north-western China. Int. J. Syst. Evol. Microbiol. 2010, 60, 958–962. [Google Scholar] [CrossRef] [Green Version]
- Martens, M.; Delaere, M.; Coopman, R.; De Vos, P.; Gillis, M.; Willems, A. Multilocus sequence analysis of Ensifer and related taxa. Int. J. Syst. Evol. Microbiol. 2007, 57, 489–503. [Google Scholar] [CrossRef] [Green Version]
- WonLaranjo, M.; Young, J.P.W.; Oliveira, S. Multilocus sequence analysis reveals multiple symbiovars within Mesorhizobium Species. Syst. Appl. Microbiol. 2012, 35, 359–367. [Google Scholar]
- Tena, W.; Wolde-Meskel, E.; Degefu, T.; Walley, F. Genetic and phenotypic diversity of rhizobia nodulating chickpea (Cicer arietinum L.) in soils from southern and central Ethiopia. Can. J. Microbiol. 2017, 63, 690–707. [Google Scholar] [CrossRef] [Green Version]
- Rai, R.; Dash, P.K.; Mohapatra, T.; Singh, A. Phenotypic and molecular characterization of indigenous rhizobia nodulating chickpea in India. Indian J. Exp. Biol. 2012, 50, 340–350. [Google Scholar] [PubMed]
- Kim, D.H.; Kaashyap, M.; Rathore, A.; Das, R.R.; Parupalli, S.; Upadhyaya, H.D.; Gopalakrishnan, S.; Gaur, P.M.; Singh, S.; Kaur, J.; et al. Phylogenetic diversity of Mesorhizobium in chickpea. J. Biosci. 2014, 39, 513–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, E.T.; Kan, F.L.; Tan, Z.Y.; Toledo, I.; Chen, W.X.; Martinez-Romero, E. Diverse Mesorhizobium plurifarium populations native to Mexican soils. Arch. Microbiol. 2003, 180, 444–454. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.W.; Park, J.Y.; Kim, J.S.; Kang, J.W. Phylogenetic analysis of the genera Bradyrhizobium, Mesorhizobium, Rhizobium and Sinorhizobium on the basis of 16S rRNA gene and internally transcribed spacer region sequences. Int. J. Syst. Evol. Microbiol. 2005, 55, 263–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.J.; Liu, T.Y.; Chen, W.F.; Wang, E.T.; Sui, X.H.; Zhang, X.X.; Li, Y.; Chen, W.X. Mesorhizobium muleiense sp. nov., nodulating with Cicer arietinum L. Int. J. Syst. Evol. Microbiol. 2012, 62, 2737–2742. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.J.; Yu, T.; Lou, K.; Mao, P.H.; Wang, E.T.; Chen, W.X. Genotypic alteration and competitive nodulation of Mesorhizobium muleiense against exotic chickpea rhizobia in alkaline soils. Syst. Appl. Microbiol. 2014, 37, 520–524. [Google Scholar] [CrossRef]
- Zhang, J.J.; Yang, X.; Guo, C.; de-Lajudie, P.; Singh, R.P.; Wang, E.; Chen, W. Mesorhizobium muleiense and Mesorhizobium gsp. nov. are symbionts of Cicer arietinum L. in alkaline soils of Gansu, Northwest China. Plant Soil. 2017, 410, 103–112. [Google Scholar] [CrossRef]
- Laranjo, M.; Machado, J.; Young, J.P.W.; Oliveira, S. High diversity of chickpea Mesorhizobium species isolated in a Portuguese agricultural region. FEMS Microbiol. Ecol. 2004, 48, 101–107. [Google Scholar] [CrossRef] [Green Version]
- Laranjo, M.; Alexandre, A.; Rivas, R.; Velázquez, E.; Young, J.P.W.; Oliveira, S. Chickpea rhizobia symbiosis genes are highly conserved across multiple Mesorhizobium species. FEMS Microbiol. Ecol. 2008, 66, 391–400. [Google Scholar] [CrossRef]
- Rivas, R.; Laranjo, M.; Mateos, P.F.; Oliveira, S.; Martinez-Molina, E.; Vel’azquez, E. Strains of Mesorhizobium amorphae and Mesorhizobium tianshanense, carrying symbiotic genes of common chickpea endosymbiotic species, constitute a novel biovar (ciceri) capable of nodulating Cicer arietinum. Lett Appl. Microbiol. 2007, 44, 412–418. [Google Scholar] [CrossRef]
- Suneja, P.; Dudeja, S.S.; Dahiya, P. Deciphering the phylogenetic relationships among rhizobia nodulating chickpea: a review. J. Appl. Biol. Biotechnol. 2016, 4, 61–70. [Google Scholar]
- Elias, N.V.; Herridge, D.F. Naturalised populations of mesorhizobia in chickpea (Cicer arietinum L.) cropping soils: effects on nodule occupancy and productivity of commercial chickpea. Plant Soil. 2015, 387, 233–249. [Google Scholar] [CrossRef]
- Laranjo, M.; Oliveira, S. Tolerance of Mesorhizobium type strains to different environmental stresses. Antonie van Leeuwenhoek. 2011, 99, 651–662. [Google Scholar] [CrossRef]
- Brígido, C.; van Tuinen, D.; Brito, I.; Alho, L.; Goss, M.J.; Carvalho, M. Management of the biological diversity of AM fungi by combination of host plant succession and integrity of extraradical mycelium. Soil Biol. Biochem. 2017, 112, 237–247. [Google Scholar] [CrossRef] [Green Version]
- Brígido, C.; Nascimento, F.X.; Duan, J.; Glick, B.R.; Oliveira, S. Expression of an exogenous 1-aminocyclopropane-1-carboxylate deaminase gene in Mesorhizobium spp. reduces the negative effects of salt stress in chickpea. FEMS Microbiol. Lett. 2013, 349, 46–53. [Google Scholar]
- Teamroong, N.; Boonkerd, N. Rhizobial production technology. In Microbial Biotechnology in Agriculture and Aquaculture; Science Publishers: Bhubaneswar, Orissa, India, 2006; Ray RC, Vo.2. [Google Scholar]
- Soe, K.M.; Yamakawa, T. Evaluation of effective Myanmar Bradyrhizobium strains isolated from Myanmar soybean and effects of coinoculation with Streptomyces griseoflavus P4 for nitrogen fixation. Soil Sci Plant. Nutr. 2013, 59, 361–370. [Google Scholar] [CrossRef]
- Wani, S.P.; Rupela, O.P.; Lee, K.K. Sustainable agriculture in a semiarid tropic through biological nitrogen fixation in grain legume. Plant Soil. 1995, 174, 29–49. [Google Scholar] [CrossRef]
- Lie, T.A.; Mulder, E.G. Breeding for symbiotic nitrogen fixation. In Plant Breeding Perspectives; Sneep, J., Hendriksen, A.J.T., Eds.; Center for Agricultural Publishing and Documentation: Wagenigen, The Netherlands, 1979; pp. 341–348. [Google Scholar]
- Ibekwe, A.M.; Angle, J.S.; Chaney, R.L.; vanBerkum, P. Enumeration and N-2 fixation potential of Rhizobium leguminosarum biovar trifolii grown in soil with varying pH values and heavy metal concentrations. Agric. Ecosyst. Environ. 1997, 61, 103–111. [Google Scholar] [CrossRef]
- Rao, D.L.N.; Giller, K.E.; Yeo, A.R.; Flowers, T.J. The effects of salinity and sodicity upon nodulation and nitrogen fixation in chickpea (Cicer arietinum). Ann. Bot. 2002, 89, 563–570. [Google Scholar] [CrossRef] [Green Version]
- Teamroong, N.; Boonkerd, N. Rhizobial Production Technology. In Microbial Biotechnology in Agriculture and Aquaculture; Ray, C.R., Ed.; Central Tuber Crops Research Institute (Regional Center), Science Publishers: Bhubaneswar, Orissa, 2006; pp. 83–86. [Google Scholar]
- Howieson, J.G.; Hara, G.W.O.; Carr, S.J. Changing roles for legumes in Mediterranean agriculture: developments from an Australian perspective. Field Crop. Res. 2000, 65, 107–122. [Google Scholar] [CrossRef]
- Ben Romdhane, S.; Tajini, F.; Trabelsi, M.; Aouani, M.E.; Mhamdi, R. Competition for nodule formation between introduced strains of Mesorhizobium ciceri and the native populations of rhizobia nodulating chickpea (Cicer arietinum) in Tunisia. World J. Microbiol. Biotechnol. 2007, 23, 1195–1201. [Google Scholar] [CrossRef]
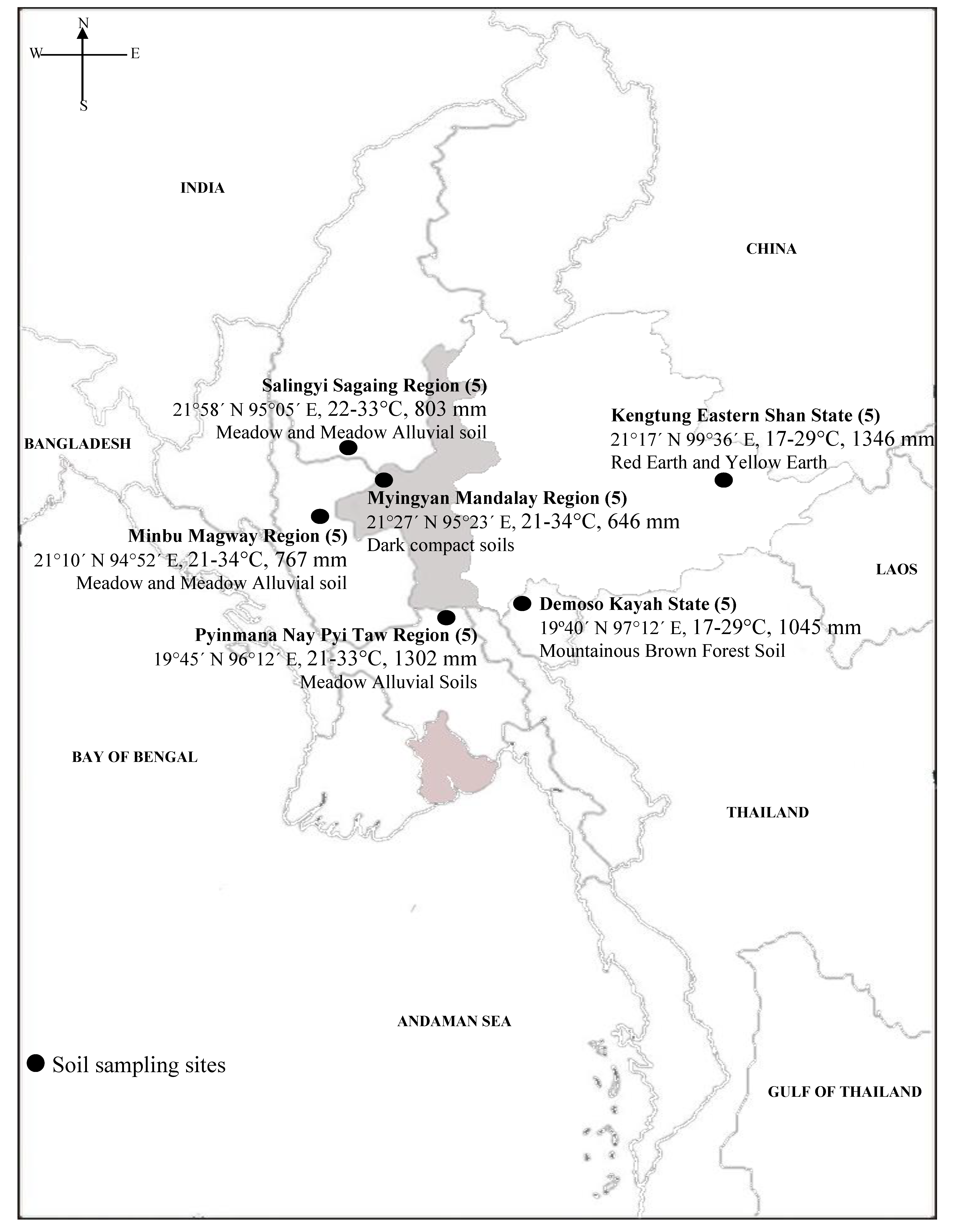
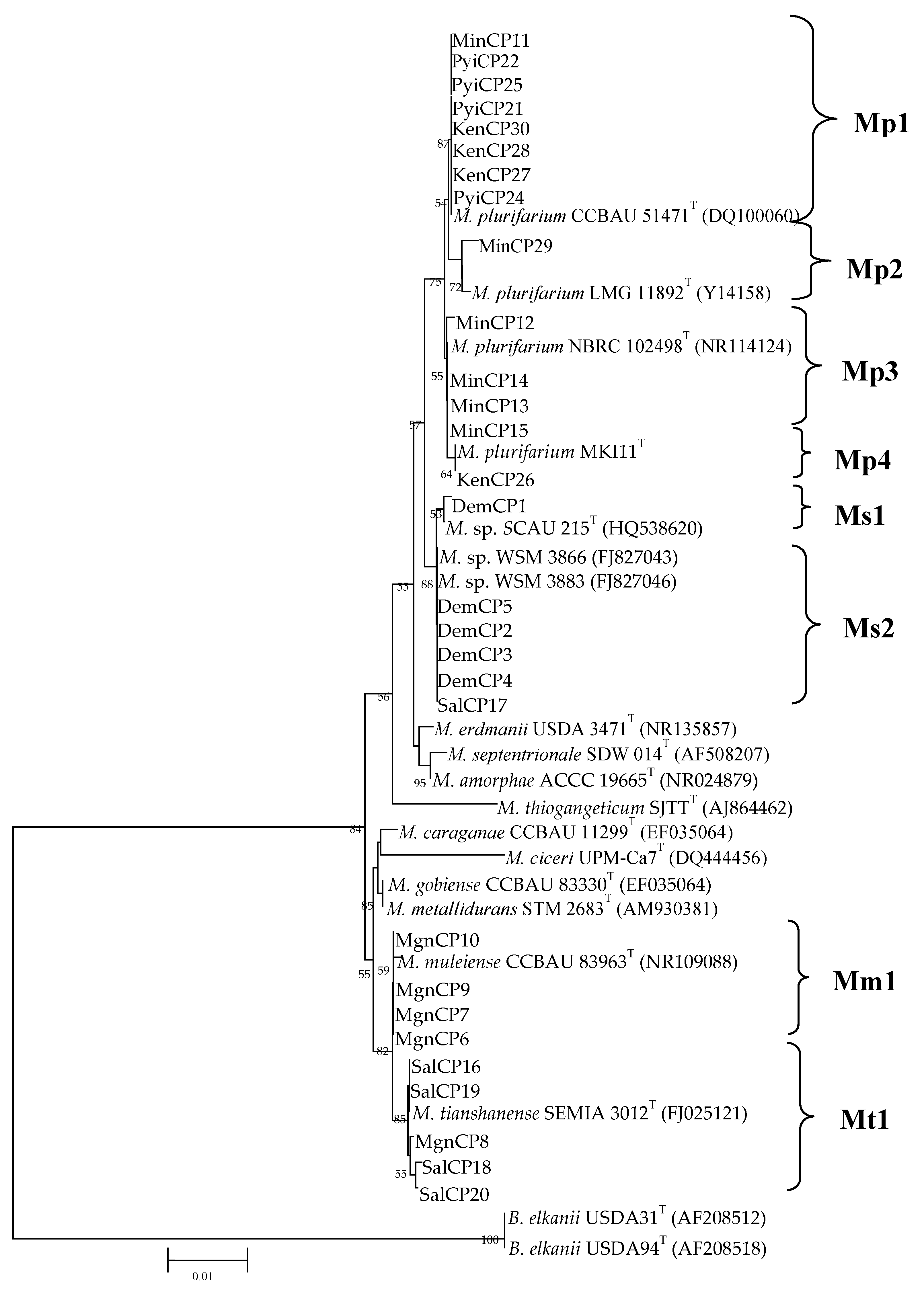
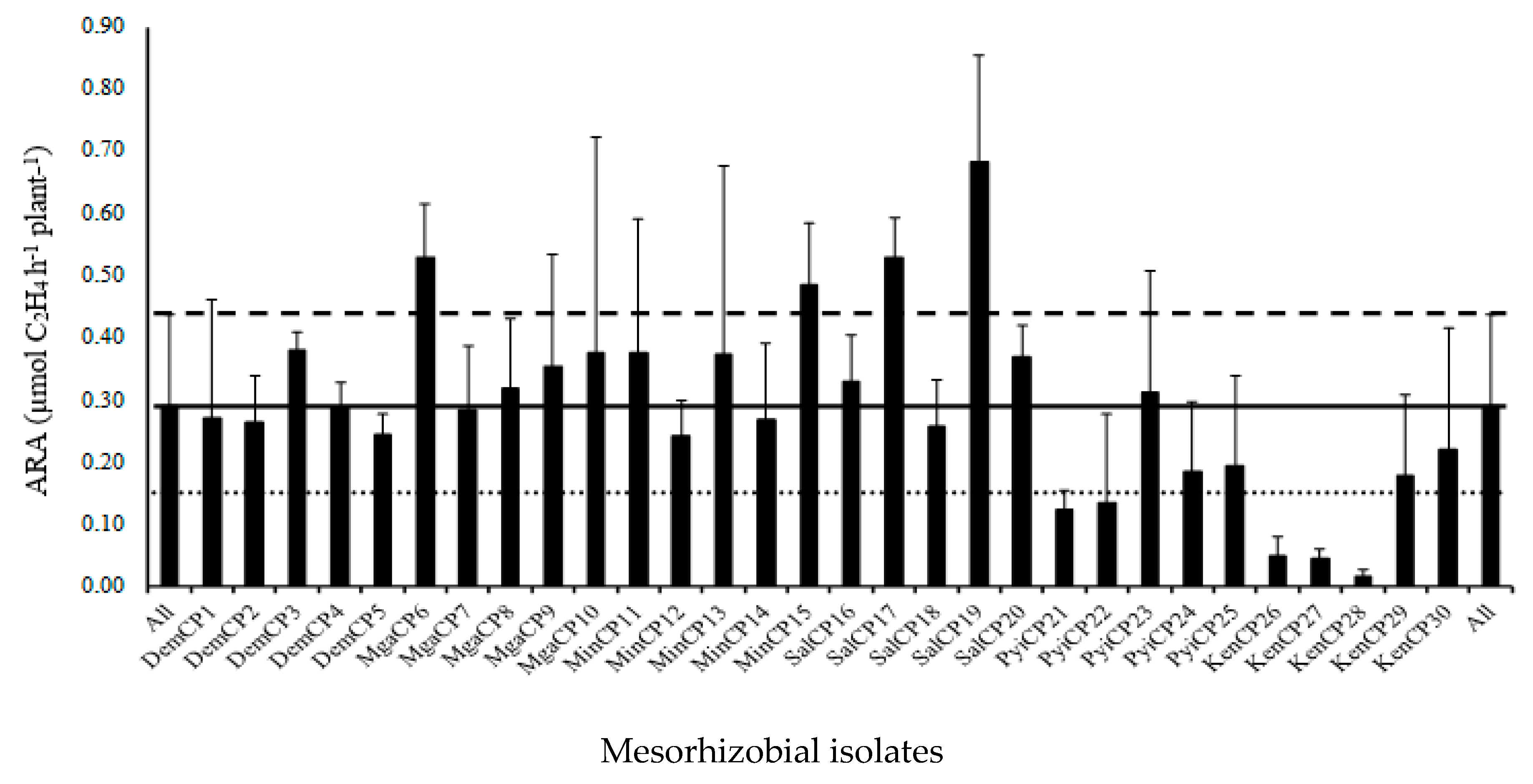
| Soil Sampling Site | Soil Classification * | Location | Climate ** (Temp; RF) |
|---|---|---|---|
| Demoso, Kayah State | Mountainous Brown Forest Soil | 19°40’ N 97°12’ E | 17-29 °C, 1045 mm |
| Myingyan, Mandalay Region | Dark Compact Soil | 21°27’ N 95°23’ E | 21-34 °C, 646 mm |
| Minbu, Magway Region | Meadow and Meadow Alluvial Soil | 21°10’ N 94°52’ E | 21-34 °C, 767 mm |
| Salingyi, Sagaing Region | Meadow and Meadow Alluvial Soil | 21°58’ N 95°05’ E | 22-33 °C, 803 mm |
| Pyinmanar, Nay Pyi Taw Region | Meadow Alluvial Soil | 19°45’ N 96°12’ E | 21-33 °C, 1302 mm |
| Kengtung, Eastern Shan State | Red Earth and Yellow Earth | 21°17’ N 99°36’ E | 17-29 °C, 1346 mm |
| Physicochemical Property | Demoso, KYS | Myingyan, MDR | Minbu, MGR | Salingyi, SGR | Pyinmanar, NPTR | Kengtung, ESS |
|---|---|---|---|---|---|---|
| Soil pH (Soil: H2O; 1:2.5) | 7.15 | 7.44 | 6.30 | 7.27 | 6.15 | 4.79 |
| Total N (%) | 0.17 | 0.06 | 0.19 | 0.06 | 0.18 | 0.16 |
| Mineralizable N (g/kg) | 1.43 | 0.32 | 1.56 | 0.48 | 1.81 | 0.51 |
| Total P2O5 (%) | 0.12 | 0.02 | 0.12 | 0.03 | 0.31 | 0.10 |
| Total K2O (%) | 1.08 | 0.18 | 1.44 | 0.41 | 1.20 | 0.77 |
| Strain Name | Genus and Species | Isolated Origin | Dendrogram Cluster | Shape | Size (mm) |
|---|---|---|---|---|---|
| DemCP1 | Mesorhizobium sp. | Demoso, Kayah State | Ms1 | UF | 2.0 |
| DemCP2 | Mesorhizobium sp. | Demoso, Kayah State | Ms2 | UF | 2.0 |
| DemCP3 | Mesorhizobium sp. | Demoso, Kayah State | Ms2 | UF | 2.0 |
| DemCP4 | Mesorhizobium sp. | Demoso, Kayah State | Ms2 | UF | 2.0 |
| DemCP5 | Mesorhizobium sp. | Demoso, Kayah State | Ms2 | UF | 2.0 |
| MgnCP6 | Mesorhizobium muleiense | Myingyan, Mandalay Region | Mm1 | UF | 1.5 |
| MgnCP7 | Mesorhizobium muleiense | Myingyan, Mandalay Region | Mm1 | UF | 1.5 |
| MgnCP8 | Mesorhizobium tianshanense | Myingyan, Mandalay Region | Mt1 | UF | 1.5 |
| MgnCP9 | Mesorhizobium muleiense | Myingyan, Mandalay Region | Mm1 | UF | 1.5 |
| MgnCP10 | Mesorhizobium muleiense | Myingyan, Mandalay Region | Mm1 | UF | 1.5 |
| MinCP11 | Mesorhizobium plurifarium | Minbu, Magway Region | Mp1 | EP | 3.0 |
| MinCP12 | Mesorhizobium plurifarium | Minbu, Magway Region | Mp3 | EP | 3.0 |
| MinCP13 | Mesorhizobium plurifarium | Minbu, Magway Region | Mp3 | EP | 3.0 |
| MinCP14 | Mesorhizobium plurifarium | Minbu, Magway Region | Mp3 | EP | 3.0 |
| MinCP15 | Mesorhizobium plurifarium | Minbu, Magway Region | Mp3 | EP | 3.0 |
| SalCP16 | Mesorhizobium tianshanense | Salingyi, Sagaing Region | Mt1 | UF | 2.0 |
| SalCP17 | Mesorhizobium sp. | Salingyi, Sagaing Region | Ms2 | UF | 2.0 |
| SalCP18 | Mesorhizobium tianshanense | Salingyi, Sagaing Region | Mt1 | UF | 2.0 |
| SalCP19 | Mesorhizobium tianshanense | Salingyi, Sagaing Region | Mt1 | UF | 2.0 |
| SalCP20 | Mesorhizobium tianshanense | Salingyi, Sagaing Region | Mt1 | UF | 2.0 |
| PyiCP21 | Mesorhizobium plurifarium | Pyinmanar, Nay Pyi Taw Region | Mp1 | EP | 2.5 |
| PyiCP22 | Mesorhizobium plurifarium | Pyinmanar, Nay Pyi Taw Region | Mp1 | EP | 2.5 |
| PyiCP23 | Mesorhizobium plurifarium | Pyinmanar, Nay Pyi Taw Region | Mp2 | EP | 2.5 |
| PyiCP24 | Mesorhizobium plurifarium | Pyinmanar, Nay Pyi Taw Region | Mp1 | EP | 2.5 |
| PyiCP25 | Mesorhizobium plurifarium | Pyinmanar, Nay Pyi Taw Region | Mp1 | EP | 2.5 |
| KenCP26 | Mesorhizobium plurifarium | Kengtung, Eastern Shan State | Mp4 | UF | 2.0 |
| KenCP27 | Mesorhizobium plurifarium | Kengtung, Eastern Shan State | Mp1 | UF | 2.0 |
| KenCP28 | Mesorhizobium plurifarium | Kengtung, Eastern Shan State | Mp1 | UF | 2.0 |
| KenCP29 | Mesorhizobium plurifarium | Kengtung, Eastern Shan State | Mp2 | UF | 2.0 |
| KenCP30 | Mesorhizobium plurifarium | Kengtung, Eastern Shan State | Mp1 | UF | 2.0 |
| Field Sites | M.plurifarium | M. sp. | M. muleiense | M. tianshanense | ||||
|---|---|---|---|---|---|---|---|---|
| % | No. Clusters | % | No. Clusters | % | No. Clusters | % | No. Clusters | |
| Demoso, Kayah State | - | - | 100 | 2 | - | - | - | - |
| Myingyan, Mandalay Region | - | - | - | - | 80 | 1 | 20 | 1 |
| Minbu, Magway Region | 100 | 2 | - | - | - | - | - | - |
| Salingyi, Sagaing Region | - | - | 20 | 1 | - | 80 | 1 | |
| Pyinmanar, Nay Pyi Taw Region | 100 | 2 | - | - | - | - | - | - |
| Kengtung, Eastern Shan State | 100 | 3 | - | - | - | - | - | - |
| Total | 50 | 20 | 13 | 17 | ||||
| Treatment | Yezin-4 | Yezin-6 | ||||||
|---|---|---|---|---|---|---|---|---|
| NDW (mg plant−1) | RDW (g plant−1) | SDW (g plant−1) | ARA (µmole C2H4 h−1 plant−1) | NDW (mg plant−1) | RDW (g plant−1) | SDW (g plant−1) | ARA (µmole C2H4 h−1 plant−1) | |
| DemCP4 | 8.77 b | 0.07 a | 0.17 a | 0.31 b | 7.13 b | 0.08 a | 0.15 a | 0.24 b |
| MgnCP6 | 14.57 ab | 0.08 a | 0.21 a | 0.54 ab | 10.27 ab | 0.08 a | 0.19 a | 0.31 ab |
| MinCP15 | 9.87 b | 0.09 a | 0.21 a | 0.46 ab | 9.50 ab | 0.09 a | 0.19 a | 0.39 ab |
| SalCP17 | 14.47 ab | 0.10 a | 0.22 a | 0.55 a | 13.23 a | 0.10 a | 0.22 a | 0.57 a |
| SalCP19 | 22.87 a | 0.09 a | 0.22 a | 0.66 a | 12.83 ab | 0.09 a | 0.20 a | 0.54 ab |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soe, K.M.; Htwe, A.Z.; Moe, K.; Tomomi, A.; Yamakawa, T. Diversity and Effectivity of Indigenous Mesorhizobium Strains for Chickpea (Cicer arietinum L.) in Myanmar. Agronomy 2020, 10, 287. https://doi.org/10.3390/agronomy10020287
Soe KM, Htwe AZ, Moe K, Tomomi A, Yamakawa T. Diversity and Effectivity of Indigenous Mesorhizobium Strains for Chickpea (Cicer arietinum L.) in Myanmar. Agronomy. 2020; 10(2):287. https://doi.org/10.3390/agronomy10020287
Chicago/Turabian StyleSoe, Khin Myat, Aung Zaw Htwe, Kyi Moe, Abiko Tomomi, and Takeo Yamakawa. 2020. "Diversity and Effectivity of Indigenous Mesorhizobium Strains for Chickpea (Cicer arietinum L.) in Myanmar" Agronomy 10, no. 2: 287. https://doi.org/10.3390/agronomy10020287
APA StyleSoe, K. M., Htwe, A. Z., Moe, K., Tomomi, A., & Yamakawa, T. (2020). Diversity and Effectivity of Indigenous Mesorhizobium Strains for Chickpea (Cicer arietinum L.) in Myanmar. Agronomy, 10(2), 287. https://doi.org/10.3390/agronomy10020287





