Phenolic Acid-Degrading Consortia Increase Fusarium Wilt Disease Resistance of Chrysanthemum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Soil Sampling
2.2. Soil Disease Suppressiveness and Fusarium Wilt Incidence in Pot Assays
2.3. Pyrosequencing and Quantification of Rhizomicrobial abundance
2.4. Detection of PA Content, Microbial degradation, Mcrobial inoculation and Colony counting
2.5. Effects of PAs on the Growth of F. oxysporum
2.6. Bioinformatics and Statistical analyses
3. Results
3.1. Soil Characteristics and Fusarium Wilt Disease Incidence
3.2. Rhizosphere Microbial Community Composition
3.3. Rhizosphere Microbial Community Abundance and Diversity
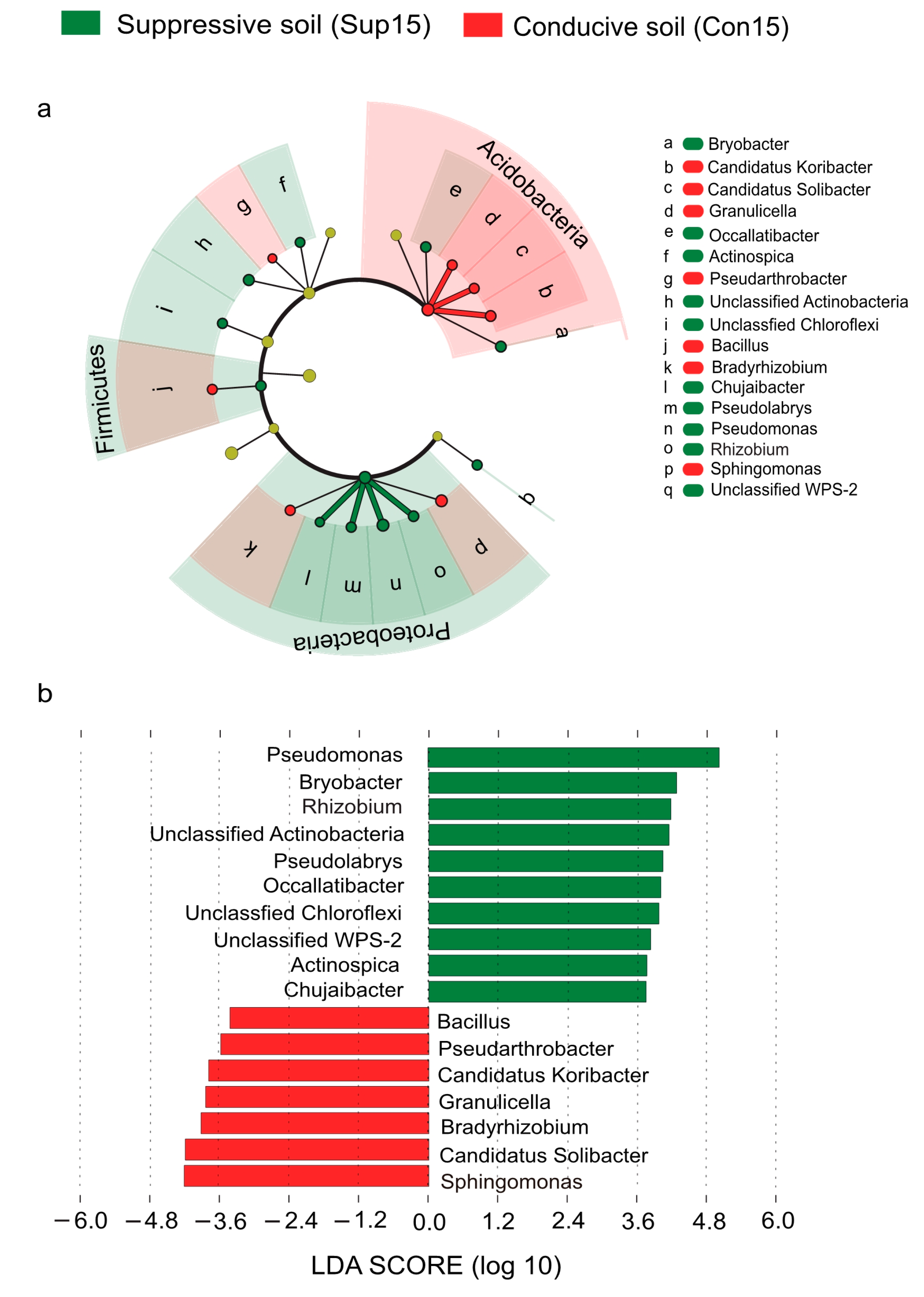
3.4. Bacterial Indicators of Disease Suppression to Fusarium Wilt Disease
3.5. Functional Selection of Consortia Associated with Soil Disease Suppressiveness
3.6. Efficient Suppression of Fusarium Wilt Disease by a PA-degrading Consortium
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Li, X.G.; Ding, C.F.; Hua, K.; Zhang, T.L.; Zhang, Y.N.; Zhao, L.; Yang, Y.R.; Liu, J.G.; Wang, X.X. Soil sickness of peanuts is attributable to modifications in soil microbes induced by peanut root exudates rather than to direct allelopathy. Soil Biol. Biochem. 2014, 78, 149–159. [Google Scholar] [CrossRef]
- Shen, Z.; Penton, C.R.; Lv, N.; Xue, C.; Yuan, X.; Ruan, Y.; Li, R.; Shen, Q. Banana Fusarium wilt disease incidence is influenced by shifts of soil microbial communities under different monoculture spans. Microb. Ecol. 2018, 75, 739–750. [Google Scholar] [CrossRef]
- Li, C.; Li, X.; Kong, W.; Wu, Y.; Wang, J. Effect of monoculture soybean on soil microbial community in the Northeast China. Plant Soil 2010, 330, 423–433. [Google Scholar] [CrossRef]
- Xiong, W.; Li, R.; Ren, Y.; Liu, C.; Zhao, Q.Y.; Wu, H.S.; Jousset, A.; Shen, Q.R. Distinct roles for soil fungal and bacterial communities associated with the suppression of vanilla Fusarium wilt disease. Soil Biol. Biochem. 2017, 101, 198–200. [Google Scholar] [CrossRef]
- Larkin, R.P. Soil health paradigms and implications for disease management. Annu. Rev. Phytopathol. 2015, 53, 199–221. [Google Scholar] [CrossRef] [PubMed]
- Kwak, M.J.; Kong, H.G.; Choi, K.; Kwon, S.K.; Song, J.Y.; Lee, J.; Lee, P.A.; Choi, S.Y.; Seo, M.; Lee, H.J.; et al. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 2018, 36, 1117. [Google Scholar] [CrossRef] [PubMed]
- Carrión, V.J.; Perez-Jaramillo, J.; Cordovez, V.; Tracanna, V.; de Hollander, M.; Ruiz-Buck, D.; Mendes, L.W.; van Ijcken, W.F.J.; Gomez-Exposito, R.; Elsayed, S.S.; et al. Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 2019, 366, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Siegel-Hertz, K.; Edel-Hermann, V.; Chapelle, E.; Terrat, S.; Raaijmakers, J.M.; Steinberg, C. Comparative microbiome analysis of a Fusarium wilt suppressive soil and a Fusarium wilt conducive soil from the Châteaurenard Region. Front. Microbiol. 2018, 9, 568. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Kruijt, M.; De Bruijn, I.; Dekkers, E.; van der Voort, M.; Schneider, J.H.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A.; et al. Deciphering the rhizosphere microbiome for disease suppressive bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef]
- Cha, J.Y.; Han, S.; Hong, H.J.; Cho, H.; Kim, D.; Kwon, Y.; Kwon, S.K.; Crüsemann, M.; Bok Lee, Y.; Kim, J.F.; et al. Microbial and biochemical basis of a Fusarium wilt-suppressive soil. ISME J. 2016, 10, 119–129. [Google Scholar] [CrossRef] [Green Version]
- Lebreton, L.; Lucas, P.; Dugas, F.; Guillerm, A.Y.; Schoeny, A.; Sarniguet, A. Changes in population structure of the soil borne fungus Gaeumannomyces graminis var. tritici during continuous wheat cropping. Environ. Microbiol. 2004, 6, 1174–1185. [Google Scholar] [CrossRef] [PubMed]
- Kyselková, M.; Kopecký, J.; Frapolli, M.; Défago, G.; Ságová-Marecková, M.; Grundmann, G.L.; Moënne-Loccoz, Y. Comparison of rhizobacterial community composition in soil suppressive or conducive to tobacco black root rot disease. ISME J. 2009, 3, 1127–1138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garbeva, P.; van Veen, J.A.; van Elsas, J.D. Microbial diversity in soil: Selection of microbial populations by plant and soil type and implications for disease suppressiveness. Annu. Rev. Phytopathol. 2004, 42, 243–270. [Google Scholar] [CrossRef] [PubMed]
- Ramette, A.; Moënne-Loccoz, Y.; Défago, G. Prevalence of fluorescent pseudomonads producing antifungal phloroglucinols and/or hydrogen cyanide in soils naturally suppressive or conducive to tobacco black root rot. FEMS Microbiol. Ecol. 2003, 44, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Manici, L.M.; Caputo, F. Fungal community diversity and soil health in intensive potato cropping systems of the east Po valley, northern Italy. Ann. Appl. Biol. 2009, 155, 245–258. [Google Scholar] [CrossRef]
- Figuerola, E.L.; Guerrero, L.D.; Türkowsky, D.; Wall, L.G.; Erijman, L. Crop monoculture rather than agriculture reduces the spatial turnover of soil bacterial communities at a regional scale. Environ. Microbiol. 2015, 17, 678–688. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Zhao, Q.; Zhao, J.; Xun, W.; Li, R.; Zhang, R.; Wu, H.; Shen, Q. Different continuous cropping spans significantly affect microbial community membership and structure in a vanilla-grown soil as revealed by deep pyrosequencing. Microb. Ecol. 2015, 70, 209–218. [Google Scholar] [CrossRef]
- Frapolli, M.; Moënne-Loccoz, Y.; Meyer, J.; Défago, G. A new DGGE protocol targeting 2,4-diacetylphloroglucinol biosynthetic gene phlD from phylogenetically contrasted biocontrol pseudomonads for assessment of disease-suppressive soils. FEMS Microbiol. Ecol. 2008, 64, 468–481. [Google Scholar] [CrossRef] [Green Version]
- Inderbitzin, P.; Ward, J.; Barbella, A.; Solares, N.; Izyumin, D.; Burman, P.; Chellemi, D.O.; Subbarao, K.V. Soil microbiomes associated with Verticillium wilt-suppressive broccoli and chitin amendments are enriched with potential biocontrol agents. Phytopathology 2018, 108, 31–43. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.R.; Jeon, C.W.; Shin, J.H.; Weller, D.M.; Thomashow, L.; Kwak, Y.S. Function and distribution of a lantipeptide in strawberry Fusarium wilt disease-suppressive soils. Mol. Plant Microbe Interact. 2019, 32, 306–312. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Wu, L.; Wang, J.; Zhu, Q.; Lin, S.; Xu, J.; Zheng, C.; Chen, J.; Qin, X.; Fang, C.; et al. Mixed phenolic acids mediated proliferation of pathogens Talaromyces helicus and Kosakonia sacchari in continuously monocultured Radix pseudostellariae rhizosphere Soil. Front. Microbiol. 2016, 7, 335. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wang, J.; Huang, W.; Wu, H.; Chen, J.; Yang, Y.; Zhang, Z.; Lin, W. Plant-microbe rhizosphere interactions mediated by Rehmannia glutinosa root exudates under consecutive monoculture. Sci. Rep. 2015, 5, 15871. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.Y.; Pan, L.P.; Li, H.H. Isolation, identification and characterization of soil microbes which degrade phenolic allelochemicals. J. Appl. Microbiol. 2010, 108, 1839–1849. [Google Scholar] [CrossRef] [PubMed]
- Chong, N.M.; Chang, C.S.; Tsai, S.C. Evolutions of microbial degradation pathways for parent xenobiotic and for its metabolites follow different schemes. Environ. Sci. Pollut. Res. Int. 2012, 19, 3276–3281. [Google Scholar] [CrossRef] [PubMed]
- Sagarkar, S.; Mukherjee, S.; Nousiainen, A.; Björklöf, K.; Purohit, H.J.; Jørgensen, K.S.; Kapley, A. Monitoring bioremediation of atrazine in soil microcosms using molecular tools. Environ. Pollut. 2013, 172, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, Y.; Zhang, K.; Wang, X.; Ma, C.; Tang, H.; Xu, P. Atrazine degradation by a simple consortium of Klebsiella sp. A1 and Comamonas sp. A2 in nitrogen enriched medium. Biodegradation 2010, 21, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zarecki, R.; Medina, S.; Ofaim, S.; Liu, X.; Chen, C.; Hu, S.; Brom, D.; Gat, D.; Porob, S. Modeling microbial communities from atrazine contaminated soils promotes the development of biostimulation solutions. ISME J. 2019, 13, 494–508. [Google Scholar] [CrossRef] [Green Version]
- de Souza, M.L.; Newcombe, D.; Alvey, S.; Crowley, D.E.; Hay, A.; Sadowsky, M.J.; Wackett, L.P. Molecular basis of a bacterial consortium: Interspecies catabolism of atrazine. Appl. Environ. Microbiol. 1998, 64, 178–184. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.; Alvey, S.; Crowley, D.E. Cooperative catabolic pathways within an atrazine-degrading enrichment culture isolated from soil. FEMS Microbiol. Ecol. 2005, 53, 265–275. [Google Scholar] [CrossRef]
- Bento, F.M.; Camargo, F.A.O.; Okeke, B.C.; Frankenberger, W.T. Comparative bioremediation of soils contaminated with diesel oil by natural attenuation, biostimulation and bioaugmentation. Bioresour. Technol. 2005, 96, 1049–1055. [Google Scholar] [CrossRef]
- Mrozik, A.; Piotrowska-Seget, Z. Bioaugmentation as a strategy for cleaning up of soils contaminated with aromatic compounds. Microbiol. Res. 2010, 165, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.F.; Li, X.L.; Xiang, S.Z.; Ma, Z.Y.; Hu, S.J.; Tu, C. Bio-organic fertilizer promotes plant growth and yield and improves soil microbial community in continuous monoculture system of Chrysanthemum morifolium cv. Chuju. Int. J. Agric. Biol. 2017, 19, 563–568. [Google Scholar] [CrossRef]
- Guo, J.J.; Liu, W.B.; Zhu, C.; Luo, G.W.; Kong, Y.L.; Ling, N.; Wang, M.; Dai, J.Y.; Shen, Q.R.; Guo, S.W. Bacterial rather than fungal community composition is associated with microbial activities and nutrient-use efficiencies in a paddy soil with short-term organic amendments. Plant Soil 2018, 424, 335–349. [Google Scholar] [CrossRef]
- Cotto, A.; Looper, J.K.; Mota, L.C.; Son, A. Quantitative polymerase chain reaction for microbial growth kinetics of mixed culture system. J. Microbiol. Biotechnol. 2015, 25, 1928–1935. [Google Scholar] [CrossRef] [PubMed]
- Klein, E.; Katan, J.; Gamliel, A. Soil suppressiveness by organic amendment to Fusarium disease in cucumber: Effect on pathogen and host. Phytoparasitica 2016, 44, 239–249. [Google Scholar] [CrossRef]
- Rosenzweig, N.; Tiedje, J.M.; Quensen, J.F.; Meng, Q.; Hao, J.J. Microbial communities associated with potato common scab-suppressive soil determined by pyrosequencing analyses. Plant Dis. 2012, 96, 718–725. [Google Scholar] [CrossRef] [Green Version]
- Van der Ent, S.; Van Hulten, M.; Pozo, M.J.; Czechowski, T.; Udvardi, M.K.; Pieterse, C.M.; Ton, J. Priming of plant innate immunity by rhizobacteria and β-aminobutyric acid: Differences and similarities in regulation. New Phytol. 2009, 183, 419–431. [Google Scholar] [CrossRef] [Green Version]
- Ling, N.; Huang, Q.; Guo, S.; Shen, Q. Paenibacillus polymyxa SQR-21 systemically affects root exudates of watermelon to decrease the conidial germination of Fusarium oxysporum f. sp. niveum. Plant Soil 2011, 341, 485–493. [Google Scholar] [CrossRef]
- Yuan, J.; Raza, W.; Shen, Q.; Huang, Q. Antifungal activity of Bacillus amyloliquefaciens NJN-6 volatile compounds against Fusarium oxysporum f. sp. cubense. Appl. Environ. Microbiol. 2012, 78, 5942–5944. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.; Chellemi, D.O.; Graham, J.H.; Rosskopf, E.N. Assessment of fungal communities in soil and tomato roots subjected to diverse land and crop management systems. Soil Biol. Biochem. 2008, 40, 1967–1970. [Google Scholar] [CrossRef]
- Zhou, X.G.; Zhang, J.H.; Pan, D.D.; Ge, X.; Jin, X.; Chen, S.C.; Wu, F.Z. p-Coumaric acid can alter the composition of cucumber rhizosphere microbial communities and induce negative plant-microbial interactions. Biol. Fertil. Soils 2018, 54, 363–372. [Google Scholar] [CrossRef]
- Zhou, X.; Wu, F. p-Coumaric acid influenced cucumber rhizosphere soil microbial communities and the growth of Fusarium oxysporum f. sp. cucumerinum Owen. PLoS ONE 2012, 7, e48288. [Google Scholar]
- Luo, J.; Ran, W.; Hu, J.; Yang, X.; Xu, Y.; Shen, Q. Application of bio-organic fertilizer significantly affected fungal diversity of soils. Soil Sci. Soc. Am. 2010, 74, 2039–2048. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, J.; Gu, T.; Zhang, W.; Shen, Q.; Yin, S.; Qiu, H. Microbial community diversities and taxa abundances in soils along a seven-year gradient of potato monoculture using high throughput pyrosequencing approach. PLoS ONE 2014, 9, e86610. [Google Scholar] [CrossRef]
- Van der Putten, W.H.; Bardgett, R.D.; Bever, J.D.; Bezemer, T.M.; Casper, B.B.; Fukami, T.; Kardol, P.; Klironomos, J.N.; Kulmatiski, A.; Schweitzer, J.A.; et al. Plant-soil feedbacks: The past, the present and future challenges. J. Ecol. 2013, 101, 265–276. [Google Scholar] [CrossRef]
- Shen, Z.; Wang, D.; Ruan, Y.; Xue, C.; Zhang, J.; Li, R.; Shen, Q. Deep 16S rRNA pyrosequencing reveals a bacterial community associated with Banana Fusarium wilt disease suppression induced by bio-organic fertilizer application. PLoS ONE 2014, 9, e98420. [Google Scholar] [CrossRef]
- Roesch, L.F.; Fulthorpe, R.R.; Riva, A.; Casella, G.; Hadwin, A.K.; Kent, A.D.; Daroub, S.H.; Camargo, F.A.; Farmerie, W.G.; Triplett, E.W. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007, 1, 283–290. [Google Scholar] [CrossRef]
- Shen, C.; Xiong, J.; Zhang, H.; Feng, Y.; Lin, X.; Li, X.; Liang, W.; Chu, H. Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai mountain. Soil Biol. Biochem. 2013, 57, 204–211. [Google Scholar] [CrossRef]
- Li, X.G.; Ding, C.F.; Zhang, T.L.; Wang, X.X. Fungal pathogen accumulation at the expense of plant beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol. Biochem. 2014, 72, 11–18. [Google Scholar] [CrossRef]
- Mazurier, S.; Corberand, T.; Lemanceau, P.; Raaijmakers, J.M. Phenazine antibiotics produced by fluorescent pseudomonads contribute to natural soil suppressiveness to Fusarium wilt. ISME J. 2009, 3, 977–991. [Google Scholar] [CrossRef]
- Ren, L.; Huo, H.; Zhang, F.; Hao, W.; Xiao, L.; Dong, C.; Xu, G. The components of rice and watermelon root exudates and their effects on pathogenic fungus and watermelon defense. Plant Signal. Behav. 2016, 11, e1187357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wintermute, E.H.; Silver, P.A. Emergent cooperation in microbial metabolism. Mol. Syst. Biol. 2010, 6, 407. [Google Scholar] [CrossRef] [PubMed]
- Johns, N.I.; Blazejewski, T.; Gomes, A.L.; Wang, H.H. Principles for designing synthetic microbial communities. Curr. Opin. Microbiol. 2016, 31, 146–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheth, R.U.; Cabral, V.; Chen, S.P.; Wang, H.H. Manipulating bacterial communities by in situ microbiome engineering. Trends Genet. 2016, 32, 189–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]







© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, C.; Ma, Z.; Lu, X.; Zhu, L.; Wang, J. Phenolic Acid-Degrading Consortia Increase Fusarium Wilt Disease Resistance of Chrysanthemum. Agronomy 2020, 10, 385. https://doi.org/10.3390/agronomy10030385
Zhou C, Ma Z, Lu X, Zhu L, Wang J. Phenolic Acid-Degrading Consortia Increase Fusarium Wilt Disease Resistance of Chrysanthemum. Agronomy. 2020; 10(3):385. https://doi.org/10.3390/agronomy10030385
Chicago/Turabian StyleZhou, Cheng, Zhongyou Ma, Xiaoming Lu, Lin Zhu, and Jianfei Wang. 2020. "Phenolic Acid-Degrading Consortia Increase Fusarium Wilt Disease Resistance of Chrysanthemum" Agronomy 10, no. 3: 385. https://doi.org/10.3390/agronomy10030385
APA StyleZhou, C., Ma, Z., Lu, X., Zhu, L., & Wang, J. (2020). Phenolic Acid-Degrading Consortia Increase Fusarium Wilt Disease Resistance of Chrysanthemum. Agronomy, 10(3), 385. https://doi.org/10.3390/agronomy10030385




