TILLING in Cereal Crops for Allele Expansion and Mutation Detection by Using Modern Sequencing Technologies
Abstract
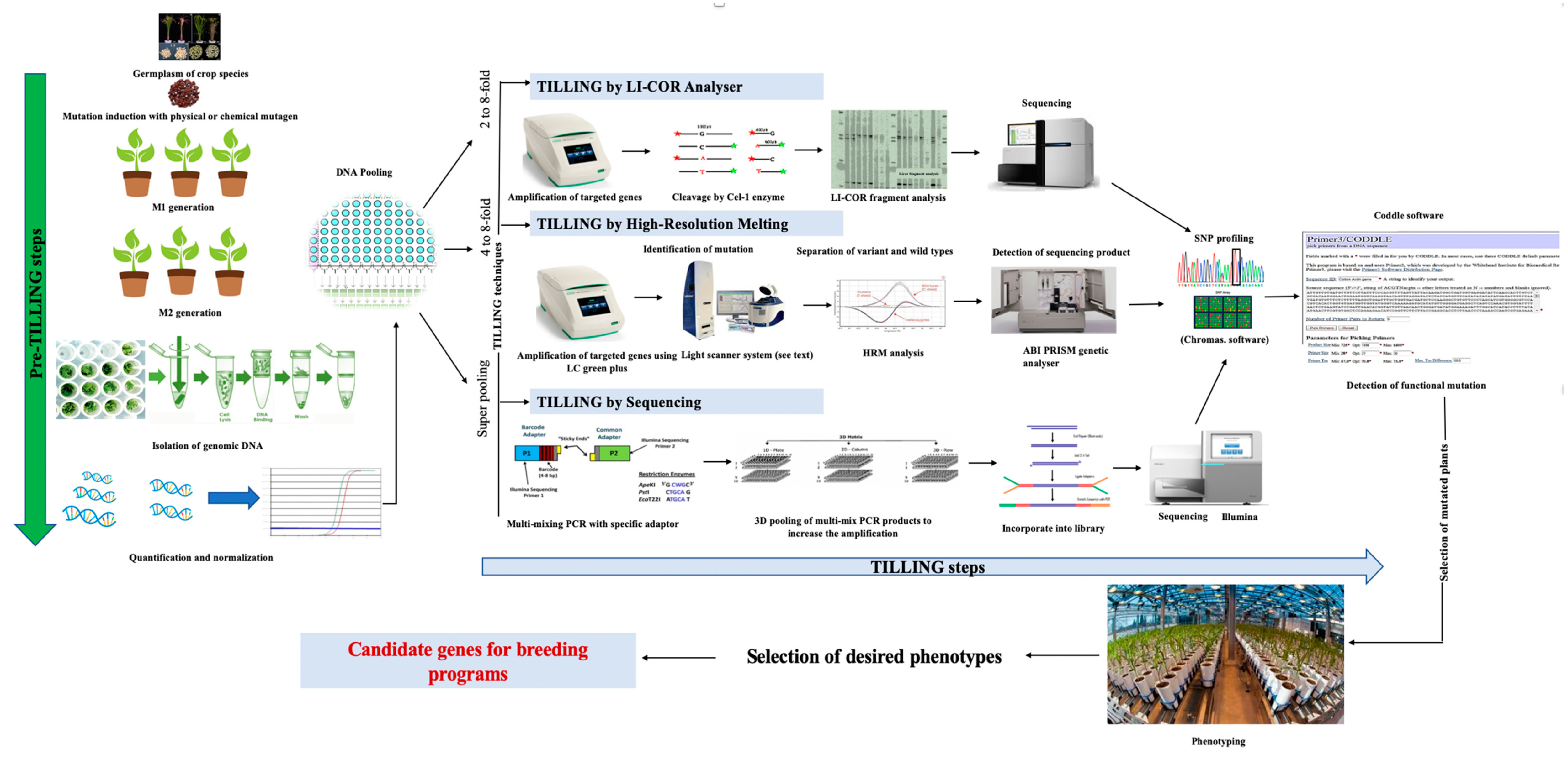
:1. Introduction
1.1. Background
1.2. TILLING and EcoTILLING
1.3. Generation of Induced Mutation for TILLING
1.4. Methods of Mutation Detection in TILLING
- The 3D pooling strategy allows for the observation of individual mutant plants and for the molecular recognition of mutation without requiring additional deconvolution of pools and additional sequencing steps;
- TbyS can be used to recognize single base alterations and their impact on specific traits;
- TbyS does not depend on fluorescent primers;
- TbyS enables flexible options for pooling techniques.
2. Application of TILLING in Cereal Crops
2.1. TILLING for Starch Synthesis
2.2. TILLING for Plant Architecture
2.3. TILLING for Disease Resistance
2.4. TILLING for Other Yield-Related Parameters
3. Prospect of TILLIING
3.1. Next-Generation Sequencing and TILLING
3.2. Importance of TbyS
3.3. Genome-Editing Technology and TILLING
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Allen, A.M.; Barker, G.L.A.; Berry, S.T.; Coghill, J.A.; Gwilliam, R.; Kirby, S.; Robinson, P.; Brenchley, R.C.; D’Amore, R.; McKenzie, N. Transcript-specific, single-nucleotide polymorphism discovery and linkage analysis in hexaploid bread wheat (Triticum aestivum L.). Plant. Biotechnol. J. 2011, 9, 1086–1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mba, C. Induced Mutations Unleash the Potentials of Plant Genetic Resources for Food and Agriculture. Agronomy 2013, 3, 200–231. [Google Scholar] [CrossRef] [Green Version]
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 327, 818. [Google Scholar] [CrossRef]
- Godfray, H.C.; Garnett, T. Food security and sustainable intensification. Philos. Trans. R. Soc. Lond. 2014, 369. [Google Scholar] [CrossRef]
- Griggs, D.; Staffordsmith, M.; Gaffney, O.; Rockström, J.; Ohman, M.C.; Shyamsundar, P.; Steffen, W.; Glaser, G.; Kanie, N.; Noble, I. Policy: Sustainable development goals for people and planet. Nature 2013, 495, 305–307. [Google Scholar] [CrossRef] [PubMed]
- Mo, Y.; Howell, T.; Vasquez-Gross, H.; de Haro, L.A.; Dubcovsky, J.; Pearce, S. Mapping causal mutations by exome sequencing in a wheat TILLING population: A tall mutant case study. Mol. Genet. Genomics 2018, 293, 463–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suprasanna, P.; Mirajkar, S.J.; Bhagwat, S.G. Induced Mutations and Crop Improvement. Plant. Biol. Biotechnol. 2015, 593–617. [Google Scholar] [CrossRef]
- Oladosu, Y.; Rafii, M.Y.; Abdullah, N.; Hussin, G.; Ramli, A.; Rahim, H.A.; Miah, G.; Usman, M. Principle and application of plant mutagenesis in crop improvement: A review. Biotechnol. Biotechnol. Equip. 2016, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Mba, C.; Afza, R.; Bado, S.; Jain, S.M.; Davey, M.R.; Anthony, P. Induced mutagenesis in plants using physical and chemical agents. Plant. Cell Cult. Essen. Methods 2010, 111–130. [Google Scholar]
- Taheri, S.; Abdullah, T.L.; Jain, S.M.; Sahebi, M.; Azizi, P. TILLING, high-resolution melting (HRM), and next-generation sequencing (NGS) techniques in plant mutation breeding. Mole. Breed 2017, 37. [Google Scholar] [CrossRef]
- Li, H.; Rasheed, A.; Hickey, L.T.; He, Z. Fast-forwarding genetic gain. Trends Plant. Sci. 2018, 23, 184–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunter, B.; Bas, M.; Kantoglu, Y.; Burak, M. Mutation breeding of sweet cherry (Prunus avium L.) var. 0900 Ziraat. Plant. Mutation Breed. Biotechnol. 2012, 453–463. [Google Scholar]
- IAEA. Mutant Variety Database. Available online: http://mvd.iaea.org/ (accessed on 2 March 2016).
- Jankowicz-Cieslak, J.; Mba, C.; Till, B.J. Mutagenesis for crop breeding and functional genomics. Biotechnol. Plant. Mutation Breed 2017, 3–18. [Google Scholar] [CrossRef]
- Newhouse, K.E.; Smith, W.A.; Starrett, M.A.; Schaefer, T.J.; Singh, B.K. Tolerance to imidazolinone herbicides in wheat. Plant. Physiol. 1992, 100, 882–886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, H.; Liu, Y.; Li, X.; Yan, Z.; Xie, Y.; Xiong, H.; Zhao, L.; Gu, J.; Zhao, S.; Liu, L. Novel mutant alleles of the starch synthesis gene TaSSIVb-D result in the reduction of starch granule number per chloroplast in wheat. BMC Genomics 2017, 18, 358. [Google Scholar] [CrossRef] [PubMed]
- Onda, Y.; Mochida, K. Exploring Genetic Diversity in Plants Using High-Throughput Sequencing Techniques. Curr. Genomics 2016, 17, 358. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.L.; Uauy, C.; Robson, F.; Till, B. TILLING in extremis. Plant. Biotechnol. J. 2012, 10, 761–772. [Google Scholar] [CrossRef]
- Mccallum, C.M.; Comai, L.; Greene, E.A.; Henikoff, S. Targeting induced local lesions in genomes (TILLING) for plant functional genomics. Plant. Physiol. 2000, 123, 439. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.P.K.; Mckeown, P.C.; Boualem, A.; Ryder, P.; Brychkova, G.; Bendahmane, A.; Sarkar, A.; Chatterjee, M.; Spillane, C. TILLING by Sequencing (TbyS) for targeted genome mutagenesis in crops. Mol. Breed. 2017, 37, 14. [Google Scholar] [CrossRef]
- Henikoff, S.; Comai, L. Single-nucleotide mutations for plant functional genomics. Annu. Rev. Plant. Biol. 2003, 54, 375. [Google Scholar] [CrossRef]
- Kurowska, M.; Daszkowskagolec, A.; Gruszka, D.; Marzec, M.; Szurman, M.; Szarejko, I.; Maluszynski, M. TILLING—A shortcut in functional genomics. J. Appl. Genet. 2011, 52, 371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashtwari, M.; Wani, A.A.; Rather, R.N. TILLING: An alternative path for crop improvement. J. Crop Improv. 2019, 33, 83–109. [Google Scholar] [CrossRef]
- Borevitz, J.O.; Liang, D.; Plouffe, D.; Chang, H.; Zhu, T.; Weigel, D.; Berry, C.C.; Winzeler, E.; Chory, J. Large-Scale Identification of Single-Feature Polymorphisms in Complex Genomes. Genome Res. 2003, 13, 513–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colbert, T.; Till, B.J.; Tompa, R.; Reynolds, S. High-throughput screening for induced point mutations. Plant. Physiol. 2001, 126, 480. [Google Scholar] [CrossRef] [PubMed]
- Jawher, M.; Arabi, M.I.E.; MirAli, N.; Till, B.J. Efficient discovery of single-nucleotide variations in Cochliobolus sativus vegetative compatibility groups by EcoTILLING. J. Plant Biochem. Physiol. 2018, 6, 2. [Google Scholar]
- Wang, J.; Sun, J.; Liu, D.; Yang, W.; Wang, D.; Tong, Y.; Zhang, A. Analysis of Pina and Pinb alleles in the micro-core collections of Chinese wheat germplasm by EcoTILLING and identification of a novel Pinb allele. J. Cereal Sci. 2008, 48, 836–842. [Google Scholar] [CrossRef] [Green Version]
- Bajaj, D.; Srivastava, R.; Nath, M.; Tripathi, S.; Bharadwaj, C.; Upadhyaya, H.D.; Tyagi, A.K.; Parida, S.K. EcoTILLING-Based Association Mapping Efficiently Delineates Functionally Relevant Natural Allelic Variants of Candidate Genes Governing Agronomic Traits in Chickpea. Front. Plant Sci. 2016, 7, 450. [Google Scholar] [CrossRef] [Green Version]
- Kadaru, S.B.; Yadav, A.S.; Fjellstrom, R.G.; Oard, J.H. Alternative EcoTILLING protocol for rapid, cost-effective single-nucleotide polymorphism discovery and genotyping in rice (Oryza sativa L.). Plant. Mol. Biol. Reporter 2006, 24, 3–22. [Google Scholar] [CrossRef]
- Comai, L.; Young, K.; Till, B.J.; Reynolds, S.H.; Greene, E.A.; Codomo, C.A.; Enns, L.C.; Johnson, J.E.; Burtner, C.; Odden, A.R. Efficient discovery of DNA polymorphisms in natural populations by EcoTILLING. Plant. J. 2004, 37, 778–786. [Google Scholar] [CrossRef]
- Liang, C.; Wang, S.Q.; Hu, Y.G. Detection of SNPs in the VRN-A1 gene of common wheat (Triticum aestivum L.) by a modified EcoTILLING method using agarose gel electrophoresis. Aus. J. Crop. Sci. 2011, 5, 318–326. [Google Scholar]
- Ma, X.; Sajjad, M.; Wang, J.; Yang, W.; Sun, J.; Li, X.; Zhang, A.; Liu, D. Diversity, distribution of Puroindoline genes and their effect on kernel hardness in a diverse panel of Chinese wheat germplasm. BMC Plant. Bio. 2017, 17, 158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rakshit, S.; Rakshit, A.; Matsumura, H.; Takahashi, Y.; Hasegawa, Y.; Ito, A.; Ishii, T.; Miyashita, N.T.; Terauchi, R. Large-scale DNA polymorphism study of Oryza sativa and O. rufipogon reveals the origin and divergence of Asian rice. Theor. Appl. Genet. 2007, 114, 731–743. [Google Scholar] [CrossRef] [PubMed]
- Raghavan, C.; Naredo, M.E.B.; Wang, H.; Atienza, G.; Liu, B.; Qiu, F.; McNally, K.L.; Leung, H. Rapid method for detecting SNPs on agarose gels and its application in candidate gene mapping. Mol. Breed. 2006, 19, 87–101. [Google Scholar] [CrossRef]
- Ochiai, K.; Shimizu, A.; Okumoto, Y.; Fujiwara, T.; Matoh, T. Suppression of a NAC-like transcription factor gene improves boron-toxicity tolerance in rice. Plant. Physiol. 2011, 156, 1457–1463. [Google Scholar] [CrossRef] [Green Version]
- Negrão, S.; Almadanim, C.; Pires, I.; McNally, K.L.; Oliveira, M.M. Use of EcoTILLING to identify natural allelic variants of rice candidate genes involved in salinity tolerance. Plant. Genet. Resour. 2011, 9, 300–304. [Google Scholar] [CrossRef]
- Yu, S.; Liao, F.; Wang, F.; Wen, W.; Li, J.; Mei, H.; Luo, L. Identification of rice transcription factors associated with drought tolerance using the EcoTILLING method. PLoS ONE 2012, 7, e30765. [Google Scholar] [CrossRef]
- Negrao, S.; Almadanim, M.C.; Pires, I.S.; Abreu, I.A.; Maroco, J.; Courtois, B.; Gregorio, G.B.; McNally, K.L.; Oliveira, M.M. New allelic variants found in key rice salt-tolerance genes: An association study. Plant. Biotechnol. J. 2013, 11, 87–100. [Google Scholar] [CrossRef] [Green Version]
- Raja, R.B.; Agasimani, S.; Jaiswal, S.; Thiruvengadam, V.; Sabariappan, R.; Chibbar, R.N.; Ram, S.G. EcoTILLING by sequencing reveals polymorphisms in genes encoding starch synthases that are associated with low glycemic response in rice. BMC Plant. Biol. 2017, 17, 13. [Google Scholar] [CrossRef] [Green Version]
- Xia, Y.; Ning, Z.; Bai, G.; Li, R.; Yan, G.; Siddique, K.H.; Baum, M.; Guo, P. Allelic variations of a light harvesting chlorophyll a/b-binding protein gene (Lhcb1) associated with agronomic traits in barley. PLoS ONE 2012, 7, e37573. [Google Scholar] [CrossRef] [Green Version]
- Xia, Y.; Li, R.; Ning, Z.; Bai, G.; Siddique, K.H.; Yan, G.; Baum, M.; Varshney, R.K.; Guo, P. Single nucleotide polymorphisms in HSP17.8 and their association with agronomic traits in barley. PLoS ONE 2013, 8, e56816. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Shi, L.; Tian, F.; Ning, H.; Wu, X.; Long, Y.; Meng, J. Assessment of FAE1 polymorphisms in three Brassica species using EcoTILLING and their association with differences in seed erucic acid contents. BMC Plant. Biol. 2010, 10, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, C.L.; Wang, G.Y.; Wang, J.B.; Yan, G.X.; Chen, B.Y.; Xu, K.; Li, J.; Gao, G.Z.; Wu, X.M.; Zhao, B.; et al. High-throughput discovery of chloroplast and mitochondrial DNA polymorphisms in Brassicaceae species by ORG-EcoTILLING. PLoS ONE 2012, 7, e47284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, J.; Cai, M.; Yan, G.; Wang, N.; Li, F.; Chen, B.; Gao, G.; Xu, K.; Li, J.; Wu, X. High-throughput multiplex cpDNA resequencing clarifies the genetic diversity and genetic relationships among Brassica napus, Brassica rapa and Brassica oleracea. Plant. Biotech. J. 2016, 14, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Upadhyaya, P.; Tyagi, K.; Sarma, S.; Tamboli, V.; Sreelakshmi, Y.; Sharma, R. Natural variation in folate levels among tomato (Solanum lycopersicum) accessions. Food Chem. 2017, 217, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Mohan, V.; Gupta, S.; Thomas, S.; Mickey, H.; Charakana, C.; Chauhan, V.S.; Sharma, K.; Kumar, R.; Tyagi, K.; Sarma, S.; et al. Tomato Fruits Show Wide Phenomic Diversity but Fruit Developmental Genes Show Low Genomic Diversity. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.D.; Sun, J.L.; Bu, S.H.; Deng, K.S.; Tao, T.; Zhang, Y.M.; Zhang, T.Z.; Du, X.M.; Zhou, B.L. EcoTILLING revealed SNPs in GhSus genes that are associated with fiber- and seed-related traits in upland cotton. Sci. Rep. 2016, 6, 29250. [Google Scholar] [CrossRef] [PubMed]
- Ibiza, V.P.; Cañizares, J.; Nuez, F. EcoTILLING in Capsicum species: Searching for new virus resistances. BMC Genomics 2010, 11, 631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marroni, F.; Pinosio, S.; Di Centa, E.; Jurman, I.; Boerjan, W.; Felice, N.; Cattonaro, F.; Morgante, M. Large-scale detection of rare variants via pooled multiplexed next-generation sequencing: Towards next-generation EcoTILLING. Plant. J. 2011, 67, 736–745. [Google Scholar] [CrossRef] [PubMed]
- Sabetta, W.; Blanco, A.; Zelasco, S.; Lombardo, L.; Perri, E.; Mangini, G.; Montemurro, C. Fad7 gene identification and fatty acids phenotypic variation in an olive collection by EcoTILLING and sequencing approaches. Plant. Physiol. Biochem. 2013, 69, 1–8. [Google Scholar] [CrossRef]
- Maghuly, F.; Jankowicz-Cieslak, J.; Pabinger, S.; Till, B.J.; Laimer, M. Geographic origin is not supported by the genetic variability found in a large living collection of Jatropha curcas with accessions from three continents. Biotechnol. J. 2015, 10, 536–551. [Google Scholar] [CrossRef] [Green Version]
- Parry, M.A.; Madgwick, P.J.; Bayon, C.; Tearall, K.; Hernandez-Lopez, A.; Baudo, M.; Rakszegi, M.; Hamada, W.; Al-Yassin, A.; Ouabbou, H. Mutation discovery for crop improvement. J. Exp. Bot. 2009, 60, 2817–2825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooper, J.L.; Till, B.J.; Laport, R.G.; Darlow, M.C.; Kleffner, J.M.; Jamai, A.; El-Mellouki, T.; Liu, S.; Ritchie, R.; Nielsen, N. TILLING to detect induced mutations in soybean. BMC Plant. Biol. 2008, 8, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, T.; Eiguchi, M.; Kumamaru, T.; Satoh, H.; Matsusaka, H.; Moriguchi, K.; Nagato, Y.; Kurata, N. MNU-induced mutant pools and high performance TILLING enable finding of any gene mutation in rice. Mol. Genet. Genom. 2008, 279, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Serrat, X.; Esteban, R.; Guibourt, N.; Moysset, L.; Nogués, S.; Lalanne, E. EMS mutagenesis in mature seed-derived rice calli as a new method for rapidly obtaining TILLING mutant populations. Plant. Methods 2014, 10, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blomstedt, C.K.; Gleadow, R.M.; O’Donnell, N.; Naur, P.; Jensen, K.; Laursen, T.; Olsen, C.E.; Stuart, P.; Hamill, J.D.; Møller, B.L. A combined biochemical screen and TILLING approach identifies mutations in Sorghum bicolor L. Moench resulting in acyanogenic forage production. Plant. Biotechnol. J. 2012, 10, 54–66. [Google Scholar] [CrossRef]
- Piron, F.; Nicolaï, M.; Minoïa, S.; Piednoir, E.; Moretti, A.; Salgues, A.; Zamir, D.; Caranta, C.; Bendahmane, A. An induced mutation in tomato eIF4E leads to immunity to two potyviruses. PLoS ONE 2010, 5, e11313. [Google Scholar] [CrossRef] [Green Version]
- Dahmani-Mardas, F.; Troadec, C.; Boualem, A.; Leveˆque, S.; Alsadon, A.A.; Aldoss, A.A.; Dogimont, C.; Bendahmane, A. Engineering melon plants with improved fruit shelf life using the TILLING approach. PLoS ONE 2010, 5, e15776. [Google Scholar] [CrossRef] [Green Version]
- Sestili, F.; Botticella, E.; Bedo, Z.; Phillips, A.; Lafiandra, D. Production of novel allelic variation for genes involved in starch biosynthesis through mutagenesis. Mol. Breed. 2010, 25, 145. [Google Scholar] [CrossRef]
- Henikoff, S.; Till, B.J.; Comai, L. TILLING. Traditional Mutagenesis Meets Functional Genomics. Plant. Physiol. 2004, 135, 630–636. [Google Scholar] [CrossRef] [Green Version]
- Tsai, H.; Howell, T.; Nitcher, R.; Missirian, V.; Watson, B.; Ngo, K.J.; Lieberman, M.; Fass, J.; Uauy, C.; Tran, R.K. Discovery of Rare Mutations in Populations: TILLING by Sequencing. Plant. Physiol. 2011, 156, 1257. [Google Scholar] [CrossRef] [Green Version]
- Acevedo-Garcia, J.; Spencer, D.; Thieron, H.; Reinstädler, A.; Hammond-Kosack, K.; Phillips, A.L.; Panstruga, R. mlo-based powdery mildew resistance in hexaploid bread wheat generated by a non-transgenic TILLING approach. Plant. Biotechnol. J. 2017, 15, 367. [Google Scholar] [CrossRef] [PubMed]
- Botticella, E.; Sestili, F.; Hernandez-Lopez, A.; Phillips, A.; Lafiandra, D. High Resolution Melting analysis for the detection of EMS induced mutations in wheat Sbella genes. BMC Plant. Biol. 2011, 11, 156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.D.; Chu, Z.Z.; Liu, X.G.; Jing, H.C.; Liu, Y.G.; Hao, D.Y. A cost-effective high-resolution melting approach using the EvaGreen dye for DNA polymorphism detection and genotyping in plants. J. Integrat. Plant. Biol. 2010, 52, 1036–1042. [Google Scholar] [CrossRef] [PubMed]
- Quail, M.A.; Smith, M.; Coupland, P.; Otto, T.D.; Harris, S.R.; Connor, T.R.; Bertoni, A.; Swerdlow, H.P.; Gu, Y. A tale of three next generation sequencing platforms: Comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 2012, 13, 341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Abernathy, B.; Zeng, Y.; Ozias-Akins, P. TILLING by sequencing to identify induced mutations in stress resistance genes of peanut (Arachis hypogaea). BMC Genomics 2015, 16, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamski, N.M.; Borrill, P.; Brinton, J.; Harrington, S.; Marchal, C.; Uauy, C. A roadmap for gene functional characterisation in wheat. PeerJ 2018, 2167–9843. [Google Scholar]
- Irshad, A.; Guo, H.; Zhang, S.; Gu, J.; Zhao, L.; Xie, Y.; Xiong, H.; Zhao, S.; Ding, Y.; Ma, Y. EcoTILLING Reveals Natural Allelic Variations in Starch Synthesis Key Gene TaSSIV and Its Haplotypes Associated with Higher Thousand Grain Weight. Genes 2019, 10, 307. [Google Scholar] [CrossRef] [Green Version]
- Uauy, C.; Wulff, B.B.; Dubcovsky, J. Combining Traditional Mutagenesis with New High-Throughput Sequencing and Genome Editing to Reveal Hidden Variation in Polyploid Wheat. Ann. Rev. Genetics 2017, 51, 435–454. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Hao, L.; Parry, M.A.; Phillips, A.L.; Hu, Y.G. Progress in TILLING as a tool for functional genomics and improvement of crops. J. Integr. Plant. Biol. 2014, 56, 425–443. [Google Scholar] [CrossRef] [Green Version]
- Szurman-Zubrzycka, M.E.; Zbieszczyk, J.; Marzec, M.; Jelonek, J.; Chmielewska, B.; Kurowska, M.M.; Krok, M.; Daszkowska-Golec, A.; Guzy-Wrobelska, J.; Gruszka, D. HorTILLUS—A Rich and Renewable Source of Induced Mutations for Forward/Reverse Genetics and Pre-breeding Programs in Barley (Hordeum vulgare L.). Front. Plant. Sci. 2018, 9, 216. [Google Scholar] [CrossRef]
- Gilchrist, E.; Haughn, G. Reverse genetics techniques: Engineering loss and gain of gene function in plants. Brief. Funct. Genomics 2010, 9, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, X.; Li, Z.; Yang, G.; Yang, H.; Feng, H.; Xu, X.; Wang, J.; Li, X.; Luo, J. Winter wheat yield estimation based on multi-source medium resolution optical and radar imaging data and the AquaCrop model using the particle swarm optimization algorithm. ISPRS J. Photogramm. Remote Sens. 2017, 126, 24–37. [Google Scholar] [CrossRef]
- Krasileva, K.V.; Vasquez-Gross, H.A.; Howell, T.; Bailey, P.; Paraiso, F.; Clissold, L.; Simmonds, J.; Ramirez-Gonzalez, R.H.; Wang, X.; Borrill, P. Uncovering hidden variation in polyploid wheat. Proc. Natl. Acad. Sci. USA 2017, 114, 913–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Zou, M.; Liu, K.; Gu, Z.; Yang, R. Effect of mild thermal treatment on the polymerization behavior, conformation and viscoelasticity of wheat gliadin. Food Chem. 2018, 239, 984–992. [Google Scholar] [CrossRef]
- Slade, A.J.; McGuire, C.; Loeffler, D.; Mullenberg, J.; Skinner, W.; Fazio, G.; Holm, A.; Brandt, K.M.; Steine, M.N.; Goodstal, J.F. Development of high amylose wheat through TILLING. BMC Plant Biol. 2012, 12, 10. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Yan, Z.; Li, X.; Xie, Y.; Xiong, H.; Liu, Y.; Zhao, L.; Gu, J.; Zhao, S.; Liu, L. Development of a High-Efficient Mutation Resource with Phenotypic Variation in Hexaploid Winter Wheat and Identification of Novel Alleles in the TaAGP.L-B1 Gene. Front. Plant. Sci. 2017, 8, 1404. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Yoon, M.-R.; Chun, A.; Tai, T.H. Identification of novel mutations in the rice starch branching enzyme I gene via TILLING by sequencing. Euphytica 2018, 214, 94. [Google Scholar] [CrossRef]
- Hwang, J.E.; Jang, D.S.; Lee, K.J.; Ahn, J.W.; Kim, S.H.; Kang, S.Y.; Kim, D.S.; Kim, J.B. Identification of gamma ray irradiation-induced mutations in membrane transport genes in a rice population by TILLING. Genes Genet. Syst. 2017, 91, 245–256. [Google Scholar] [CrossRef] [Green Version]
- Kusumi, K.; Hirotsuka, S.; Kumamaru, T.; Iba, K. Increased leaf photosynthesis caused by elevated stomatal conductance in a rice mutant deficient in SLAC1, a guard cell anion channel protein. J. Exp. Bot. 2012, 63, 5635–5644. [Google Scholar] [CrossRef] [Green Version]
- Caldwell, D.G.; McCallum, N.; Shaw, P.; Muehlbauer, G.J.; Marshall, D.F.; Waugh, R. A structured mutant population for forward and reverse genetics in Barley (Hordeum vulgare L.). Plant. J. 2004, 40, 143–150. [Google Scholar] [CrossRef]
- Marzec, M.; Gruszka, D.; Tylec, P.; Szarejko, I. Identification and functional analysis of the HvD14 gene involved in strigolactone signaling in Hordeum vulgare. Physiol. Plant. 2016, 158, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Bagge, M.; Xia, X.; Lübberstedt, T. Functional markers in wheat. Curr. Opin. Plant. Biol. 2007, 10, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Chawade, A.; Bräutigam, M.; Larsson, M.; Nakash, M.A.; Olsson, O.; Sikora, P.; Chen, T.; Vivekanand, V. Development and characterization of an oat TILLING-population and identification of mutations in lignin and β-glucan biosynthesis genes. BMC Plant. Biol. 2010, 10, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Till, B.J.; Reynolds, S.H.; Weil, C.; Springer, N.; Burtner, C.; Young, K.; Bowers, E.; Codomo, C.A.; Enns, L.C.; Odden, A.R. Discovery of induced point mutations in maize genes by TILLING. BMC Plant. Biol. 2004, 4, 12. [Google Scholar] [CrossRef] [Green Version]
- Till, B.J.; Cooper, J.; Tai, T.H.; Colowit, P.; Greene, E.A.; Henikoff, S.; Comai, L. Discovery of chemically induced mutations in rice by TILLING. BMC Plant. Biol. 2007, 7, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.-I.; Tai, T.H. Identification of novel rice low phytic acid mutations via TILLING by sequencing. Mol. Breed. 2014, 34, 1717–1729. [Google Scholar] [CrossRef]
- Gottwald, S.; Bauer, P.; Komatsuda, T.; Lundqvist, U.; Stein, N. TILLING in the two-rowed barley cultivar’Barke’reveals preferred sites of functional diversity in the gene HvHox1. BMC Res. Notes 2009, 2, 258. [Google Scholar] [CrossRef] [Green Version]
- Sparla, F.; Falini, G.; Botticella, E.; Pirone, C.; Talamè, V.; Bovina, R.; Salvi, S.; Tuberosa, R.; Sestili, F.; Trost, P. New starch phenotypes produced by TILLING in barley. PLoS ONE 2014, 9, e107779. [Google Scholar] [CrossRef] [Green Version]
- King, R.; Bird, N.; Ramirez-Gonzalez, R.; Coghill, J.A.; Patil, A.; Hassani-Pak, K.; Uauy, C.; Phillips, A.L. Mutation scanning in wheat by exon capture and next-generation sequencing. PLoS ONE 2015, 10, e0137549. [Google Scholar] [CrossRef] [Green Version]
- Chen, A.; Dubcovsky, J. Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genet. 2012, 8, e1003134. [Google Scholar] [CrossRef] [Green Version]
- Uauy, C.; Paraiso, F.; Colasuonno, P.; Tran, R.K.; Tsai, H.; Berardi, S.; Comai, L.; Dubcovsky, J. A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant. Biol. 2009, 9, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colasuonno, P.; Incerti, O.; Lozito, M.L.; Simeone, R.; Gadaleta, A.; Blanco, A. DHPLC technology for high-throughput detection of mutations in a durum wheat TILLING population. BMC Genet. 2016, 17, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rawat, N.; Sehgal, S.K.; Joshi, A.; Rothe, N.; Wilson, D.L.; McGraw, N.; Vadlani, P.V.; Li, W.; Gill, B.S. A diploid wheat TILLING resource for wheat functional genomics. BMC Plant. Biol. 2012, 12, 205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Simmonds, J.; Pan, Q.; Davidson, D.; He, F.; Battal, A.; Akhunova, A.; Trick, H.N.; Uauy, C.; Akhunov, E. Gene editing and mutagenesis reveal inter-cultivar differences and additivity in the contribution of TaGW2 homoeologues to grain size and weight in wheat. Theor. Appl. Genet. 2018, 131, 2463–2475. [Google Scholar] [CrossRef] [Green Version]
- Moehs, C.P.; Austill, W.J.; Holm, A.; Large, T.A.; Loeffler, D.; Mullenberg, J.; Schnable, P.S.; Skinner, W.; van Boxtel, J.; Wu, L. Development of reduced gluten wheat enabled by determination of the genetic basis of the lys3a low hordein barley mutant. Plant. Physiol. 2018, 179, 1692–1703. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Guo, H.; Wang, Y.; Xie, Y.; Zhao, L.; Gu, J.; Zhao, S.; Zhao, B.; Wang, G.; Liu, L. Identification of novel alleles induced by EMS-mutagenesis in key genes of kernel hardness and starch biosynthesis in wheat by TILLING. Genes Genom. 2017, 39, 387–395. [Google Scholar] [CrossRef]
- Severune, H.; Daniela, S.; Simon, G.K.; Bettina, K.; Thomas, W.; Gerhard, H.; Mirjam, N.F.; James, B.; Thomas, P.; Milena, O.; et al. The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated recepter-like kinase. Proc. Natl. Acad. Sci. USA 2015, 112, 8780–8785. [Google Scholar]
- Stolarek, M.; Gruszka, D.; Braszewska-Zalewska, A.; Maluszynski, M. Alleles of newly identified barley gene HvPARP3 exhibit changes in efficiency of DNA repair. DNA Repair 2015, 28, 116–130. [Google Scholar] [CrossRef]
- Mendiondo, G.M.; Gibbs, D.J.; Szurman-Zubrzycka, M.; Korn, A.; Marquez, J.; Szarejko, I.; Maluszynski, M.; King, J.; Axcell, B.; Smart, K. Enhanced waterlogging tolerance in barley by manipulation of expression of the N-end rule pathway E3 ligase PROTEOLYSIS 6. Plant. Biotechnol. J. 2016, 14, 40–50. [Google Scholar] [CrossRef]
- Hu, X. TILLING-based analysis of disease resistance genes in barley. J. Shandong Agric. Univ. 2012. [Google Scholar]
- Van Nimwegen, K.J.; van Soest, R.A.; Veltman, J.A.; Nelen, M.R.; van der Wilt, G.J.; Vissers, L.E.; Grutters, J.P. Is the $1000 genome as near as we think? A cost analysis of next-generation sequencing. Clin. Chem. 2016, 62, 1458–1464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant. Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothberg, J.M.; Hinz, W.; Rearick, T.M.; Schultz, J.; Mileski, W.; Davey, M.; Leamon, J.H.; Johnson, K.; Milgrew, M.J.; Edwards, M. An integrated semiconductor device enabling non-optical genome sequencing. Nature 2011, 475, 348. [Google Scholar] [CrossRef] [PubMed]
- Wetterstrand, K.A. DNA sequencing costs: Data from the NHGRI Genome Sequencing Program (GSP). Nat. Human Res. Instit. 2013. [Google Scholar]
- Rigola, D.; van Oeveren, J.; Janssen, A.; Bonné, A.; Schneiders, H.; van der Poel, H.J.; van Orsouw, N.J.; Hogers, R.C.; de Both, M.T.; van Eijk, M.J. High-throughput detection of induced mutations and natural variation using KeyPoint™ technology. PLoS ONE 2009, 4, e4761. [Google Scholar] [CrossRef] [Green Version]
- Tasai, H.; Tyson, H.; Rebecca, N.; Victor, M.; Brian, W.; Kathie, N.; Meric, N.; Joseph, F.; Cristobal, U. Discovery of rare mutations in populations: TILLING by sequencing. Plant. Physiol. 2011. [Google Scholar] [CrossRef] [Green Version]
- Henry, I.M.; Nagalakshmi, U.; Lieberman, M.C.; Ngo, K.J.; Krasileva, K.V.; Vasquez-Gross, H.; Akhunova, A.; Akhunov, E.; Dubcovsky, J.; Tai, T.H. Efficient genome-wide detection and cataloging of EMS-induced mutations using exome capture and next-generation sequencing. Plant. Cell 2014, 26, 1382–1397. [Google Scholar] [CrossRef] [Green Version]
- Reddy, T.V.; Dwivedi, S.; Sharma, N.K. Development of TILLING by sequencing platform towards enhanced leaf yield in tobacco. Ind. Crops Prod. 2012, 40, 324–335. [Google Scholar] [CrossRef]
- Krothapalli, K.; Buescher, E.M.; Li, X.; Brown, E.; Chapple, C.; Dilkes, B.P.; Tuinstra, M.R. Forward genetics by genome sequencing reveals that rapid cyanide release deters insect herbivory of Sorghum bicolor. Genetics 2013, 195, 309–318. [Google Scholar] [CrossRef] [Green Version]
- Elahi, N.; Duncan, R.W.; Stasolla, C. Decreased seed oil production in FUSCA3 Brassica napus mutant plants. Plant. Physiol. Biochem. 2015, 96, 222–230. [Google Scholar] [CrossRef]
- Minoia, S.; Boualem, A.; Marcel, F.; Troadec, C.; Quemener, B.; Cellini, F.; Petrozza, A.; Vigouroux, J.; Lahaye, M.; Carriero, F. Induced mutations in tomato SlExp1 alter cell wall metabolism and delay fruit softening. Plant. Sci. 2016, 242, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhu, Q.; Chen, Y.; Liu, Y.-G. CRISPR/Cas9 platforms for genome editing in plants: Developments and applications. Mol. Plant. 2016, 9, 961–974. [Google Scholar] [CrossRef] [Green Version]
- Petolino, J.F.; Srivastava, V.; Daniell, H. Editing Plant Genomes: A new era of crop improvement. Plant. Biotechnol. J. 2016, 14, 435–436. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Wang, Y.; Li, J.; Gao, C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 2014, 9, 2395. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xu, R.; Xie, C.; Huang, C.; Liao, H.; Xu, Y.; Wang, J.; Li, W.-X. Large-scale evaluation of maize germplasm for low-phosphorus tolerance. PLoS ONE 2015, 10, e0124212. [Google Scholar] [CrossRef]
- Xu, Y.; Li, P.; Zou, C.; Lu, Y.; Xie, C.; Zhang, X.; Prasanna, B.M.; Olsen, M.S. Enhancing genetic gain in the era of molecular breeding. J. Exp. Bot. 2017, 68, 2641–2666. [Google Scholar] [CrossRef]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef]
- Wang, W.; Pan, Q.; He, F.; Akhunova, A.; Chao, S.; Trick, H.; Akhunov, E. Transgenerational CRISPR-Cas9 activity facilitates multiplex gene editing in allopolyploid wheat. CRISPR J. 2018, 1, 65–74. [Google Scholar] [CrossRef] [Green Version]
- Avni, R.; Nave, M.; Barad, O.; Baruch, K.; Twardziok, S.O.; Gundlach, H.; Hale, I.; Mascher, M.; Spannagl, M.; Wiebe, K. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 2017, 357, 83–97. [Google Scholar] [CrossRef] [Green Version]
| Species | Ploidy Level | Genotypes | Gene Name | No. of Genes | SNPs/Haplotypes | Methodology | Genome Size | Trait | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Arabidopsis thaliana | 2x | 192 | DMMT2, DRM1C7, PIF2, AtWR | 5 | 55 (haplotypes) | CEL-1-PAGE | 125 MB | [30] | |
| Triticum aestivum | 6x | 214 | VRN-A1 | 1 | CJE/Agarose | 17 GB | Vernalization | [31] | |
| 1787 | Pin a, Pin b | 2 | 15 | CEL-1-PAGE | Kernel hardness | [32] | |||
| Oryza sativa | 2x | 48 | 14 | CEL-1-PAGE/UL | 400–430 MB | [33] | |||
| 48 | MYB1, TPP, ADF | 3 | Agarose gel | [34] | |||||
| 45 | 87 | - | CJE/Agarose gel | Boron toxicity | [35] | ||||
| 375 | OSCP17 | 1 | 38 | CJE/Agarose gel | Salt tolerance | [36] | |||
| 95 | 19 | 14 | CEL-1-PAGE | Drought tolerance | [37] | ||||
| 392 | osCPK17, osRMC, osNHX1, osHKTI;5, SalT | 5 | 69 | CEL-1-PAGE | Salt resistant | [38] | |||
| 512 | GBSSI, SSI, SSIIa, SSIIIa, SBEIa, SBEIIb | 6 | 23 | EcoTbyS | Starch synthesis | [39] | |||
| Hordeum vulgare | 2x | 292 | Lhcb1 | 1 | 23 | CEL-1-PAGE | 5.3 GB | Chlorophyll protein | [40] |
| 210 | HSP17.8 | 1 | 11 | CEL-1-PAGE | Heat shock protein | [41] | |||
| Brassica sp. | 2x 4x | 117 | FAE1-A8, FAE1-C3 | 2 | 18 | CEL-1-PAGE | 488–1544 MB 157 MB | Erucic acid content | [42] |
| 187 | accD, matK, rbcL, atp6 | 4 | 60 | CEL-1-PAGE | Organelle genome | [43] | |||
| 676 | Chloroplast DNA | 1 | 538 | EcoTbyS | Chloroplast DNA | [44] | |||
| Solanum lycopersicum | 2x | 49 | GCH1, ADCS, ADCL1, ADCL2, FPGSp, FPGSm, GGH1, GGH2, GGH3 | 9 | CEL-1-PAGE | 950 MB | Folate biosynthesis | [45] | |
| 127 | ACS2, CoP1, CYC-B, MSH2, NAC-NOR, PHoT1, PHYA, PHYB, PSY1 | 9 | 54 | CEL-1-PAGE | Plant development | [46] | |||
| Gossypium hirsutum | 4x | 277 | GhSus1At, GhSus1Dt, GhSus3At, GhSus4Dt, GhSus5Dt, GhSus6At, GhSus7Dt, GhSus8Dt | 8 | 24 | CEL-1-PAGE | 1724 MB | Sucrose synthesis | [47] |
| Capsicum annum | 2x | 233 | eIF4E, I1F(iso)4E, eIF(iso)4G, eIF4G | 4 | 62 | CEL-1-PAGE | 3.48 GB | Virus resistance | [48] |
| Populus nigra | 2x | 768 | CAD4, HCT1, C3H3, CCR7, 4CL3 | 5 | 84 | TbyS | Lignin biosynthesis | [49] | |
| Glycine max | 2x | 25 | Gy1, Gy2, Gy3, Gy4, Gy5 | 5 | - | Agarose gel | 1.15 GB | Seed protein | [29] |
| Olea europea | 2x | 96 | fad7 | 1 | 3 (haplotypes) | CEL-1-PAGE | Fatty acid enzyme | [50] | |
| Beta vulgaris | 2x | 268 | BTC1, BVFL1, BvFT1 | 3 | 21 | CEL-1-PAGE | 714–758 MB | Winter hardiness | |
| Jatropha curcas | 2x | 907 | AF, EUO6, EFO3, DQ98, EU10, EU22, EU39, EU23, EU21, DQ15, DQ66, SUSY11 | 12 | 86 | CEL-1-PAGE | 320.5 MB | Oil & stress tolerance | [51] |
| Cicer arietinum | 2x | 192 | 1133 | Agarose gel | 738 MB | Seed weight | [28] |
| Species | Ploidy | Mutagen | M2 Size | MF (Kb) | Mutation Detection Technology | Trait | Reference |
|---|---|---|---|---|---|---|---|
| Maize | 2x | EMS | 750 | 1/485 | CEL-1- PAGE | Chromomethylase | [85] |
| Rice | 2x | EMS | - | 1/1000 | CEL-1- PAGE | [32] | |
| 768 | 1/294 | CEL-1-PAGE | [86] | ||||
| 6912 | 1/451 | CEL-I -Agarose gel | [33] | ||||
| EMS | 2048 | 1/293 | TILLING by sequencing | Phytic acid metabolism | [87] | ||
| Barley | 2x | EMS | 9216 | 1/1000 | dHPLC | Floral parts regulation | [81] |
| EMS | 10,279 | 1/500 | CEL-1-PAGE | Fungus immunity | [88] | ||
| NaN3 | 5600 | 1/374 | CEL-I-Agarose gel | Starch metabolism | [89] | ||
| Wheat | 6x 4x | EMS | 2020 | 1/26 | HRM | Resistance against powdery mildew | [62] |
| EMS | 4500 | 1/35,000 | TILLING by sequencing | [90] | |||
| EMS | 10,000 | 1/24 | CEL-1, PAGE | Quality of starch | [76] | ||
| EMS | 4500 | 1/84 | CEL-1-PAGE, HRM | Quality of starch | [63] | ||
| EMS | 2610 | 1/34;1/47 | Agarose gel, PAGE | Development of spike | [91] | ||
| EMS | 8000 | 1/40 | CEL-1-PAGE | Starch quality | [92] | ||
| EMS | 3992 | HRM | Starch metabolism | [59] | |||
| EMS | 1140 | 1/77 | CEL I- Agarose gel, dHPLC | Carotenoid metabolism | [93] | ||
| EMS | 1532 | 1/92 | CEL-I | Waxy and lignin | [94] | ||
| EMS | 733 | Exome sequencing | Plant height | [6] | |||
| EMS | TILLING by Sequencing, CEL-1- PAGE | Thousand grain weight | [95] | ||||
| EMS | 10,000 | TILLING by Electrophoresis | Gluten content | [96] | |||
| EMS | 1122 | CEL-1-PAGE | Kernel hardness and starch | [97] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Irshad, A.; Guo, H.; Zhang, S.; Liu, L. TILLING in Cereal Crops for Allele Expansion and Mutation Detection by Using Modern Sequencing Technologies. Agronomy 2020, 10, 405. https://doi.org/10.3390/agronomy10030405
Irshad A, Guo H, Zhang S, Liu L. TILLING in Cereal Crops for Allele Expansion and Mutation Detection by Using Modern Sequencing Technologies. Agronomy. 2020; 10(3):405. https://doi.org/10.3390/agronomy10030405
Chicago/Turabian StyleIrshad, Ahsan, Huijun Guo, Shunlin Zhang, and Luxiang Liu. 2020. "TILLING in Cereal Crops for Allele Expansion and Mutation Detection by Using Modern Sequencing Technologies" Agronomy 10, no. 3: 405. https://doi.org/10.3390/agronomy10030405
APA StyleIrshad, A., Guo, H., Zhang, S., & Liu, L. (2020). TILLING in Cereal Crops for Allele Expansion and Mutation Detection by Using Modern Sequencing Technologies. Agronomy, 10(3), 405. https://doi.org/10.3390/agronomy10030405






