SNP in DFR1 Coding Sequence Is Tightly Associated with Anthocyanin Accumulation in Cabbage (B. oleracea var. capitata f. alba) at Low Temperature
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Analysis of BoDFR1 Gene and Promoter Sequences
2.3. Expression Analysis of BoDFR1
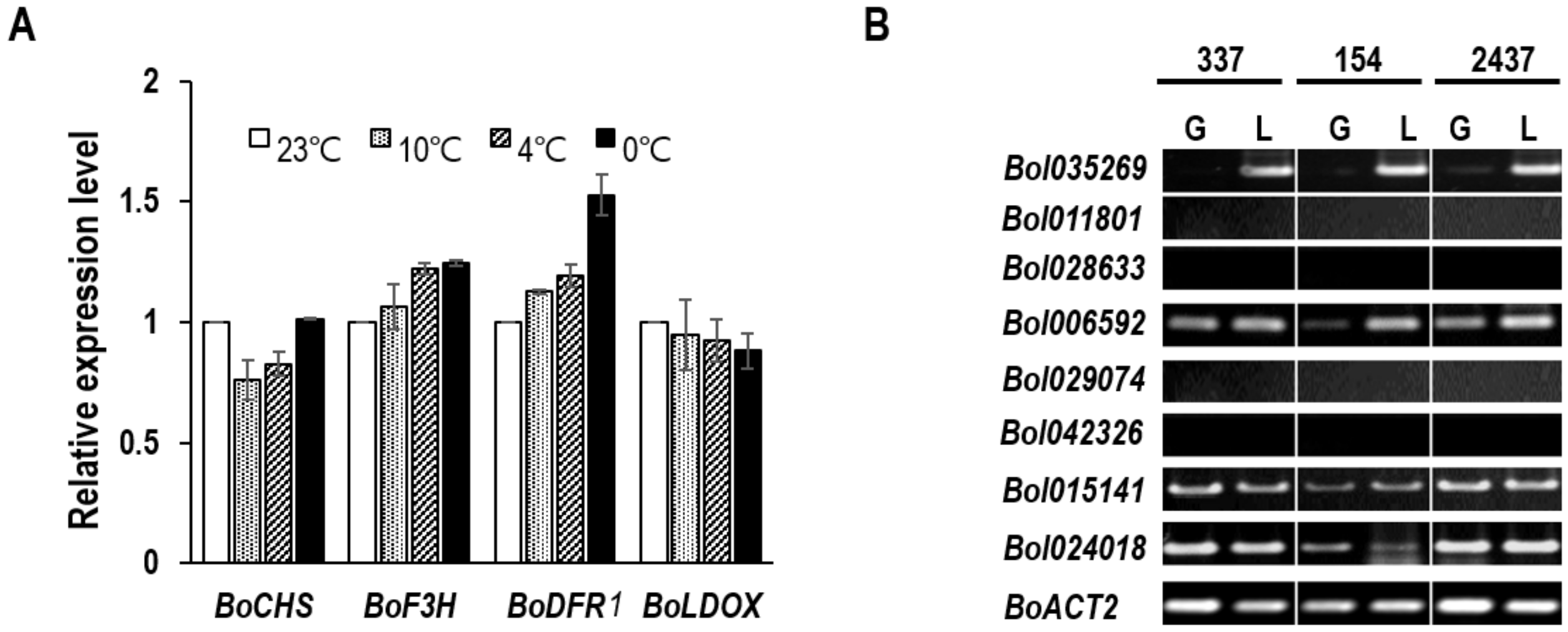
2.4. Expression Analysis of Anthocyanin Biosynthesis-Related Genes
2.5. Anthocyanin Extraction and Quantification
2.6. Establishment of PCR-Based Assay
2.7. High Resolution Melt (HRM) Analysis
2.8. Recombinant DFR1 Protein Expression
2.9. Enzyme Activity Assay
2.10. Statistical Analysis
3. Results
3.1. Identification of BoDFR1 as the Key Gene Responsible for Anthocyanin Accumulation under LT Conditions
3.2. Analysis of the BoDFR1 Gene Sequence Isolated from HAAs and LAAs
3.3. In Vitro Enzyme Activity Assay of BoDFR1 and BoDFR1v
3.4. Relationship between BoDFR1 and Other Anthocyanin Biosynthesis-Related Genes
3.5. Development of a Molecular Marker to Distinguish between BoDFR1 and BoDFR1v
4. Discussion
4.1. Selection of HAA and LAA Lines
4.2. BoDFR1 Sequence Analysis and SNP Detection
4.3. Regulation of BoDFR1 Expression
4.4. Molecular Marker Development and Application
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [Green Version]
- Bradshaw, H.D.; Schemske, D.W. Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature 2003, 426, 176–178. [Google Scholar] [CrossRef]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Mouradov, A.; Spangenberg, G. Flavonoids: A metabolic network mediating plants adaptation to their real estate. Front. Plant Sci. 2014, 5, 620. [Google Scholar] [CrossRef]
- Lorenc-Kukuła, K.; Jafra, S.; Oszmiański, J.; Szopa, J. Ectopic expression of anthocyanin 5-O-glucosyltransferase in potato tuber causes increased resistance to bacteria. J. Agric. Food Chem. 2005, 53, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Castellarin, S.D.; Gaspero, G.D. Transcriptional control of anthocyanin biosynthetic genes in extreme A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. phenotypes for berry pigmentation of naturally occurring grapevines. BMC Plant Biol. 2007, 7, 46. [Google Scholar] [CrossRef] [Green Version]
- Buer, C.S.; Imin, N.; Djordjevic, M.A. Flavonoids: New roles for old molecules. J. Integr. Plant Biol. 2010, 52, 98–111. [Google Scholar] [CrossRef] [PubMed]
- Schulz, E.; Tohge, T.; Zuther, E.; Fernie, A.R.; Hincha, D.K. Natural variation in flavonol and anthocyanin metabolism during cold acclimation in Arabidopsis thaliana accessions. Plant Cell Environ. 2015, 38, 1658–1672. [Google Scholar] [CrossRef] [PubMed]
- Schulz, E.; Tohge, T.; Zuther, E.; Fernie, A.R.; Hincha, D.K. Flavonoids are determinants of freezing tolerance and cold acclimation in Arabidopsis thaliana. Sci. Rep. 2016, 6, 34027. [Google Scholar] [CrossRef]
- Wu, Q.; Su, N.; Zhang, X.; Liu, Y.; Cui, J.; Liang, Y. Hydrogen peroxide, nitric oxide and UV RESISTANCE LOCUS8 interact to mediate UV-B-induced anthocyanin biosynthesis in radish sprouts. Sci. Rep. 2016, 6, 29164. [Google Scholar] [CrossRef]
- Naing, A.H.; Park, K.I.; Ai, T.N.; Chung, M.Y.; Han, J.S.; Kang, Y.-W.; Lim, K.-B.; Kil Kim, C. Overexpression of snapdragon Delila (Del) gene in tobacco enhances anthocyanin accumulation and abiotic stress tolerance. BMC Plant Biol. 2017, 17, 65. [Google Scholar] [CrossRef] [Green Version]
- Naing, A.H.; Ai, T.N.; Lim, K.B.; Lee, I.J.; Kim, C.K. Overexpression of Rosea1 from snapdragon enhances anthocyanin accumulation and abiotic stress tolerance in transgenic tobacco. Front. Plant Sci. 2018, 9, 1070. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Zhang, X.W.; Xu, H.F.; Bi, S.Q.; Hao, Y.-J.; Hao, Y.-J. An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol. J. 2019, 18, 337–353. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Giusti, M.M. Anthocyanins: Natural colorants with health-promoting properties. Annu. Rev. Food Sci. Technol. 2010, 1, 163–187. [Google Scholar] [CrossRef] [PubMed]
- Bártiková, H.; Skálová, L.; Dršata, J.; Boušová, I. Interaction of anthocyanin with drug-metabolizing and antioxidant enzymes. Curr. Med. Chem. 2013, 20, 4665–4679. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, L.; Wang, D.; Huo, Y.; Ji, B. Anthocyanin-rich extracts from blackberry, wild blueberry, strawberry, and chokeberry: Antioxidant activity and inhibitory effect on oleic acid-induced hepatic steatosis in vitro. J. Sci. Food Agric. 2016, 96, 2494–2503. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ma, H.; Huang, H.; Li, D.; Yao, S. Natural anthocyanins from phytoresources and their chemical researches. Nat. Prod. Res. 2013, 27, 456–469. [Google Scholar] [CrossRef]
- Li, D.; Zhang, Y.; Liu, Y.; Sun, R.; Xia, M. Purified anthocyanin supplementation reduces dyslipidemia, enhances antioxidant capacity, and prevents insulin resistance in diabetic patients. J. Nutr. 2015, 145, 742–748. [Google Scholar] [CrossRef]
- Fang, J. Bioavailability of anthocyanins. Drug Metab. Rev. 2014, 46, 508–520. [Google Scholar] [CrossRef]
- Cerlettim, C.; de Curtis, A.; Bracone, F.; Digesù, C.; Morganti, A.G.; Iacoviello, L.; De Gaetano, G.; Donati, M. Dietary anthocyanins and health: Data from FLORA and ATHENA EU projects. Br. J. Clin. Pharmacol. 2017, 83, 103–106. [Google Scholar] [CrossRef] [Green Version]
- Huang, P.C.; Kuo, W.W.; Shen, C.Y.; Chen, Y.F.; Lin, Y.M.; Ho, T.J.; Padma, V.V.; Lo, J.-F.; Huang, C.-Y. Anthocyanin attenuates doxorubicin-induced cardiomyotoxicity via estrogen receptor-α/β and stabilizes HSF1 to inhibit the IGF-IIR apoptotic pathway. Int. J. Mol. Sci. 2016, 7, 1588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, B.W.; Gong, C.C.; Song, H.F.; Cui, Y.Y. Effect of anthocyanins on the prevention and treatment of cancer. Br. J. Pharmacol. 2017, 174, 1226–1243. [Google Scholar] [CrossRef] [Green Version]
- Smeriglio, A.; Barreca, D.; Bellocco, E.; Trombetta, D. Chemistry, pharmacology and health benefits of anthocyanins. Phytother. Res. 2016, 30, 1265–1286. [Google Scholar] [CrossRef]
- Ma, L.; Sun, Z.; Zeng, Y.; Luo, M.; Yang, J. Molecular mechanism and health role of functional ingredients in blueberry for chronic disease in human beings. Int. J. Mol. Sci. 2018, 19, 2785. [Google Scholar] [CrossRef] [Green Version]
- Sandoval-Ramírez, B.A.; Catalán, Ú.; Fernández-Castillejo, S.; Rubió, L.; Macià, A.; Solà, R. Anthocyanin tissue bioavailability in animals: Possible implications for human health. A systematic review. J. Agric. Food Chem. 2018, 66, 11531–11543. [Google Scholar] [CrossRef] [Green Version]
- Krga, I.; Milenkovic, D. Anthocyanins: From sources and bioavailability to cardiovascular health benefits and molecular mechanisms of action. J. Agr. Food Chem. 2019, 67, 1771–1783. [Google Scholar] [CrossRef]
- Wu, Z.G.; Jiang, W.; Mantri, N.; Bao, X.Q.; Chen, S.L.; Tao, Z.M. Transcriptome analysis reveals flavonoid biosynthesis regulation and simple sequence repeats in yam (Dioscorea alata L.) tubers. BMC Genom. 2015, 16, 346. [Google Scholar] [CrossRef] [Green Version]
- Dubos, C.; Gourrierec, J.L.; Baudry, A.; Huep, G.; Lanet, E.; Debeaujon, I.; Routaboul, J.-M.; Alboresi, A.; Weisshaar, B.; Lepiniec, L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008, 55, 940–953. [Google Scholar] [CrossRef]
- Das, P.K.; Shin, D.H.; Choi, S.B.; Park, Y.I. Sugar-hormone cross-talk in anthocyanin biosynthesis. Mol. Cells. 2012, 34, 501–507. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trend Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Zhang, F.; Gonzalez, A.; Zhao, M.; Payne, C.T.; Lloyd, A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 2003, 130, 4859–4869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Zhang, B.; Hu, Z.; Zhang, Y.; Li, Y.; Zhou, S.; Chen, G. A putative functional MYB transcription factor induced by low temperature regulates anthocyanin biosynthesis in purple kale (Brassica oleracea var. acephala f. tricolor). Plant Cell Rep. 2012, 31, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Hayashi, M.; Goto, F.; Sato, S.; Soga, T.; Nishioka, T.; Tomita, M.; Kawai-Yamada, M.; Uchimiya, H. Evaluation of metabolic alteration in transgenic rice overexpressing dihydroflavonol 4-reductase. Ann. Bot. 2006, 98, 819–825. [Google Scholar] [CrossRef] [Green Version]
- Xie, D.Y.; Sharma, S.B.; Paiva, N.L.; Ferreira, D.; Dixon, R.A. Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 2003, 299, 396–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, D.Y.; Jackson, L.A.; Cooper, J.D.; Ferreira, D.; Paiva, N.L. Molecular and biochemical analysis of two cDNA clones encoding dihydroflavonol-4-reductase from Medicago truncatula. Plant Physiol. 2004, 134, 979–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Y.; Chiu, L.W.; Li, L. Transcriptional regulation of anthocyanin biosynthesis in red cabbage. Planta 2009, 230, 1141–1153. [Google Scholar] [CrossRef]
- Chiu, L.W.; Zhou, X.; Burke, S.; Wu, X.; Prior, R.L.; Li, L. The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiol. 2010, 154, 1470–1480. [Google Scholar] [CrossRef] [Green Version]
- Chiu, L.W.; Li, L. Characterization of the regulatory network of BoMYB2 in controlling anthocyanin biosynthesis in purple cauliflower. Planta 2012, 236, 1153–1164. [Google Scholar] [CrossRef]
- Xie, Q.; Hu, Z.; Zhang, Y.; Taian, S.; Wang, Z.; Zhao, Z.; Yang, Y.; Chen, G. Accumulation and molecular regulation of anthocyanin in purple tumorous stem mustard (Brassica juncea var. tumida Tsen et Lee). J. Agric. Food Chem. 2014, 62, 7813–7821. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, G.; Dong, T.; Pan, Y.; Zhao, Z.; Tian, S.; Hu, Z. Anthocyanin accumualtion and transcriptional regulation of anthocyanin biosynthesis in purple box choy (Brassica rapa var. chinensis). J. Agric. Food Chem. 2014, 62, 12366–12376. [Google Scholar] [CrossRef]
- Park, W.T.; Kim, J.K.; Park, S.; Lee, S.W.; Li, X.; Kim, Y.B.; Uddin, M.R.; Park, N.I.; Kim, S.-J.; Park, S.U. Metabolic profiling of glucosinoltes, anthocyanins, carotenoids, and other secondary metabolites in kohlrabi (Brassica oleracea var. gonglylodes). J. Agric. Food Chem. 2012, 6, 8111–8116. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hu, Z.; Zhu, M.; Zhu, Z.; Wang, Z.; Tian, S.; Chen, G. Anthocyanin accumulation and molecular analysis of correlated genes in purple kohlrabi (Brassica oleracea var. gongylodes L.). J. Agric. Food Chem. 2015, 63, 4160–4169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Z.; Xu, S.Z.; Cheng, Y.W.; Ya, H.Y.; Han, J.M. Transcriptome analysis and anthocyanin-related genes in red leaf lettuce. Genet. Mol. Res. 2016, 29, 15. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Han, Z.Y.; Zhang, J.; Hu, Y.; Song, T.; Yao, Y. The balance of expression of dihydroflavonol 4-reductase and flavonol synthase regulates flavonoid biosynthesis and red foliage coloration in crabapples. Sci. Rep. 2015, 5, 12228. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zheng, S.; Liu, Z.; Wang, L.; Bi, Y. Both HY5 and HYH are necessary regulators for low temperature-induced anthocyanin accumulation in Arabidopsis seedlings. J. Plant Physiol. 2011, 168, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Sugaya, S.; Gemma, H. Decreased anthocyanin biosynthesis in grape berries grown under elevated night temperature condition. Sci. Hortic. 2005, 105, 319–330. [Google Scholar] [CrossRef]
- Piero, A.R.L.; Puglisi, I.; Rapisarda, P.; Petrone, G. Anthocyanins accumulation and related gene expression in red orange fruit induced by low temperature storage. J. Agric. Food Chem. 2005, 53, 9083–9088. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.U.; Park, J.I.; Jung, H.J.; Yang, T.J.; Hur, Y.; Nou, I.S. Characterization of dihydroflavonol 4-reductase (DFR) genes and their association with cold and freezing stress in Brassica rapa. Gene 2014, 550, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.B.; Li, S.; Zhang, R.F.; Zhao, J.; Chen, Y.C.; Zhao, Q.; Yao, Y.; You, C.-X.; Zhang, X.; Hao, Y.-J. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 2012, 35, 1884–1897. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Li, Y.J.; Zhang, F.J.; Zhang, G.Z.; Jiang, X.Y.; Yu, H.M.; Hou, B.-K. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017, 89, 85–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peters, D.; Constabel, C.P. Molecular analysis of herbivore-induced condensed tannin synthesis: Cloning, expression of dihydroflavonol reductase from trembling aspen (Populus tremuloides). Plant J. 2002, 32, 701–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, K.; Iwasaka, R.; Shimada, N.; Ayabe, S.; Aoki, T.; Sakuta, M. Transcriptional control of the dihydroflavonol 4-reductase multigene family in Lotus japonicus. J. Plant Res. 2010, 123, 801–805. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Qiu, J.; Chen, F.; Lv, X.; Fu, C.; Zhao, D.; Hua, X.; Zhao, Q. Molecular characterization and expression analysis of dihydroflavonol 4-reductase (DFR) gene in Saussurea medusa. Mol. Biol. Rep. 2012, 39, 2991–2999. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Cai, X.; Shan, X.; Gao, R.; Yang, S.; Han, T.; Wang, S.; Wang, L.; Gao, X. Dihydroflavonol 4-reductase genes from Freesia hybrida play important and partially overlapping roles in the biosynthesis of flavonoids. Front. Plant Sci. 2017, 8, 428. [Google Scholar] [CrossRef] [Green Version]
- Bernhardt, J.; Stich, K.; Schwarz-Sommer, Z.; Saedler, H.; Wienand, U. Molecular analysis of a second functional A1 gene (dihydroflavonol 4-reductase) in Zea mays. Plant J. 1998, 14, 483–488. [Google Scholar] [CrossRef]
- Huang, Y.; Gou, J.; Jia, Z.; Yang, L.; Sun, Y.; Xiao, X.; Song, F.; Luo, K. Molecular cloning and characterization of two genes encoding dihydroflavonol-4-reductase from Populus trichocarpa. PLoS ONE 2012, 7, e30364. [Google Scholar] [CrossRef] [Green Version]
- Inagaki, Y.; Johzuka-Hisatomi, Y.; Mori, T.; Takahashi, S.; Hayakawa, Y.; Peyachoknagul, S.; Ozeki, Y.; Iida, S. Genomic organization of the genes encoding dihydroflavonol 4-recutase for flower pigmentation in the Japanse and common morning glories. Gene 1999, 226, 181–188. [Google Scholar] [CrossRef]
- Himi, E.; Noda, K. Isolation and location of three homoeologous dihydroflavonol-4-reductase (DFR) genes of wheat and their tissue-dependent expression. J. Exp. Bot. 2004, 55, 365–375. [Google Scholar] [CrossRef]
- Kim, S.; Baek, D.; Cho, D.Y.; Lee, E.T.; Yoon, M.K. Identification of two novel inactive DFR-A alleles responsible for failure to produce anthocyanin and development of a simple PCR-based molecular marker for bulb color selection in onion (Allium cepa L.). Theor. Appl. Genet. 2009, 118, 1391–1399. [Google Scholar] [CrossRef] [PubMed]
- Hua, C.; Linling, L.; Shuiyuan, C.; Fuliang, C.; Feng, X.; Honghui, Y.; Conghua, W. Molecular cloning and characterization of three genes encoding dihydroflavonol-4-reductase from Ginkgo biloba in anthocyanin biosynthetic pathway. PLoS ONE 2013, 8, e72017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimada, N.; Sasaki, R.; Sato, S.; Kaneko, T.; Tabata, S.; Aoki, T.; Ayabe, S.-I.A. A comprehensive analysis of six dihydroflavonol 4-reductases encoded by a gene cluster of the Lotus japonicus genome. J. Exp. Bot. 2005, 56, 2573–2585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Yoo, K.S.; Pike, L.M. Development of a PCR-based marker utilizing a deletion mutation in the dihydroflavonol 4-reductase (DFR) gene responsible for the lack of anthocyanin production in yellow onions (Allium cepa). Theor. Appl. Genet. 2005, 10, 588–595. [Google Scholar] [CrossRef]
- De Jong, W.S.; De Jong, D.M.; De Jong, H.; Kalazich, J.; Bodis, M. An allele of dihydroflavonol 4-reductase associated with the ability to produce red anthocyanin pigments in potato (Solanum tuberosum L.). Theor. Appl. Genet. 2003, 107, 1375–1383. [Google Scholar] [CrossRef]
- Peukert, M.; Weise, S.; Röder, M.S.; Matthies, I.E. Development of SNP markers for genes of the phenylpropanoid pathway and their association to kernel and malting traits in barley. BMC Genet. 2013, 14, 97. [Google Scholar] [CrossRef] [Green Version]
- McClean, P.E.; Lee, R.K.; Miklas, P.N. Sequence diversity analysis of dihydrofavonol 4-reductase intron 1 in common bean. Genome 2004, 47, 266–280. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△CT 682 method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Lee, H.S.; Wicker, L. Anthocyanin pigments in the skin of lychee fruit. J. Food Sci. 1991, 56, 466–468. [Google Scholar] [CrossRef]
- Giusti, M.M.; Wrolstad, R.E. Characterization and measurement of anthocyanins by UV-visible spectroscopy. In Current Protocols in Food Analytical Chemistry; Wrolstadt, R.E., Ed.; Wiley Press: New York, NY, USA, 2001; pp. F1.2.1–F1.2.13. [Google Scholar]
- Hayashi, K.; Hashimoto, N.; Daigen, M.; Ashikawa, I. Development of PCR-based SNP markers for rice blast resistance genes. Theor. Appl. Genet. 2004, 108, 1212–1220. [Google Scholar] [CrossRef]
- Gaudet, M.; Fara, A.G.; Beritognolo, I.; Sabatti, M. Allele-specific PCR in SNP genotyping. Methods Mol. Biol. 2009, 578, 415–424. [Google Scholar]
- Hayashi, M.; Takahashi, H.; Tamura, K.; Huang, J.; Yu, L.H.; Kawai-Yamada, M.; Tezuka, T.; Uchimiya, H. Enhanced dihydroflavonol-4-reductase activity and NAD homeostasis leading to cell death tolerance in transgenic rice. Proc. Natl. Acad. Sci. USA 2005, 102, 7020–7025. [Google Scholar] [CrossRef] [Green Version]
- Urao, T.; Katagiri, T.; Mizoguchi, T.; Yamaguchi-Shinozaki, K.; Hayashida, N.; Shinozaki, K. Two genes that encode Ca2+-dependent protein-kinases are induced by drought and high-salt stresses in Arabidopsis thaliana. Mol. Genet. Genom. 1994, 244, 331–340. [Google Scholar] [CrossRef]
- Menkens, A.E.; Schindler, U.; Cashmore, A.R. The G-box: A ubiquitous regulatory DNA element in plants bound by the GBF family of bzip proteins. Trends Biochem. Sci. 1995, 20, 506–510. [Google Scholar] [CrossRef]
- Stålberg, K.; Ellerström, M.; Ezcurra, I.; Ablov, S.; Rask, L. Disruption of an overlapping E box/ABRE motif abolished high transcription of the napA storage protein promoter in transgenic Brassica napus seeds. Planta 1996, 199, 515–519. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003, 15, 63–78. [Google Scholar] [CrossRef] [Green Version]
- Baranowskij, N.; Frohberg, C.; Prat, S.; Willmitzer, L. A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. EMBO J. 1994, 13, 5383–5392. [Google Scholar] [CrossRef]
- Johnson, E.T.; Ryu, S.; Yi, H.K.; Shin, B.; Cheong, H.; Choi, G. Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase. Plant J. 2001, 25, 325–333. [Google Scholar] [CrossRef]
- Petit, P.; Granier, T.; d’Estaintot, B.L.; Manigand, C.; Bathany, K.; Schmitter, J.M.; Lauvergeat, V.; Hamdi, S.; Gallois, B. Crystal structure of grape dihydroflavonol4-reductase, a key enzyme in flavonoid biosynthesis. J. Mol. Biol. 2007, 368, 1345–1357. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Jeong, C.Y.; Kang, G.H.; Yoo, S.D.; Hong, S.W.; Lee, H. MYBD employed by HY5 increases anthocyanin accumulation via repression of MYBL2 in Arabidopsis. Plant J. 2015, 84, 1192–1205. [Google Scholar] [CrossRef] [Green Version]
- Chalker-Scott, L. Environmental significance of anthocyanins in plant stress responses. Photochem. Photobiol. 1999, 70, 1–9. [Google Scholar] [CrossRef]
- Guo, N.; Han, S.; Zong, M.; Wang, G.; Zheng, S.; Liu, F. Identification and differential expression analysis of anthocyanin biosynthetic genes in leaf color variants of ornamental kale. BMC Genom. 2019, 20, 564. [Google Scholar] [CrossRef] [Green Version]
- Shi, M.Z.; Xie, D.Y. Features of anthocyanin biosynthesis in pap1-D and wild-type Arabidopsis thaliana plants grown in different light intensity and culture media conditions. Planta 2010, 231, 1385–1400. [Google Scholar] [CrossRef]
- Liu, X.P.; Gao, B.Z.; Han, F.Q.; Fang, Z.Y.; Yang, L.M.; Zhuang, M.; Lv, H.; Liu, Y.; Li, Z.; Cai, C.; et al. Genetics and fine mapping of a purple leaf gene, BoPr, in ornamental kale (Brassica oleracea L. var. acephala). BMC Genom. 2017, 18, 230. [Google Scholar] [CrossRef] [Green Version]
- Espley, R.V.; Brendolise, C.; Chagné, D.; Kutty-Amma, S.; Green, S.; Volz, R.; Putterill, J.; Schouten, H.J.; Gardiner, S.E.; Hellens, R.P.; et al. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell. 2009, 21, 168–183. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Lee, H.J.; Jung, Y.J.; Kang, K.K.; Tyagi, W.; Kovach, M.; Sweeney, M.; McCouch, S.; Cho, Y.-G. Functional properties of an alternative, tissue-specific promoter for rice NADPH-dependent dihydroflavonol reductase. PLoS ONE 2017, 12, e0183722. [Google Scholar] [CrossRef] [Green Version]
- Petridis, A.; Döll, S.; Nichelmann, L.; Bilger, W.; Mock, H.P. Arabidopsis thaliana G2-LIKE FLAVONOID REGULATOR and BRASSINOSTEROID ENHANCED EXPRESSION1 are low-temperature regulators of flavonoid accumulation. N. Phytol. 2016, 211, 912–925. [Google Scholar] [CrossRef] [Green Version]
- Debeaujon, I.; Nesi, N.; Perez, P.; Devic, M.; Grandjean, O.; Caboche, M.; Lepiniec, L. Proanthocyanidin-accumulating cells in Arabidopsis testa: Regulation of differentiation and role in seed development. Plant Cell. 2003, 15, 2514–2531. [Google Scholar] [CrossRef] [Green Version]
- Baudry, A.; Heim, M.A.; Dubreucq, B.; Caboche, M.; Weisshaar, B.; Lepiniec, L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanindin biosynthesis in Arabidopsis thaliana. Plant J. 2004, 39, 366–380. [Google Scholar] [CrossRef]
- Baudry, A.; Caboche, M.; Lepiniec, L. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 2006, 46, 768–779. [Google Scholar] [CrossRef]
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Dang, P.; Liu, L.; He, C. Cold acclimation by the CBF–COR pathway in a changing climate: Lessons from Arabidopsis thaliana. Plant Cell Rep. 2019, 3, 511–519. [Google Scholar] [CrossRef] [Green Version]










| Genotype ID or Name | Characteristic † | Genetic Status | Use § | Seed Source |
|---|---|---|---|---|
| 337 | HAA | Inbred line | NTS, RT-PCR | Asia Seed Co. |
| 2437 | HAA | .. | NTS, RT-PCR | Asia Seed Co. |
| 154 | HAA | .. | NTS, RT-PCR, qRT-PCR | Asia Seed Co. |
| 09WH45 | LAA | .. | NTS, qRT-PCR | Asia Seed Co. |
| 2409 | LAA | .. | NTS, qRT-PCR | Asia Seed Co. |
| 842 | LAA | .. | NTS | Asia Seed Co. |
| Speed King | LAA | F1 cultivar | NTS, qRT-PCR | Takii Seed Co. |
| BN106 | Cold tolerance | Inbred line | qRT-PCR | Asia Seed Co. |
| 7S4-63 | Purple cabbage | .. | NTS | Asia Seed Co. |
| 7S4-51 | Purple cabbage | .. | NTS | Asia Seed Co. |
| CT-415 | HAA | F1 cultivar | NTS, qRT-PCR | Asia Seed Co. |
| T-523 | HAA | F1 cultivar | NTS, qRT-PCR | Takii Seed Co. |
| T-530 | HAA | .. | NTS, qRT-PCR | Takii Seed Co. |
| Golden Cross | LAA | .. | NTS, qRT-PCR | Takii Seed Co. |
| Use | SNP Position (bp) | Primer Orientation | Target SNP | Primer Name | Primer Sequence (5′→3′) | PCR Reaction Conditions | Product Size (bp) | ||
|---|---|---|---|---|---|---|---|---|---|
| Template DNA (ng) | Annealing Temperature (°C) | No. of Cycles | |||||||
| PCR Analysis | 423 | Forward | C | BoDFR 423C-F1 | GTTATACTATGAACCTTTTTCTTATTACAC | 5 | 60 | 27 | 266 |
| T | BoDFR 423T-F1 | GTTATACTATGAACCTTTTTCTTATTACAT | 5 | 60.5 | 27 | 266 | |||
| Reverse | BoDFR 423CT-R1 | CTGTCATCTTCTTGGACATGATAAAGT | |||||||
| 628 | Forward | C | BoDFR 628C-F2 | ATGATGAAAACGATTGGAGTGATATC | 5 | 60 | 27 | 295 | |
| T | BoDFR 628T-F2 | ATGATGAAAACGATTGGAGTGATATT | 5 | 61 | 27 | 295 | |||
| Reverse | BoDFR 628CT-R1 | GCGGTAATAAGGCTAGG | |||||||
| 1118 | Forward | T | BoDFR1-1118T-F2 | TTTGTTCCTCTCACGATGCAACGATACT | 7.5 | 62.7 | 27 | 197 | |
| A | BoDFR1-1118A-F1 | GTTTGTTCCTCTCACGATGCAACGATTTA | 7.5 | 62.7 | 27 | 198 | |||
| Reverse | BoDFR1-1118TA-R | CTCTTTAGATTCTCATCCACTCCTTCAAA | |||||||
| HRM | 1118 | BoDFR-HRM-aF12 | TCCACTTGGACGACTTATGCAATGCT | 66.3 | |||||
| BoDFR-HRM-aR1 | ATCTTACGTTGAAGGCACGTTATATTCTG | 66.2 | |||||||
| BoDFR-HRM-pF1 | GCAACGATTCATACTATCTC | 54.3 | |||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, H.; Park, J.-I.; Hwang, B.-H.; Yi, H.; Kim, H.; Hur, Y. SNP in DFR1 Coding Sequence Is Tightly Associated with Anthocyanin Accumulation in Cabbage (B. oleracea var. capitata f. alba) at Low Temperature. Agronomy 2020, 10, 602. https://doi.org/10.3390/agronomy10040602
Song H, Park J-I, Hwang B-H, Yi H, Kim H, Hur Y. SNP in DFR1 Coding Sequence Is Tightly Associated with Anthocyanin Accumulation in Cabbage (B. oleracea var. capitata f. alba) at Low Temperature. Agronomy. 2020; 10(4):602. https://doi.org/10.3390/agronomy10040602
Chicago/Turabian StyleSong, Hayoung, Jong-In Park, Byung-Ho Hwang, Hankuil Yi, HyeRan Kim, and Yoonkang Hur. 2020. "SNP in DFR1 Coding Sequence Is Tightly Associated with Anthocyanin Accumulation in Cabbage (B. oleracea var. capitata f. alba) at Low Temperature" Agronomy 10, no. 4: 602. https://doi.org/10.3390/agronomy10040602
APA StyleSong, H., Park, J.-I., Hwang, B.-H., Yi, H., Kim, H., & Hur, Y. (2020). SNP in DFR1 Coding Sequence Is Tightly Associated with Anthocyanin Accumulation in Cabbage (B. oleracea var. capitata f. alba) at Low Temperature. Agronomy, 10(4), 602. https://doi.org/10.3390/agronomy10040602





