Grain Quality and Allelic Variation of the Badh2 Gene in Thai Fragrant Rice Landraces
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Seed Morphological Characterization
2.3. Grain Qualities
2.4. DNA Extraction and Badh2 Sequences
2.5. Statistical Analysis
3. Results
3.1. Seed Morphological Variation of Thai Fragrant Rice Landraces
3.2. Grain Quality
3.3. Badh2 Sequence Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sakthivel, K.; Sundaram, R.M.; Shobha Rani, N.; Balachandran, S.M.; Neeraja, C.N. Genetic and Molecular Basis of Fragrance in Rice. Biotechnol. Adv. 2009, 27, 468–473. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Singh, U.S.; Khush, G.S. Aromatic Rices; Oxford and India Book House Publishing Co. Pvt. Ltd.: New Delhi, India, 2000. [Google Scholar]
- FAO Rice Price Update. Available online: http://www.fao.org/economic/est/publications/rice-publications/the-fao-rice-price-update/en (accessed on 13 April 2020).
- TREA. Thai Rice Exporters Association. Available online: http://www.thairiceexporters.or.th/price.htm (accessed on 13 April 2020).
- Buttery, R.G.; Ling, L.C.; Juliano, B.O.; Turnbaugh, J.G. Cooked Rice Aroma and 2-Acetyl-1-Pyrroline. J. Agric. Food Chem. 1983, 31, 823–826. [Google Scholar] [CrossRef]
- Ahn, S.N.; Bollich, C.N.; Tanksley, S.D. RFLP Tagging of a Gene for Aroma in Rice. Appl. Genet. 1992, 84, 825–828. [Google Scholar] [CrossRef]
- Yi, M.; Nwe, K.T.; Vanavichit, A.; Chai-arree, W.; Toojinda, T. Marker Assisted Backcross Breeding to Improve Cooking Quality Traits in Myanmar Rice Cultivar Manawthukha. Field Crop. Res. 2009, 113, 178–186. [Google Scholar] [CrossRef]
- Fitzgerald, T.L.; Waters, D.L.E.; Brooks, L.O.; Henry, R.J. Fragrance in Rice (Oryza Sativa) Is Associated with Reduced Yield under Salt Treatment. Environ. Exp. Bot. 2010, 68, 292–300. [Google Scholar] [CrossRef]
- Bradbury, L.M.T.; Fitzgerald, T.L.; Henry, R.J.; Jin, Q.; Waters, D.L.E. The Gene for Fragrance in Rice. Plant Biotechnol. J. 2005, 3, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Itani, T.; Tamaki, M.; Hayata, Y.; Fushimi, T.; Hashizume, K. Variation of 2-Acetyl-1-Pyrroline Concentration in Aromatic Rice Grains Collected in the Same Region in Japan and Factors Affecting Its Concentration. Plant Prod. Sci. 2004, 7, 178–183. [Google Scholar] [CrossRef]
- Vanavichit, A.; Kamolsukyeunyong, W.; Siangliw, M.; Siangliw, J.L.; Traprab, S.; Ruengphayak, S.; Chaichoompu, E.; Saensuk, C.; Phuvanartnarubal, E.; Toojinda, T.; et al. Thai Hom Mali Rice: Origin and Breeding for Subsistence Rainfed Lowland Rice System. Rice 2018, 11, 20. [Google Scholar] [CrossRef]
- Kovach, M.J.; Calingacion, M.N.; Fitzgerald, M.A.; McCouch, S.R. The Origin and Evolution of Fragrance in Rice (Oryza Sativa L.). Proc. Natl. Acad. Sci. USA 2009, 106, 14444–14449. [Google Scholar] [CrossRef]
- Singh, V.; Singh, A.K.; Mohapatra, T.; Ellur, R.K. Pusa Basmati 1121—A Rice Variety with Exceptional Kernel Elongation and Volume Expansion after Cooking. Rice 2018, 11, 19. [Google Scholar] [CrossRef]
- Promotional and Public Relations Activities on Thai Hom Mali Rice Standard. Available online: http://www.dft.go.th/th-th/Service-DFT/Service-Information/DATA-Group-Product/Detail-DATA-Group-Product/ArticleId/7494/dft-Thai-jasmine-rice-22 (accessed on 1 April 2020).
- Thai Ministry of Commerce. Thailand Standards for Rice. Available online: http://www.dft.go.th/th-th/ShareDocument1/ArticleId/8860/-Thailand-Standards-for-Rice (accessed on 13 April 2020).
- Shi, W.; Yang, Y.; Chen, S.; Xu, M. Discovery of a New Fragrance Allele and the Development of Functional Markers for the Breeding of Fragrant Rice Varieties. Mol. Breed. 2008, 22, 185–192. [Google Scholar] [CrossRef]
- Sakthivel, K.; Rani, N.S.; Pandey, M.K.; Sivaranjani, A.K.P.; Neeraja, C.N.; Balachandran, S.M.; Madhav, M.S.; Viraktamath, B.C.; Prasad, G.S.V.; Sundaram, R.M. Development of a Simple Functional Marker for Fragrance in Rice and Its Validation in Indian Basmati and Non-Basmati Fragrant Rice Varieties. Mol. Breed. 2009, 24, 185–190. [Google Scholar] [CrossRef]
- Gaur, A.; Wani, S.; Deepika, P.; Bharti, N.; Malav, A.; Shikari, A.; Bhat, A. Understanding the Fragrance in Rice. Rice Res. Open Access 2016, 4. [Google Scholar] [CrossRef]
- Bradbury, L.M.T.; Henry, R.J.; Jin, Q.; Reinke, R.F.; Waters, D.L.E. A Perfect Marker for Fragrance Genotyping in Rice. Mol. Breed. 2005, 16, 279–283. [Google Scholar] [CrossRef]
- Amarawathi, Y.; Singh, R.; Singh, A.K.; Singh, V.P.; Mohapatra, T.; Sharma, T.R.; Singh, N.K. Mapping of Quantitative Trait Loci for Basmati Quality Traits in Rice (Oryza Sativa L.). Mol. Breed. Mol. Breed. New Strat. Plant Improv. 2008, 21, 49–65. [Google Scholar] [CrossRef]
- Shao, G.N.; Tang, A.; Tang, S.Q.; Luo, J.; Jiao, G.A.; Wu, J.L.; Hu, P.S. A New Deletion Mutation of Fragrant Gene and the Development of Three Molecular Markers for Fragrance in Rice. Plant Breed. 2011, 130, 172–176. [Google Scholar] [CrossRef]
- Shao, G.; Tang, S.; Chen, M.; Wei, X.; He, J.; Luo, J.; Jiao, G.; Hu, Y.; Xie, L.; Hu, P. Haplotype Variation at Badh2, the Gene Determining Fragrance in Rice. Genomics 2013, 101, 157–162. [Google Scholar] [CrossRef]
- Shi, Y.; Zhao, G.; Xu, X.; Li, J. Discovery of a New Fragrance Allele and Development of Functional Markers for Identifying Diverse Fragrant Genotypes in Rice. Mol. Breed. 2014, 33, 701–708. [Google Scholar] [CrossRef]
- Ootsuka, K. Genetic Polymorphisms in Japanese Fragrant Landraces and Novel Fragrant Allele Domesticated in Northern Japan. Breed. Sci. 2014, 64, 115–124. [Google Scholar] [CrossRef]
- He, Q.; Yu, J.; Kim, T.-S.; Cho, Y.-H.; Lee, Y.-S.; Park, Y.-J. Resequencing Reveals Different Domestication Rate for BADH1 and BADH2 in Rice (Oryza Sativa). PLoS ONE 2015, 10, e0134801. [Google Scholar] [CrossRef]
- Myint, K.M.; Arikit, S.; Wanchana, S.; Yoshihashi, T.; Choowongkomon, K.; Vanavichit, A. A PCR-Based Marker for a Locus Conferring the Aroma in Myanmar Rice (Oryza Sativa L.). Appl. Genet. 2012, 125, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Oo, K.S.; Kongjaimun, A.; Khanthong, S.; Yi, M.; Myint, T.T.; Korinsak, S.; Siangliw, J.L.; Myint, K.M.; Vanavichit, A.; Malumpong, C.; et al. Characterization of Myanmar Paw San Hmwe Accessions Using Functional Genetic Markers. Rice Sci. 2015, 22, 53–64. [Google Scholar] [CrossRef]
- Chan, S. Malys Angkor Crowned World’s Best Rice. Available online: https://www.khmertimeskh.com/540271/malys-angkor-crowned-worlds-best-rice/ (accessed on 13 April 2020).
- Bairagi, S.; Mohanty, S.; Custodio, M.C. Consumers’ Preferences for Rice Attributes in Cambodia: A Choice Modeling Approach. J. Agribus. Dev. Emerg. Econ. 2019, 9, 94–108. [Google Scholar] [CrossRef]
- Saetan, J. Seed Development and Maturation and Panicle Position on Seed Quality of Upland Rice Cv. Leum Pua. Master’s Thesis, Prince of Songkla University, Songkla, Thailand, 2017. [Google Scholar]
- TRKB. Rice Variety. Available online: http://www.ricethailand.go.th/rkb3/Varieties.htm (accessed on 13 April 2020).
- Sadabpod, K.; Tongyonk, L.; Kangsadalampai, K. Effect of Hom Nil Rice and Black Glutinous Rice Extracts during Treatment of Chicken Extract with Sodium Nitrite Using Ames Test. J. Health Sci. Med. Res. 2014, 32, 139–149. [Google Scholar]
- KURDI. Hom-nil, A Black Non-Glutinous Rice with High Nutritional Quality; Kasetsart University Research and Development Institute: KURDI. Available online: https://www3.rdi.ku.ac.th/?p=27452 (accessed on 13 April 2020).
- Chan-in, P.; Jamjod, S.; Yimyam, N.; Pusadee, T. Diversity of BADH2 Alleles and Microsatellite Molecules in Highland Fragrant Rice Landraces. J. Agric. 2019, 35, 23–35. [Google Scholar]
- Tejakum, P.; Khumto, S.; Jamjod, S.; Yimyam, N.; Pusadee, T. Yield, Grain Quality and Fragrance of a Highland Fragrant Rice Landrace Variety, Bue Ner Moo. Khon Kaen Agric. J. 2019, 47, 317–326. [Google Scholar] [CrossRef]
- Sarkarung, S.; Somrith, B.; Chitrakorn, S. Aromatic Rice of Thailand. In Aromatic Rices; Singh, R.K., Singh, U.S., Khush, G.S., Eds.; Mohan Primlani for Oxford and India Book House Publishing Co. Pvt. Ltd.: New Delhi, India, 2000; pp. 180–183. [Google Scholar]
- The Rice Trader: TRT World’s Best Rice News. Available online: https://thericetrader.com/news/ (accessed on 13 April 2020).
- Arunmas, P. Farmers Urge Action after Top Rice Fails to Win Prize. Available online: https://www.bangkokpost.com/business/1796249/farmers-urge-action-after-top-rice-fails-to-win-prize (accessed on 30 December 2019).
- Harakotr, B.; Prompoh, K.; Boonyuen, S.; Suriharn, B.; Lertrat, K. Variability in Nutraceutical Lipid Content of Selected Rice (Oryza Sativa L. Spp. Indica) Germplasms. Agronomy 2019, 9, 823. [Google Scholar] [CrossRef]
- Harlan, J.R. Crops and Man. Syst. Bot. 1977, 2, 227. [Google Scholar] [CrossRef]
- Londo, J.P.; Chiang, Y.-C.; Hung, K.-H.; Chiang, T.-Y.; Schaal, B.A. Phylogeography of Asian Wild Rice, Oryza Rufipogon, Reveals Multiple Independent Domestications of Cultivated Rice, Oryza Sativa. Proc. Natl. Acad. Sci. USA 2006, 103, 9578–9583. [Google Scholar] [CrossRef]
- Pusadee, T.; Jamjod, S.; Chiang, Y.-C.; Rerkasem, B.; Schaal, B.A. Genetic Structure and Isolation by Distance in a Landrace of Thai Rice. Proc. Natl. Acad. Sci. USA 2009, 106, 13880–13885. [Google Scholar] [CrossRef]
- Khush, G.S.; Toenniessen, G.H. Rice Biotechnology; International Rice Research Institute: Los Baños, Philippines, 1991. [Google Scholar]
- Dowling, N.G.; Greenfield, S.M.; Fischer, K.S. Sustainability of Rice in the Global Food System; International Rice Research Institute: Los Baños, Philippines, 1998. [Google Scholar]
- Pusadee, T.; Oupkaew, P.; Rerkasem, B.; Jamjod, S.; Schaal, B.A. Natural and Human-mediated Selection in a Landrace of Thai Rice (Oryza Sativa). Ann. Appl. Biol. 2014, 165, 280–292. [Google Scholar] [CrossRef]
- Govindaraj, M.; Vetriventhan, M.; Srinivasan, M. Importance of Genetic Diversity Assessment in Crop Plants and Its Recent Advances: An Overview of Its Analytical Perspectives. Genet. Res. Int. 2015, 2015, e431487. [Google Scholar] [CrossRef] [PubMed]
- IBPGR-IRRI Rice Advisory Committee and International Board for Plant Genetic Resources. Descriptors for Rice, Oryza Sativa L.; International Rice Research Institute: Los Baños, Philippines, 1980. [Google Scholar]
- Matsuo, T. Genecological Studies on Cultivated Rice. Bull. Natl. Inst. Agric. Sci. Jpn. D 1952, 3, 1–111. [Google Scholar]
- Shannon, C.E. A Mathematical Theory of Communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Bhattacharya, K.R. Gelatinization Temperature of Rice Starch and Its Determination. In Chemical Aspects of rice Grain Grain Quality; International Rice Research Institute: Los Baños, Philippines, 1979; pp. 231–250. [Google Scholar]
- Mariotti, M.; Fongaro, L.; Catenacci, F. Alkali Spreading Value and Image Analysis. J. Cereal Sci. 2010, 52, 227–235. [Google Scholar] [CrossRef]
- Perez, C.M.; Juliano, B.O. Modification of the Simplified Amylose Test for Milled Rice. Starch-Stärke 1978, 30, 424–426. [Google Scholar] [CrossRef]
- Sood, B.C.; Siddiq, E.A. A Rapid Technique for Scent Determination in Rice. Indian J. Genet. Plant Breed. 1978, 38, 268–275. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A Software for Comprehensive Analysis of DNA Polymorphism Data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-Joining Networks for Inferring Intraspecific Phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Heidhues, F.; Rerkasem, B. IRRI’s Upland Rice Research Follow-up Review; Giar Science Council: Rome Italy, 2006. [Google Scholar]
- Xiongsiyee, V.; Rerkasem, B.; Veeradittakit, J.; Saenchai, C.; Lordkaew, S.; Prom-u-thai, C.T. Variation in Grain Quality of Upland Rice from Luang Prabang Province, Lao PDR. Rice Sci. 2018, 25, 94–102. [Google Scholar] [CrossRef]
- Jamjod, S.; Yimyam, N.; Lordkaew, S.; Prom-u-thai, C.; Rerkasem, B. Characterization of On-Farm Rice Germplasm in an Area of the Crop’s Center of Diversity. Chiang Mai Univ. J. Nat. Sci. 2017, 16, 85–98. [Google Scholar] [CrossRef][Green Version]
- Juliano, B.O.; Duff, B. Rice Grain Quality as an Emerging Priority in National Rice Breeding Programmes. In Rice Grain Marketing and Quality Issues; International Rice Research Institute: Manila, Philippines, 1991; pp. 55–64. [Google Scholar]
- Bollich, C.N. Release of New Rice Cultivar Jasmine 85 in USA. Int. Rice Res. Newsl. 1989, 14, 12. [Google Scholar]
- Marchetti, M.A.; Bollich, C.N.; Webb, B.D.; Jackson, B.R.; McClung, A.M.; Scott, J.E.; Hung, H.H. Registration of ‘Jasmine 85′Rice. Crop. Sci. 1998, 38, 896. [Google Scholar] [CrossRef]
- Sha, X.Y.; Linscombe, S.D.; Jodari, F.; Chu, Q.R.; Groth, D.E.; Blanche, S.B.; Harrell, D.L.; White, L.M.; Oard, J.H.; Chen, M.H.; et al. Registration of ‘Jazzman’ Aromatic Long-Grain Rice. J. Plant Regist. 2011, 5, 304–308. [Google Scholar] [CrossRef]
- Seekhaw, P.; Mahatheeranont, S.; Sookwong, P.; Luangkamin, S.; Neonplab, A.N.L.; Puangsombat, P. Phytochemical Constituents of Thai Dark Purple Glutinous Rice Bran Extract [Cultivar Luem Pua (Oryza Sativa L.)]. Chiang Mai J. Sci. 2018, 45, 1383–1395. [Google Scholar]
- Suwanarit, A.; Kreetapirom, S.; Buranakarn, S.; Varanyanond, W.; Tungtrakul, P.; Somboonpong, S.; Rattapat, S.; Ratanasupa, S.; Romyen, P.; Wattanapayapkul, S.; et al. Effects of nitrogen fertilizer on grain qualities of Khaw Dauk Mali-105 aromatic rice. J. The Kasetsart (Nat. Sci.) 1996, 30, 458–474. [Google Scholar]
- Leesawatwong, M.; Jamjod, S.; Rerkasem, B.; Pinjai, S. Determinants of a Premium-Priced, Special-Quality Rice. Int. Rice Res. Notes 2003, 28, 33. [Google Scholar]
- Yoshihashi, T.; Huong, N.T.T.; Inatomi, H. Precursors of 2-Acetyl-1-Pyrroline, a Potent Flavor Compound of an Aromatic Rice Variety. J. Agric. Food Chem. 2002, 50, 2001–2004. [Google Scholar] [CrossRef]
- Yoshihashi, T.; Nguyen, T.T.H.; Kabaki, N. Area Dependency of 2-Acetyl-1-Pyrroline Content in an Aromatic Rice Variety, Khao Dawk Mali 105. Jpn. Agric. Res. Q. Jarq 2004, 38, 105–109. [Google Scholar] [CrossRef]
- Golam, F.; Norzulaani, K.; Jennifer, A.H.; Subha, B.; Zulqarnain, M.; Osman, M.; Nazia, A.M.; Zulqarnian, M.; Mohammad, O. Evaluation of Kernel Elongation Ratio and Aroma Association in Global Popular Aromatic Rice Cultivars in Tropical Environment. Afr. J. Agric. Res. 2010, 5, 1515–1522. [Google Scholar]
- Wakte, K.; Zanan, R.; Hinge, V.; Khandagale, K.; Nadaf, A.; Henry, R. Thirty-Three Years of 2-Acetyl-1-Pyrroline, a Principal Basmati Aroma Compound in Scented Rice (Oryza Sativa L.): A Status Review. J. Sci. Food Agric. 2016. [Google Scholar] [CrossRef]
- Gay, F.; Maraval, I.; Roques, S.; Gunata, Z.; Boulanger, R.; Audebert, A.; Mestres, C. Effect of Salinity on Yield and 2-Acetyl-1-Pyrroline Content in the Grains of Three Fragrant Rice Cultivars (Oryza Sativa L.) in Camargue (France). Field Crop. Res. 2010, 117, 154–160. [Google Scholar] [CrossRef]
- Li, M.; Ashraf, U.; Tian, H.; Mo, Z.; Pan, S.; Anjum, S.A.; Duan, M.; Tang, X. Manganese-Induced Regulations in Growth, Yield Formation, Quality Characters, Rice Aroma and Enzyme Involved in 2-Acetyl-1-Pyrroline Biosynthesis in Fragrant Rice. Plant Physiol. Biochem. 2016, 103, 167–175. [Google Scholar] [CrossRef]

| Rice Samples | Source | Code | Rice Variety Name | Source (Province) |
|---|---|---|---|---|
| Rice landrace | North | BNM1 | Buer Ner Moo | Chiang Mai |
| BNM2 | Buer Ner Moo | Chiang Mai | ||
| BNM3 | Buer Ner Moo | Chiang Mai | ||
| BNM4 | Buer Ner Moo | Chiang Mai | ||
| BNM5 | Buer Ner Moo | Chiang Mai | ||
| BNM6 | Buer Ner Moo | Chiang Mai | ||
| BNM7 | Buer Ner Moo Pho Phi | Chiang Mai | ||
| BNM8 | Buer Ner Moo Pho Phi | Chiang Mai | ||
| BNM9 | Buer Ner Moo Phardo | Chiang Mai | ||
| Northeast | EL | E-Leuang | Loei | |
| KH | Kaow Hawm | Loei | ||
| HS | Hawm Sa-ngium | Loei | ||
| NDLP | Niaw Dam Luem Pua | Loei | ||
| NU | Niaw Ubon | Loei | ||
| PS | Pla Sew | Loei | ||
| PLD | Phayaa Luem Dang | Loei | ||
| SKH | Sew Kliang Hawm | Loei | ||
| South | BH62 | Baow Hawm 62 | Songkhla | |
| BH96 | Baow Hawm96 | Songkhla | ||
| HB | Hawm Baow | Songkhla | ||
| HBK | Hawm Bang Kaew | Songkhla | ||
| HNK | Hawm Na Kaow | Songkhla | ||
| Breeding Line | BNM-CMU | Buer Ner Moo-CMU | Department of Agriculture | |
| JPD | Jow Pluak Dam | Department of Agriculture | ||
| Elite rice | KDML105 | Khao Dawk Mali 105 | Department of Agriculture | |
| (as check varieties) | PTT1 | Pathum Thani 1 | Department of Agriculture | |
| SPR1 | Suphan Buri 1 | Department of Agriculture |
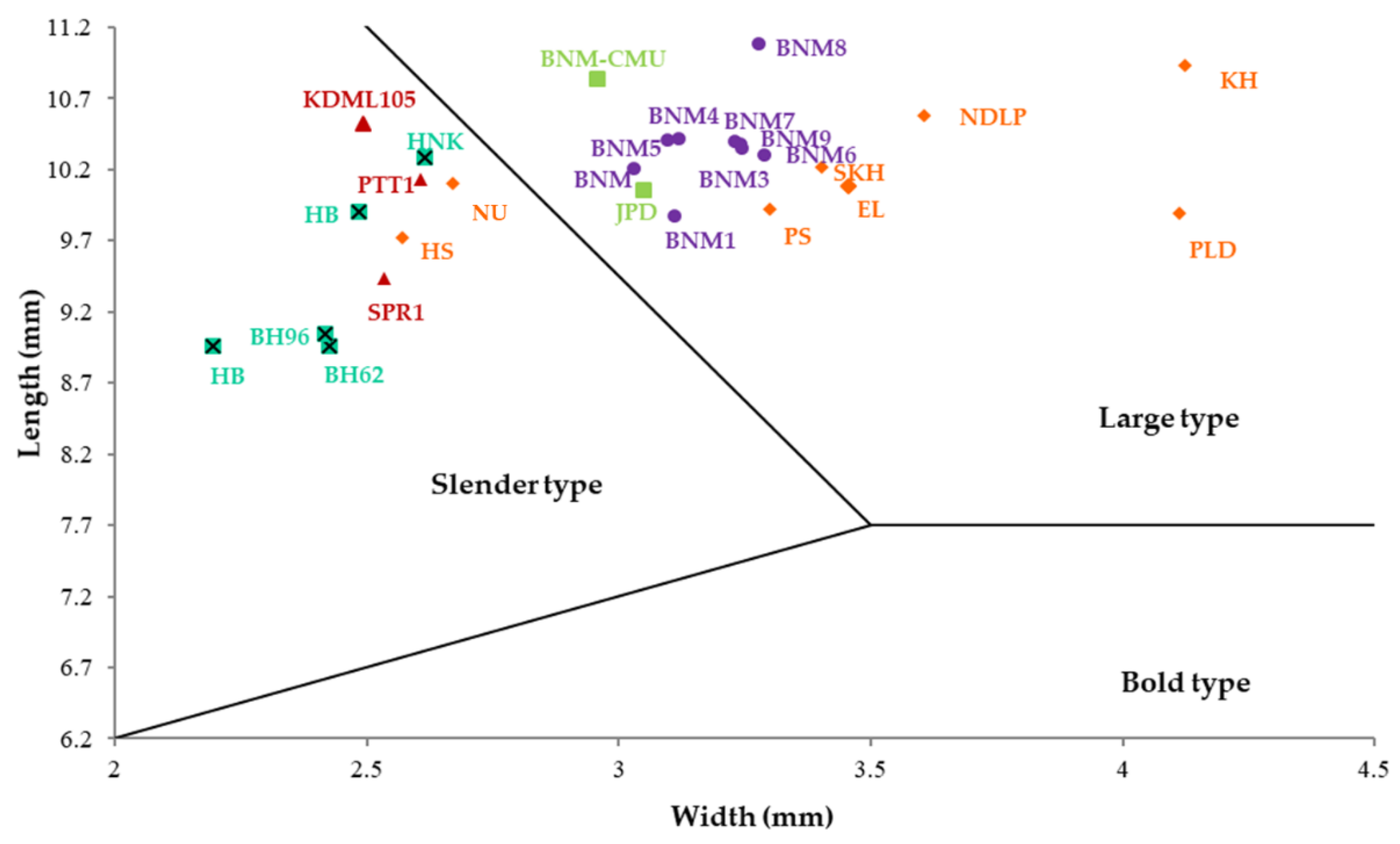
| Varieties | Grain Length (mm) | Grain Width (mm) | Grain Thickness (mm) | Grain Shape # |
|---|---|---|---|---|
| BNM1 | 9.87 | 3.11 | 2.18 | Large type |
| BNM2 | 10.20 | 3.03 | 2.10 | Large type |
| BNM3 | 10.34 | 3.25 | 2.21 | Large type |
| BNM4 | 10.41 | 3.12 | 2.15 | Large type |
| BNM5 | 10.41 | 3.10 | 2.19 | Large type |
| BNM6 | 10.29 | 3.29 | 2.21 | Large type |
| BNM7 | 10.40 | 3.23 | 2.19 | Large type |
| BNM8 | 11.08 | 3.28 | 2.18 | Large type |
| BNM9 | 10.38 | 3.24 | 2.14 | Large type |
| EL | 11.08 | 3.50 | 2.22 | Slender type |
| HS | 9.98 | 2.51 | 1.79 | Large type |
| KH | 11.25 | 3.91 | 2.45 | Slender type |
| NDLP | 11.07 | 3.53 | 2.15 | Slender type |
| NU | 10.65 | 2.62 | 1.94 | Large type |
| PLD | 11.22 | 4.04 | 2.51 | Slender type |
| PS | 10.72 | 3.09 | 2.16 | Slender type |
| SKH | 10.79 | 3.13 | 1.89 | Slender type |
| BH62 | 9.23 | 2.50 | 1.77 | Large type |
| BH96 | 9.06 | 2.40 | 1.90 | Large type |
| HB | 9.08 | 2.12 | 1.70 | Slender type |
| HBK | 9.91 | 2.48 | 1.75 | Large type |
| HNK | 9.86 | 2.64 | 1.94 | Large type |
| BNM-CMU | 10.83 | 2.96 | 2.12 | Large type |
| JPD | 10.05 | 3.05 | 2.09 | Large type |
| KDML105 | 10.52 | 2.49 | 1.88 | Slender type |
| PTT1 | 10.56 | 2.49 | 1.94 | Slender type |
| SPR1 | 9.60 | 2.38 | 2.06 | Slender type |
| Mean | 10.33 | 2.98 | 2.07 | |
| SD | 0.62 | 0.48 | 0.20 | |
| CV (%) | 6.16 | 5.77 | 6.46 | |
| F-test | *** | *** | *** | |
| LSD (0.05) | 0.32 | 0.09 | 0.07 |
| Varieties | Husk Color | Pericarp Traits | Awning | ||
|---|---|---|---|---|---|
| Phenotype | H’ | Phenotype | H’ | ||
| BNM1 | Straw and brown furrows on straw | 0.135 | Colorless | Short and partly awned | 0.135 |
| BNM2 | Straw and brown furrows on straw | 0.098 | Colorless | Short and partly awned | 0.227 |
| BNM3 | Straw | 0 | Colorless | Short and partly awned | 0.135 |
| BNM4 | Straw | 0 | Colorless | Short and partly awned | 0.168 |
| BNM5 | Straw | 0 | Colorless | Short and partly awned | 0.456 |
| BNM6 | Straw and brown furrows on straw | 0.098 | Colorless | Absent | 0 |
| BNM7 | Straw and brown furrows on straw | 0.168 | Colorless | Short and partly awned | 0.325 |
| BNM8 | Straw | 0 | Colorless | Short and partly awned | 0.423 |
| BNM9 | Straw | 0 | Colorless | Short and partly awned | 0.254 |
| EL | Straw | 0 | Colorless | Absent | 0 |
| HS | Brown furrows on straw | 0 | Colorless | Absent | 0 |
| KH | Straw | 0 | Colorless | Absent | 0 |
| NDLP | Brown furrows on straw | 0 | Purple | Absent | 0 |
| NU | Straw | 0 | Colorless | Absent | 0 |
| PLD | Straw | 0 | Colorless | Absent | 0 |
| PS | Straw | 0 | Colorless | Short awned | 0 |
| SKH | Straw | 0 | Colorless | Absent | 0 |
| BH62 | Straw | 0 | Colorless | Absent | 0 |
| BH96 | Straw | 0 | Colorless | Short awned | 0 |
| HB | Straw and brown furrows on straw | 0.991 | Colorless | Short and partly awned | 0.683 |
| HBK | Straw and brown furrows on straw | 0.637 | Colorless | Absent | 0 |
| HNK | Straw and brown furrows on straw | 0.673 | Colorless | Short and partly awned | 0.598 |
| BNM-CMU | Straw | 0 | Colorless | Short and partly awned | 0.135 |
| JPD | Black | 0 | Colorless | Absent | 0 |
| KDML105 | Straw | 0 | Colorless | Absent | 0 |
| PTT1 | Straw | 0 | Colorless | Short and partly awned | 0.199 |
| SPR1 | Straw | 0 | Colorless | Absent | 0 |
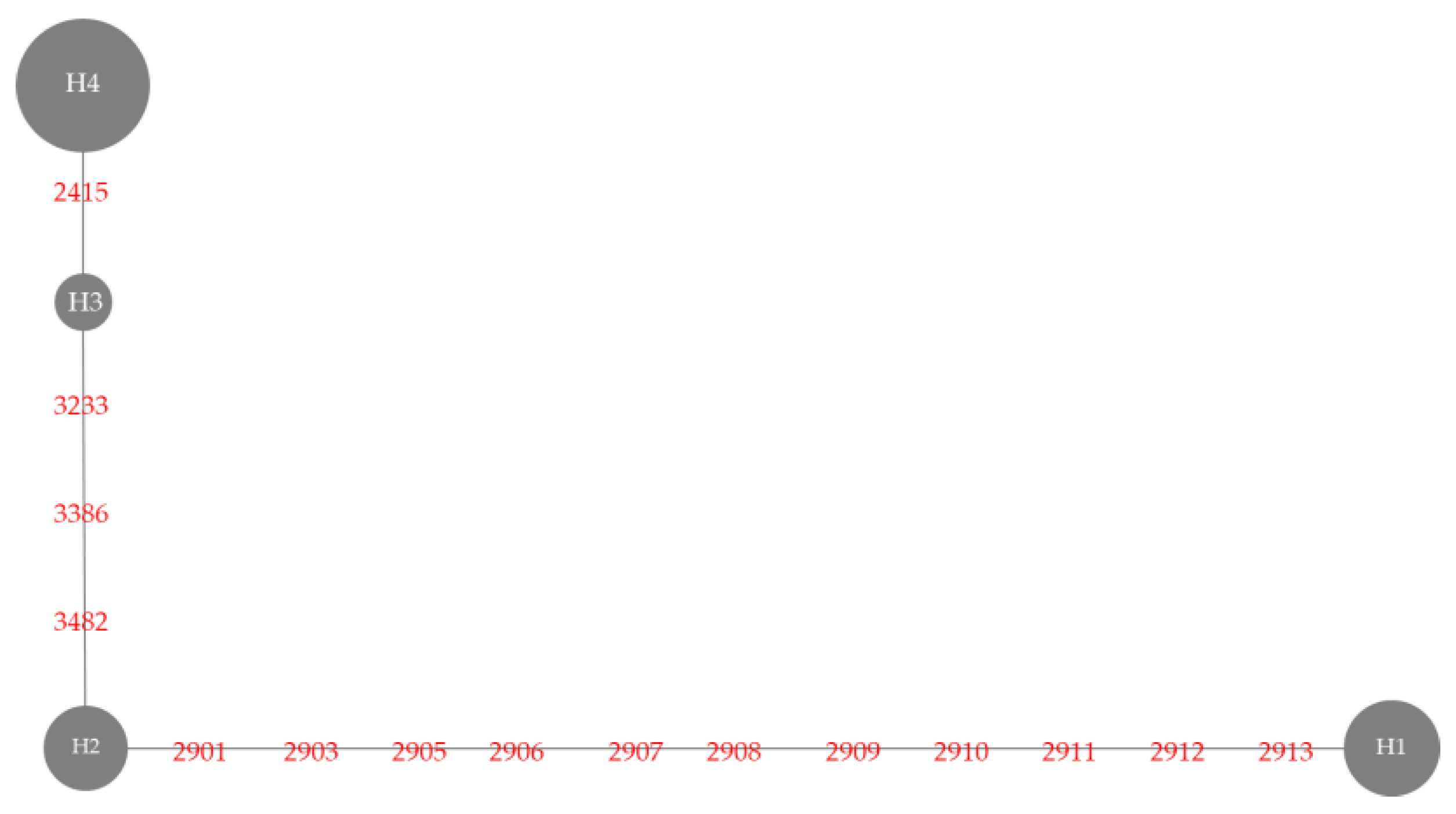
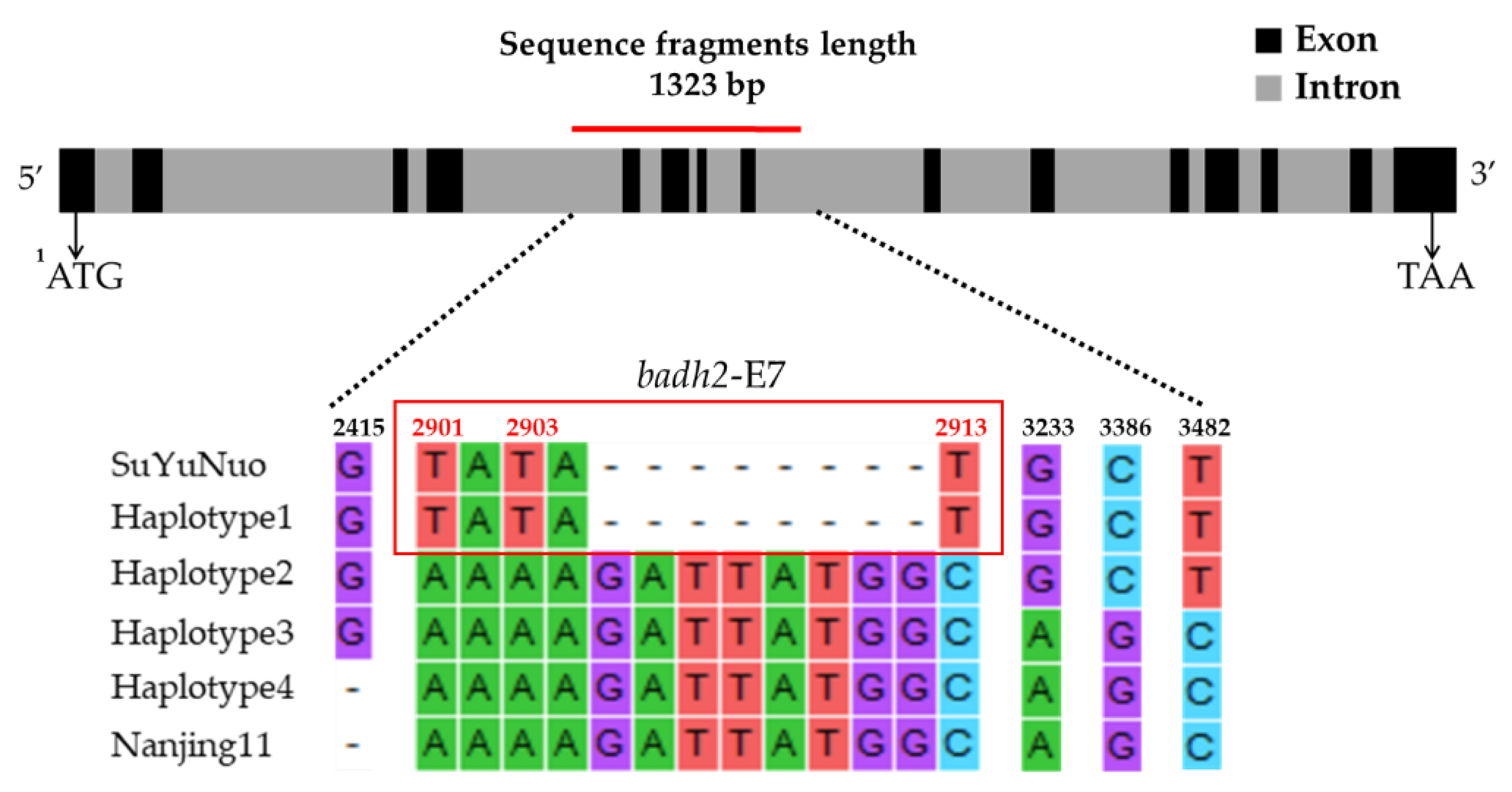
| Haplotype | Variety | Nucleotide Position | Sensory Test # | Gelatinization Temperature | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2415 | 2901 | 2903 | 2905-2912 | 2913 | 3233 | 3386 | 3482 | ||||
| Haplotype 1 | BNM4 | Insertion G | T | T | 8 deletion | T | G | C | T | Moderately aromatic | Medium |
| KH | Insertion G | T | T | 8 deletion | T | G | C | T | Strongly aromatic | High | |
| NDLP | Insertion G | T | T | 8 deletion | T | G | C | T | Strongly aromatic | High | |
| PLD | Insertion G | T | T | 8 deletion | T | G | C | T | Strongly aromatic | High | |
| BNM-CMU | Insertion G | T | T | 8 deletion | T | G | C | T | Moderately aromatic | Medium | |
| KDML105 | Insertion G | T | T | 8 deletion | T | G | C | T | Strongly aromatic | Low | |
| PTT1 | Insertion G | T | T | 8 deletion | T | G | C | T | Moderately aromatic | Low | |
| SuYuNuoδ | insertion G | T | T | 8 deletion | T | G | C | T | ND | ND | |
| Haplotype 2 | BH62 | Insertion G | A | A | No deletion | C | G | C | T | Slightly aromatic | High |
| BH96 | Insertion G | A | A | No deletion | C | G | C | T | Slightly aromatic | High | |
| HS | Insertion G | A | A | No deletion | C | G | C | T | Slightly aromatic | High | |
| JPD | Insertion G | A | A | No deletion | C | G | C | T | Slightly aromatic | Medium | |
| PS | Insertion G | A | A | No deletion | C | G | C | T | Slightly aromatic | High | |
| SKH | Insertion G | A | A | No deletion | C | G | C | T | Moderately aromatic | High | |
| Haplotype 3 | EL | Insertion G | A | A | No deletion | C | A | G | C | Non-aromatic | Low |
| HBK | Insertion G | A | A | No deletion | C | A | G | C | Non-aromatic | High | |
| HNK | Insertion G | A | A | No deletion | C | A | G | C | Non-aromatic | High | |
| NU | Insertion G | A | A | No deletion | C | A | G | C | Non-aromatic | Medium | |
| Haplotype 4 | BNM1 | - | A | A | No deletion | C | A | G | C | Non-aromatic | Low |
| BNM2 | - | A | A | No deletion | C | A | G | C | Non-aromatic | Low | |
| BNM3 | - | A | A | No deletion | C | A | G | C | Non-aromatic | Medium | |
| BNM5 | - | A | A | No deletion | C | A | G | C | Non-aromatic | Medium | |
| BNM6 | - | A | A | No deletion | C | A | G | C | Non-aromatic | Medium | |
| BNM7 | - | A | A | No deletion | C | A | G | C | Non-aromatic | Medium | |
| BNM8 | - | A | A | No deletion | C | A | G | C | Non-aromatic | Medium | |
| BNM9 | - | A | A | No deletion | C | A | G | C | Non-aromatic | Medium | |
| HB | - | A | A | No deletion | C | A | G | C | Non-aromatic | Low | |
| SPR1 | - | A | A | No deletion | C | A | G | C | Non-aromatic | High | |
| Nanjing11 δδ | - | A | A | No deletion | C | A | G | C | ND | ND | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chan-in, P.; Jamjod, S.; Yimyam, N.; Rerkasem, B.; Pusadee, T. Grain Quality and Allelic Variation of the Badh2 Gene in Thai Fragrant Rice Landraces. Agronomy 2020, 10, 779. https://doi.org/10.3390/agronomy10060779
Chan-in P, Jamjod S, Yimyam N, Rerkasem B, Pusadee T. Grain Quality and Allelic Variation of the Badh2 Gene in Thai Fragrant Rice Landraces. Agronomy. 2020; 10(6):779. https://doi.org/10.3390/agronomy10060779
Chicago/Turabian StyleChan-in, Phukjira, Sansanee Jamjod, Narit Yimyam, Benjavan Rerkasem, and Tonapha Pusadee. 2020. "Grain Quality and Allelic Variation of the Badh2 Gene in Thai Fragrant Rice Landraces" Agronomy 10, no. 6: 779. https://doi.org/10.3390/agronomy10060779
APA StyleChan-in, P., Jamjod, S., Yimyam, N., Rerkasem, B., & Pusadee, T. (2020). Grain Quality and Allelic Variation of the Badh2 Gene in Thai Fragrant Rice Landraces. Agronomy, 10(6), 779. https://doi.org/10.3390/agronomy10060779






