Host Specificity of Endophytic Fungi from Stem Tissue of Nature Farming Tomato (Solanum lycopersicum Mill.) in Japan
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Tomato Plant Samples
2.2. Fungal Endophyte Isolation
2.3. Endophyte Identification
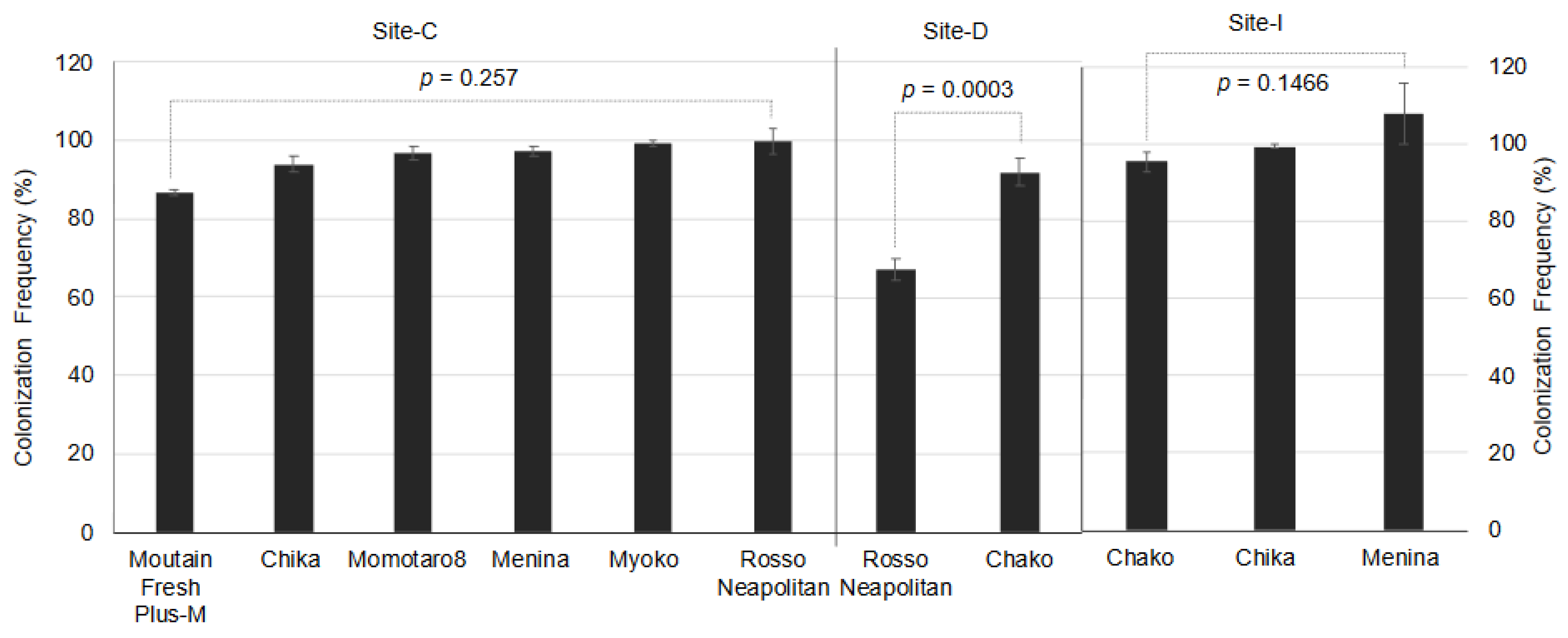
2.4. Colonization and Isolation Frequency
2.5. Diversity and Community Composition
3. Results
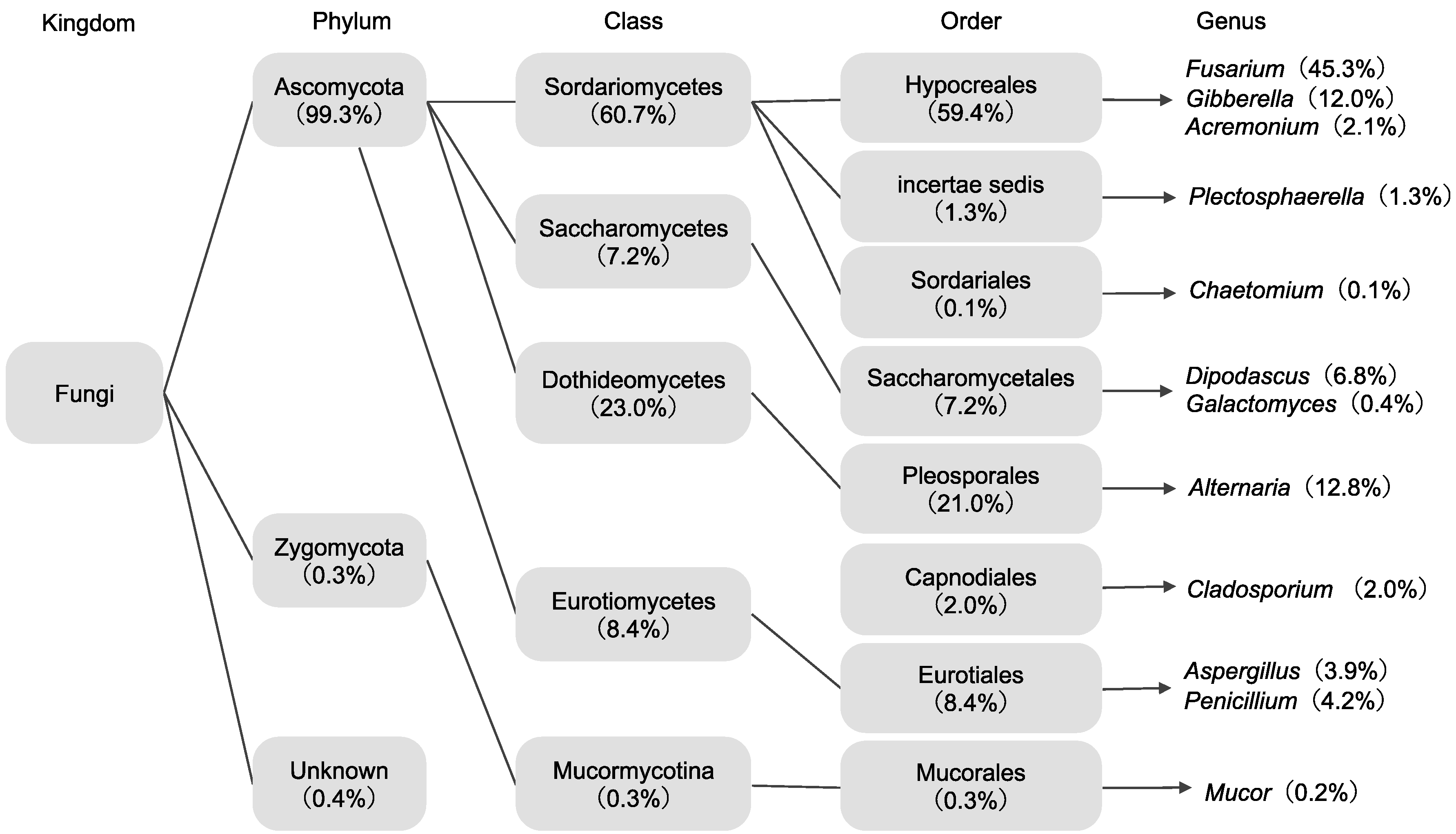
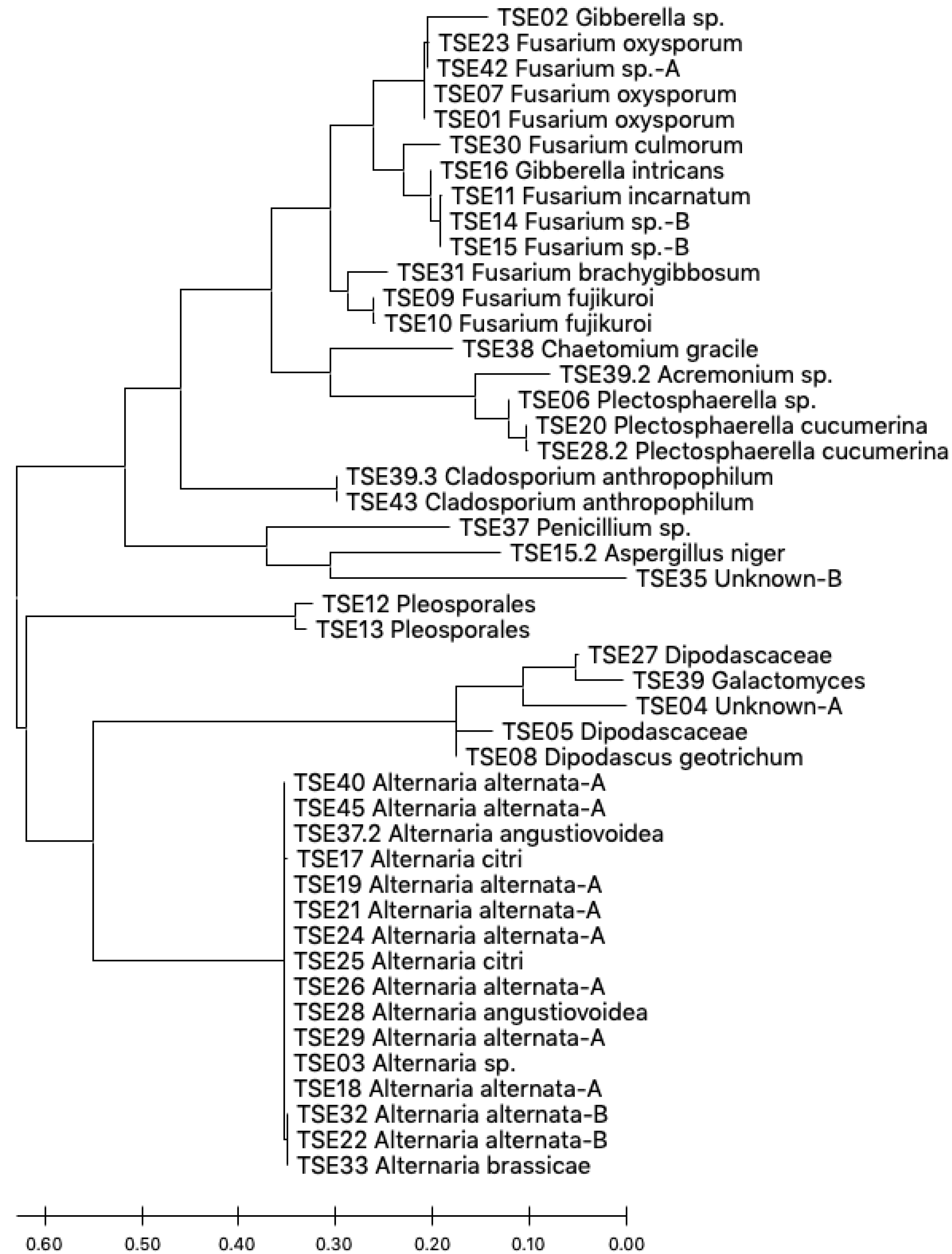
3.1. Isolation and Identification of Endophytes
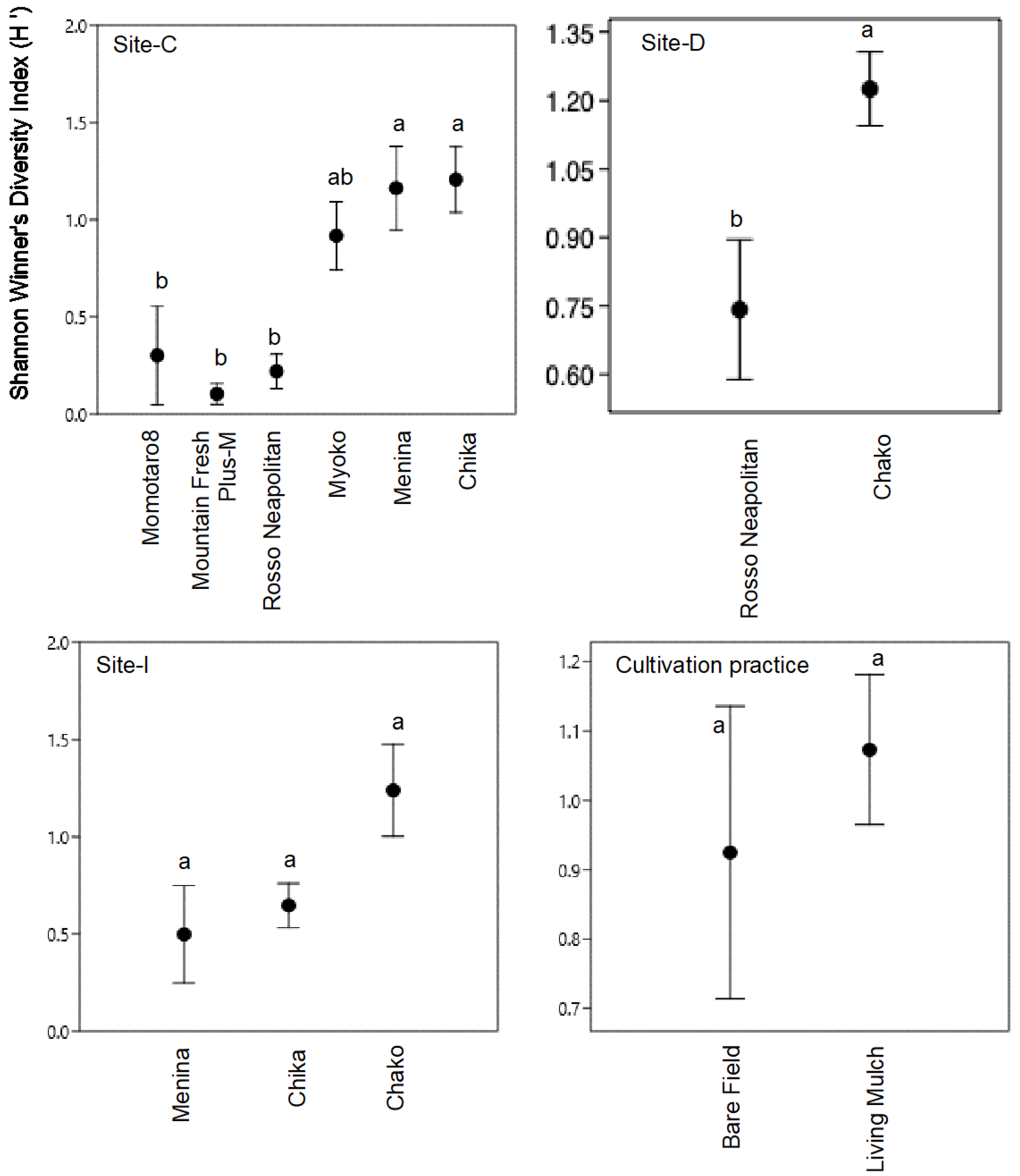
3.2. Diversity and Species Richness of Endophyte Communities (α-Diversity)
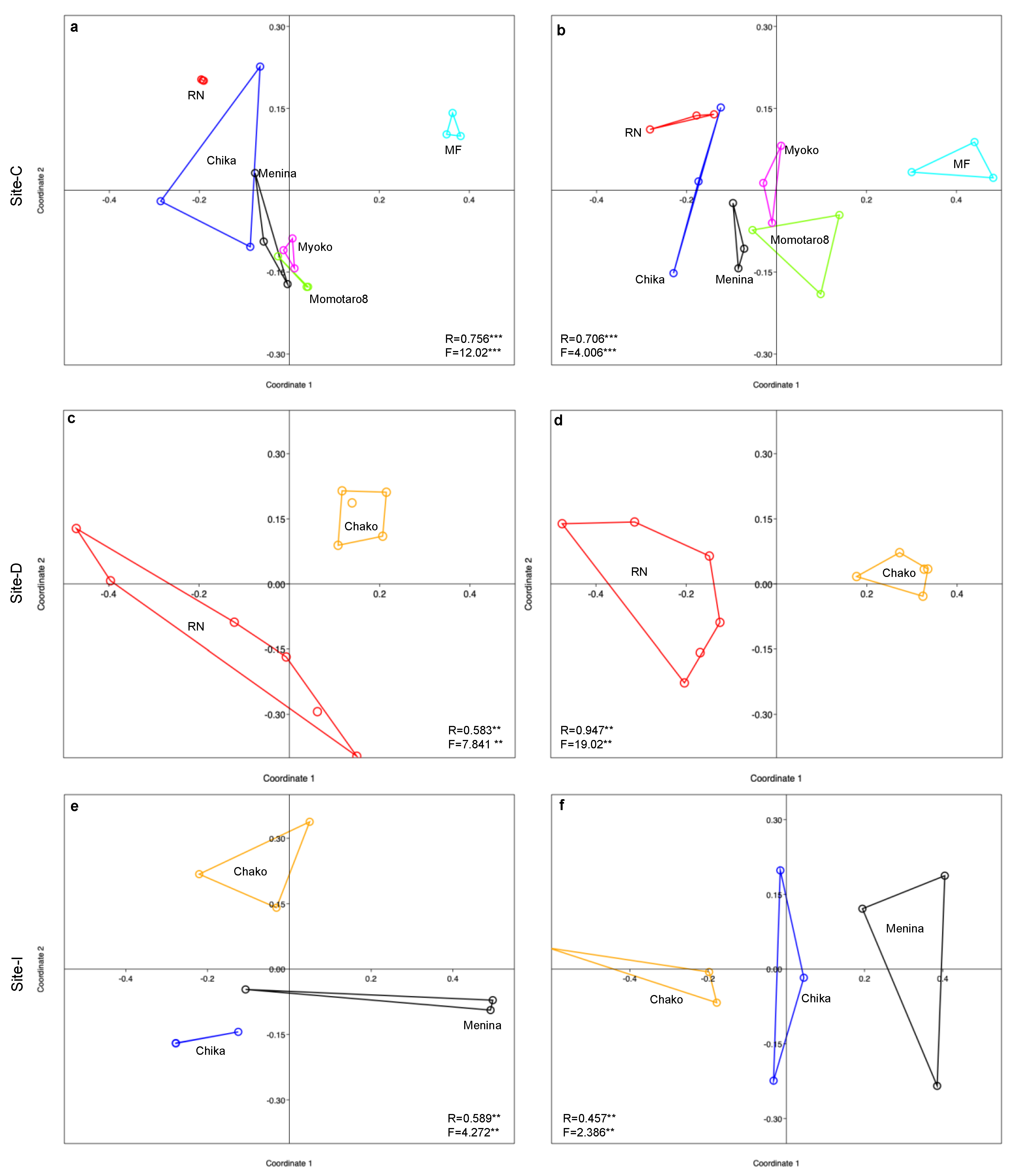
3.3. Variation in Endophyte Communities across Location, Host and Tissue (β-Diversity)
4. Discussion
4.1. Composition of Endophytes
4.2. Diversity and Structuring of the Endophyte Community
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hardoim, P.; Van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gazis, R.; Chaverri, P. Diversity of fungal endophytes in leaves and stems of wild rubber trees (Hevea brasiliensis) in Peru. Fungal Ecol. 2010, 3, 240–254. [Google Scholar] [CrossRef]
- U’Ren, J.M.; Dalling, J.W.; Gallery, R.E.; Maddison, D.R.; Davis, E.C.; Gibson, C.M.; Arnold, A.E. Diversity and evolutionary origins of fungi associated with seeds of a neotropical pioneer tree: A case study for analysing fungal environmental samples. Mycol. Res. 2009, 113, 432–449. [Google Scholar] [CrossRef] [PubMed]
- Arnold, A.E. Understanding the diversity of foliar endophytic fungi: Progress, challenges, and frontiers. Fungal Biol. Rev. 2007, 21, 51–66. [Google Scholar] [CrossRef]
- Dastogeer, K.M.G.; Li, H.; Sivasithamparam, K.; Jones, M.; Wylie, S.J. Host Specificity of Endophytic Mycobiota of Wild Nicotiana Plants from Arid Regions of Northern Australia. Microb. Ecol. 2017, 75, 74–87. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, J.; Wang, G.; Chen, J. Host identity and phylogeny shape the foliar endophytic fungal assemblages of Ficus. Ecol. Evol. 2019, 9, 10472–10482. [Google Scholar] [CrossRef] [Green Version]
- Fang, K.; Miao, Y.-F.; Chen, L.; Zhou, J.; Yang, Z.-P.; Dong, X.-F.; Zhang, H.-B. Tissue-Specific and Geographical Variation in Endophytic Fungi of Ageratina adenophora and Fungal Associations with the Environment. Front. Microbiol. 2019, 10, 2919. [Google Scholar] [CrossRef] [Green Version]
- Petrini, O. Fungal Endophytes of Tree Leaves. In Microbial Ecology of Leaves; Andrews, J.H., Hirano, S.S., Eds.; Springer: New York, NY, USA, 1991; pp. 179–197. [Google Scholar]
- Martínez-Medina, A.; Roldan, A.; Albacete, A.; Pascual, J. The interaction with arbuscular mycorrhizal fungi or Trichoderma harzianum alters the shoot hormonal profile in melon plants. Phytochemestry 2011, 72, 223–229. [Google Scholar] [CrossRef]
- Yadav, A.N.; Verma, P.; Kour, D.; Rana, K.L.; Kumar, V.; Singh, B.; Chauahan, V.S.; Sugitha, T.; Saxena, A.K.; Dhaliwal, H.S. Plant microbiomes and its beneficial multifunctional plant growth promoting attributes. Int. J. Environ. Sci. Nat. Resour. 2017, 3, 1–8. [Google Scholar] [CrossRef]
- Rodriguez, R.J.; Redman, R.S.; Henson, J.M. The Role of Fungal Symbioses in the Adaptation of Plants to High Stress Environments. Mitig. Adapt. Strat. Glob. Chang. 2004, 9, 261–272. [Google Scholar] [CrossRef]
- Azad, K.; Kaminskyj, S. A fungal endophyte strategy for mitigating the effect of salt and drought stress on plant growth. Symbiosis 2016, 68, 73–78. [Google Scholar] [CrossRef]
- Cui, J.-L.; Wang, Y.-N.; Jiao, J.; Gong, Y.; Wang, J.-H.; Wang, M. Fungal endophyte-induced salidroside and tyrosol biosynthesis combined with signal cross-talk and the mechanism of enzyme gene expression in Rhodiola crenulata. Sci. Rep. 2017, 7, 12540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dastogeer, K.M.G.; Li, H.; Sivasithamparam, K.; Jones, M.; Du, X.; Ren, Y.; Wylie, S.J. Metabolic responses of endophytic Nicotiana benthamiana plants experiencing water stress. Environ. Exp. Bot. 2017, 143, 59–71. [Google Scholar] [CrossRef]
- Dastogeer, K.M.G.; Li, H.; Sivasithamparam, K.; Jones, M.; Wylie, S.J. Fungal endophytes and a virus confer drought tolerance to Nicotiana benthamiana plants through modulating osmolytes, antioxidant enzymes and expression of host drought responsive genes. Environ. Exp. Bot. 2018, 149, 95–108. [Google Scholar] [CrossRef] [Green Version]
- Nair, D.N.; Padmavathy, S. Impact of endophytic microorganisms on plants, environment and humans. Sci. World J. 2014, 2014, 250693. [Google Scholar] [CrossRef] [Green Version]
- Higginbotham, S.J.; Arnold, A.E.; Ibáñez, A.; Spadafora, C.; Coley, P.D.; Kursar, T.A. Bioactivity of Fungal Endophytes as a Function of Endophyte Taxonomy and the Taxonomy and Distribution of Their Host Plants. PLoS ONE 2013, 8, e73192. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, D.C.; Battista, L.J.; Arnold, A.E. Fungal endophytes of aquatic macrophytes: Diverse host-generalists characterized by tissue preferences and geographic structure. Microb. Ecol. 2014, 67, 735–747. [Google Scholar] [CrossRef]
- Kremen, C.; Miles, A. Ecosystem Services in Biologically Diversified versus Conventional Farming Systems: Benefits, Externalities, and Trade-Offs. Ecol. Soc. 2012, 17, 40. [Google Scholar] [CrossRef]
- Shaikh, N.F.; Gachande, B.D. Effect of organic bio-booster and inorganic inputs on rhizosphere mycoflora population and species diversity of wheat. Int. J. Sci. Res. 2015, 4, 295–302. [Google Scholar]
- Xia, Y.; Sahib, M.R.; Amna, A.; Opiyo, S.O.; Zhao, Z.; Gao, Y.G. Culturable endophytic fungal communities associated with plants in organic and conventional farming systems and their effects on plant growth. Sci. Rep. 2019, 9, 1669. [Google Scholar] [CrossRef]
- Hartman, K.; Van Der Heijden, M.G.; Wittwer, R.A.; Banerjee, S.; Walser, J.-C.; Schlaeppi, K. Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome 2018, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Lupatini, M.; Korthals, G.W.; De Hollander, M.; Janssens, T.K.S.; Kuramae, E.E. Soil Microbiome Is More Heterogeneous in Organic Than in Conventional Farming System. Front. Microbiol. 2017, 7, 2064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okada, M. The Basis of Paradise–Kyusei Nature Farming. Atami (Japan); Sekai Kyusei Kyo Press: Atami, Japan, 1993; pp. 331–393. [Google Scholar]
- Okada, M. Health and the New Civilization; Church of World Messianity: Los Angeles, CA, USA, 1991; p. 84. [Google Scholar]
- Xu, H.-L. Nature Farming. Res. Signpost 2006, 37, 661. [Google Scholar] [CrossRef]
- Hsieh, S.C. Concept and Practice of Natural Farming in the Subtropics. In Proceedings of the Sustainable Farming: New Technology for Survival, Dusit Resort, Pattaya, Bangkok, Thailand, 11–14 May 1993. [Google Scholar]
- Hemenway, T. Gaia’s Garden: A Guide to Home-Scale Permaculture; Chelsea Green Publishing: White River Junction, VT, USA, 2009. [Google Scholar]
- Mollison, B. Permaculture: A Designer’s Manual; Tagari Publications: Sisters Creek, Australia, 1988. [Google Scholar]
- Turner, N.; Rateaver, B. Fertility Farming; Faber and Faber Ltd.: London, UK, 1951. [Google Scholar]
- Khadse, A.; Rosset, P. Zero Budget Natural Farming in India—From inception to institutionalization. Agroecol. Sustain. Food Syst. 2019, 43, 848–871. [Google Scholar] [CrossRef]
- Naachimuthu, K.P. Sustainable Agriculture—The Indian Way. J. Rural Ind. Dev. 2015, 3, 25–32. [Google Scholar]
- Fukuoka, M. The Natural Way of Farming: The Theory and Practice of Green Philosophy; Bookventure: Ishpeming, MI, USA, 1985. [Google Scholar]
- Derpsch, R.; Friedrich, T.; Kassam, A.; Li, H. Current status of adoption of no-till farming in the world and some of its main benefits. Int. J. Agric. Biol. Eng. 2010, 3, 1–25. [Google Scholar]
- Xu, H.-L. Nature Farming: History, principles and perspectives. J. Crop Prod. 2001, 3, 1–10. [Google Scholar] [CrossRef]
- Van Quyen, N.; Sharma, S. Relative effect of organic and conventional farming on growth, yield and grain quality of scented rice and soil fertility 1: Relative wirkung von organischem und konventionellem ackerbau auf wachstum, ertrag und kornqualität von reis und auf die bodenqualität. Arch. Agron. Soil Sci. 2003, 49, 623–629. [Google Scholar]
- Xia, Y.; DeBolt, S.; Dreyer, J.; Scott, D.; Williams, M.A. Characterization of culturable bacterial endophytes and their capacity to promote plant growth from plants grown using organic or conventional practices. Front. Plant Sci. 2015, 6, 490. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Monaim, M.F.; Abdel-Gaid, M.A.; Zayan, S.A.; Nassef, D.M. Enhancement of Growth Parameters and Yield Components in Eggplant using Antagonism of Trichoderma spp. Against Fusarium Wilt Disease. Int. J. Phytopathol. 2014, 3, 33–40. [Google Scholar] [CrossRef] [Green Version]
- Berg, G.; Grube, M.; Schloter, M.; Smalla, K. The plant microbiome and its importance for plant and human health. Front. Microbiol. 2014, 5, 491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulz, B.; Wanke, U.; Draeger, S.; Aust, H.-J. Endophytes from herbaceous plants and shrubs: Effectiveness of surface sterilization methods. Mycol. Res. 1993, 97, 1447–1450. [Google Scholar] [CrossRef]
- Strobel, G.A.; Hess, W.M.; Ford, E.; Sidhu, R.S.; Yang, X. Taxol from fungal endophytes and the issue of biodiversity. J. Ind. Microbiol. Biotechnol. 1996, 17, 417–423. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- Abarenkov, K.; Nilsson, R.H.; Larsson, K.-H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; Pennanen, T.; et al. The UNITE Database for Molecular Identification of Fungi—Recent Updates and Future Perspectives. New Phytol. 2010, 186, 281–285. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Quinn, G.P.; Keough, M.J. Multidimensional Scaling and Cluster Analysis. In Experimental Design and Data Analysis for Biologists; Quinn, G.P., Keough, M.J., Eds.; Cambridge University Press: Cambridge, UK, 2002; pp. 473–493. [Google Scholar]
- Wolda, H. Similarity indices, sample size and diversity. Oecologia 1981, 50, 296–302. [Google Scholar] [CrossRef]
- Gauch, H.G. Multivariate Analysis in Community Ecology; Cambridge University Press: Cambridge, UK, 1982; p. 312. [Google Scholar]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. Past: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Hibbett, D.S.; Binder, M.; Bischoff, J.F.; Blackwell, M.; Cannon, P.F.; Eriksson, O.E.; Huhndorf, S.; James, T.; Kirk, P.M.; Lücking, R.; et al. A higher-level phylogenetic classification of the Fungi. Mycol. Res. 2007, 111, 509–547. [Google Scholar] [CrossRef]
- Aamir, M.; Rai, K.K.; Zehra, A.; Kumar, S.; Yadav, M.; Shukla, V.; Upadhyay, R.S. Fungal endophytes: Classification, diversity, ecological role, and their relevance in sustainable agriculture. In Microbial Endophytes; Kumar, A., Singh, V.K., Eds.; Woodhead Publishing: Cambridge, MA, USA, 2020; pp. 291–323. [Google Scholar]
- Dastogeer, K.M.G.; Wylie, S.J. Plant–Fungi Association: Role of Fungal Endophytes in Improving Plant Tolerance to Water Stress. In Plant-Microbe Interactions in Agro-Ecological Perspectives; Singh, D.P., Singh, H.B., Prabha, R., Eds.; Springer Nature Singapore Pte Ltd.: Singapore, 2017; pp. 143–159. [Google Scholar]
- Rodriguez, R.J.; White, J.F., Jr.; Arnald, A.E.; Redman, R.S. Fungal endophytes: Diversity and functional roles. New Phytol. 2009, 182, 314–330. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Rani, V.; Saini, R.; Verma, M.L. Bioprospecting and Biotechnological Applications of Microbial Endophytes. In Microbial Technology for Health and Environment; Arora, P.K., Ed.; Springer: Singapore, 2020; pp. 191–228. [Google Scholar]
- Hyde, K.D.; Soytong, K. The Fungal Endophyte Dilemma. Fungal Divers. 2008, 33, 163–173. [Google Scholar]
- Ownley, B.H.; Griffin, M.R.; Klingeman, W.E.; Gwinn, K.D.; Moulton, J.K.; Pereira, R.M. Beauveria bassiana: Endophytic colonization and plant disease control. J. Invertebr. Pathol. 2008, 98, 267–270. [Google Scholar] [CrossRef] [PubMed]
- Arnold, A.E.; Lutzoni, F. Diversity and host range of foliar fungal endophytes: Are tropical leaves biodiversity hotspots? Ecology 2007, 88, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Pinruan, U.; Rungjindamai, N.; Choeyklin, R.; Lumyong, S.; Hyde, K.D.; Jones, E.B.G. Occurrence and diversity of basidiomycetous endophytes from the oil palm, Elaeis guineensis in Thailand. Fungal Divers. 2010, 41, 71–88. [Google Scholar] [CrossRef]
- Márquez, S.S.; Bills, G.F.; Acuña, L.D.; Zabalgogeazcoa, I. Endophytic mycobiota of leaves and roots of the grass Holcus lanatus. Fungal Divers. 2010, 41, 115–123. [Google Scholar] [CrossRef]
- Cai, L.; Hyde, K.D.; Taylor, P.W.J.; Weir, B.S.; Waller, J.; Abang, M.M.; Zhang, J.Z.; Yang, Y.L.; Phoulivong, S.; Liu, Z.Y.; et al. A polyphasic approach for studying Colletotrichum. Fungal Divers. 2009, 39, 183–204. [Google Scholar]
- Blaalid, R.; Kumar, S.; Nilsson, R.H.; Abarenkov, K.; Kirk, P.M.; Kauserud, H.; Nilsson, R.H. ITS1 versus ITS2 as DNA metabarcodes for fungi. Mol. Ecol. Resour. 2013, 13, 218–224. [Google Scholar] [CrossRef] [Green Version]
- De Beeck, M.O.; Lievens, B.; Busschaert, P.; Declerck, S.; Vangronsveld, J.; Colpaert, J.V. Comparison and Validation of Some ITS Primer Pairs Useful for Fungal Metabarcoding Studies. PLoS ONE 2014, 9, e97629. [Google Scholar] [CrossRef] [Green Version]
- Dastogeer, K.M.G.; Li, H.; Sivasithamparam, K.; Jones, M.; Wylie, S.J. A simple and rapid in vitro test for large-scale screening of fungal endophytes from drought-adapted Australian wild plants for conferring water deprivation tolerance and growth promotion in Nicotiana benthamiana seedlings. Arch. Microbiol. 2017, 199, 1357–1370. [Google Scholar] [CrossRef]
- Slippers, B.; Wingfield, M. Botryosphaeriaceae as endophytes and latent pathogens of woody plants: Diversity, ecology and impact. Fungal Biol. Rev. 2007, 21, 90–106. [Google Scholar] [CrossRef]
- Photita, W.; Lumyong, S.; Lumyong, P.; McKenzie, E.H.C.; Hyde, K.D. Are some endophytes of Musa acuminata latent pathogens? Fungal Divers. 2004, 16, 131–140. [Google Scholar]
- Saikkonen, K.; Faeth, S.H.; Helander, M.; Sullivan, T.J. Fungal endophytes: A Continuum of Interactions with Host Plants. Annu. Rev. Ecol. Syst. 1998, 29, 319–343. [Google Scholar] [CrossRef]
- Stone, J.; Petrini, O. Endophytes of forest trees: A model for fungus-plant interactions. In Plant Relationships Part B; Springer: Berlin/Heidelberg, Germany, 1997; pp. 129–140. [Google Scholar]
- Murphy, B.R.; Nieto, L.M.; Doohan, F.M.; Hodkinson, T.R. Profundae diversitas: The uncharted genetic diversity in a newly studied group of fungal root endophytes. Mycology 2015, 6, 139–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Costa Stuart, A.K.; Stuart, R.M.; Pimentel, I.C. Effect of agrochemicals on endophytic fungi community associated with crops of organic and conventional soybean (Glycine max L. Merril). Agric. Nat. Resour. 2018, 52, 388–392. [Google Scholar] [CrossRef]
- Radic, T.; Likar, M.; Hančević, K.; Bogdanović, I.; Pasković, I. Occurrence of root endophytic fungi in organic versus conventional vineyards on the Croatian coast. Agric. Ecosyst. Environ. 2014, 192, 115–121. [Google Scholar] [CrossRef]
- Gond, S.K.; Verma, V.C.; Kumar, A.; Kumar, V.; Kharwar, R.N. Study of endophytic fungal community from different parts of Aegle marmelos Correae (Rutaceae) from Varanasi (India). World J. Microbiol. Biotechnol. 2007, 23, 1371–1375. [Google Scholar] [CrossRef]
- Bae, H.; Roberts, D.P.; Lim, H.-S.; Strem, M.D.; Park, S.-C.; Ryu, C.-M.; Melnick, R.; Bailey, B.A. EndophyticTrichodermaIsolates from Tropical Environments Delay Disease Onset and Induce Resistance AgainstPhytophthora capsiciin Hot Pepper Using Multiple Mechanisms. Mol. Plant-Microbe Interact. 2011, 24, 336–351. [Google Scholar] [CrossRef] [Green Version]
- Cannon, P.F.; Simmons, C.M. Diversity and host preference of leaf endophytic fungi in the Iwokrama Forest Reserve, Guyana. Mycology 2002, 94, 210–220. [Google Scholar] [CrossRef]
- Harman, G. Multifunctional fungal plant symbionts: New tools to enhance plant growth and productivity. New Phytol. 2011, 189, 647–649. [Google Scholar] [CrossRef]
- Paparu, P.; Dubois, T.; Coyne, D.; Viljoen, A. Dual inoculation of Fusarium oxysporumendophytes in banana: Effect on plant colonization, growth and control of the root burrowing nematode and the banana weevil. Biocontrol Sci. Technol. 2009, 19, 639–655. [Google Scholar] [CrossRef]
- Larran, S.; Monaco, C.; Alippi, H. Endophytic fungi in leaves of Lycopersicon esculentum Mill. World J. Microbiol. Biotechnol. 2001, 17, 181–184. [Google Scholar] [CrossRef]
- Küngas, K.; Bahram, M.; Põldmaa, K. Host tree organ is the primary driver of endophytic fungal community structure in a hemiboreal forest. FEMS Microbiol. Ecol. 2019, 96. [Google Scholar] [CrossRef] [PubMed]
- Christian, N.; Sedio, B.E.; Florez-Buitrago, X.; Ramírez-Camejo, L.A.; Rojas, E.I.; Mejía, L.C.; Palmedo, S.; Rose, A.; Schroeder, J.W.; Herre, E.A. Host affinity of endophytic fungi and the potential for reciprocal interactions involving host secondary chemistry. Am. J. Bot. 2020, 107, 219–228. [Google Scholar] [CrossRef] [Green Version]
- Murphy, B.R.; Hodkinson, T.R. Endophyte Ecology, Diversity and Utilisation; Taylor & Francis: Abingdon, UK, 2018. [Google Scholar]
- Colwell, R.K.; Chao, A.; Gotelli, N.J.; Lin, S.-Y.; Mao, C.X.; Chazdon, R.; Longino, J.T. Models and estimators linking individual-based and sample-based rarefaction, extrapolation and comparison of assemblages. J. Plant Ecol. 2012, 5, 3–21. [Google Scholar] [CrossRef] [Green Version]
- U’Ren, J.M.; Lutzoni, F.; Miadlikowska, J.; Laetsch, A.D.; Arnold, A.E. Host and geographic structure of endophytic and endolichenic fungi at a continental scale. Am. J. Bot. 2012, 99, 898–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, M.K.; Arnold, A.E.; Johnson, N.C. Factors influencing communities of foliar fungal endophytes in riparian woody plants. Fungal Ecol. 2013, 6, 365–378. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, Q.; Lu, X.; Okane, I.; Kakishima, M. Endophytic fungal community in stems and leaves of plants from desert areas in China. Mycol. Prog. 2012, 11, 781–790. [Google Scholar] [CrossRef]
- Kumar, D.S.S.; Hyde, K.D. Biodiversity and tissue-recurrence of endophytic fungi in Tripterygium wilfordii. Fungal Divers. 2004, 17, 69–90. [Google Scholar]
- Magurran, A.E. Measuring Biological Diversity; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Higgins, K.L.; Coley, P.D.; Kursar, T.A.; Arnold, A.E. Culturing and direct PCR suggest prevalent host generalism among diverse fungal endophytes of tropical forest grasses. Mycology 2011, 103, 247–260. [Google Scholar] [CrossRef] [Green Version]
- Zimmerman, N.B.; Vitousek, P.M. Fungal endophyte communities reflect environmental structuring across a Hawaiian landscape. Proc. Natl. Acad. Sci. USA 2012, 109, 13022–13027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben Chobba, I.; Elleuch, A.; Ayadi, I.; Khannous, L.; Namsi, A.; Cerqueira, F.; Drira, N.; Gharsallah, N.; Vallaeys, T. Fungal diversity in adult date palm (Phoenix dactylifera L.) revealed by culture-dependent and culture-independent approaches. J. Zhejiang Univ. Sci. B 2013, 14, 1084–1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Site | Cultivation Practice | Cultivar |
|---|---|---|
| Site-C | Living Mulch (Grass Cultivation) | Chika, Menina, Momotaro8, Mountain Fresh Plus-M, Myoko, Rosso Neapolitan |
| Site-D | Living Mulch (Grass Cultivation) | Chako, Rosso Neapolitan |
| Site-D | Bare Field | Chako, Rosso Neapolitan |
| Site-I | Green Manuring | Chako, Chika, Menina |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammad Golam Dastogeer, K.; Oshita, Y.; Yasuda, M.; Kanasugi, M.; Matsuura, E.; Xu, Q.; Okazaki, S. Host Specificity of Endophytic Fungi from Stem Tissue of Nature Farming Tomato (Solanum lycopersicum Mill.) in Japan. Agronomy 2020, 10, 1019. https://doi.org/10.3390/agronomy10071019
Mohammad Golam Dastogeer K, Oshita Y, Yasuda M, Kanasugi M, Matsuura E, Xu Q, Okazaki S. Host Specificity of Endophytic Fungi from Stem Tissue of Nature Farming Tomato (Solanum lycopersicum Mill.) in Japan. Agronomy. 2020; 10(7):1019. https://doi.org/10.3390/agronomy10071019
Chicago/Turabian StyleMohammad Golam Dastogeer, Khondoker, Yutaro Oshita, Michiko Yasuda, Makoto Kanasugi, Eri Matsuura, Qicong Xu, and Shin Okazaki. 2020. "Host Specificity of Endophytic Fungi from Stem Tissue of Nature Farming Tomato (Solanum lycopersicum Mill.) in Japan" Agronomy 10, no. 7: 1019. https://doi.org/10.3390/agronomy10071019
APA StyleMohammad Golam Dastogeer, K., Oshita, Y., Yasuda, M., Kanasugi, M., Matsuura, E., Xu, Q., & Okazaki, S. (2020). Host Specificity of Endophytic Fungi from Stem Tissue of Nature Farming Tomato (Solanum lycopersicum Mill.) in Japan. Agronomy, 10(7), 1019. https://doi.org/10.3390/agronomy10071019






