Waxy Gene-Orthologs in Wheat × Thinopyrum Amphidiploids
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. DNA Isolation, Wx Cloning and Sequencing
2.3. Sequence Data Analysis
2.4. Amylose Content Analysis
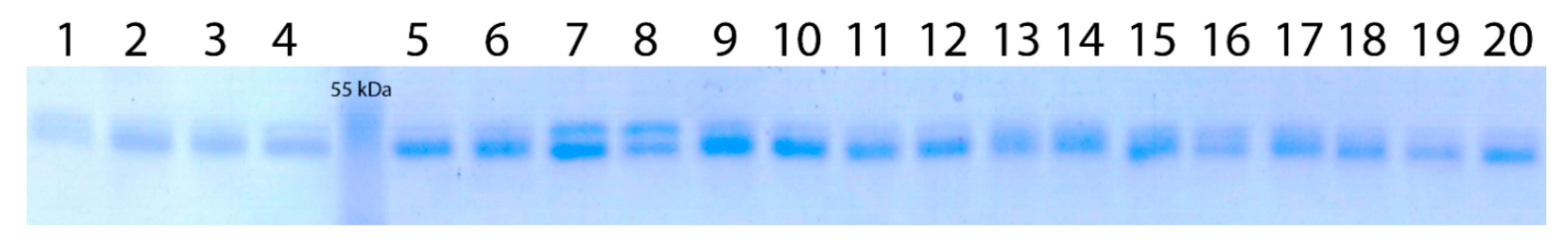
2.5. Protein Electrophoresis
3. Results
3.1. Analysis of Amylose Content in Starch in Wheat–Wheatgrass Hybrids
3.2. Analysis of Wx-Th Gene Expression Products in WWGH Accessions
3.3. Development and Verification of the PCR-Marker for Wx-Th
3.4. Study of Allele Polymorphism for the Wx-Th Gene in Partial Wheat–Wheatgrass Hybrids
3.5. Sequencing and Characterization of the Nucleotide Sequence of the Wx Gene in Wild Relatives of Wheat
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kim, W.; Johnson, J.W.; Graybosch, R.A.; Gaines, C.S. Physicochemical properties and end-use quality of wheat starch as a function of waxy protein alleles. J. Cereal Sci. 2003, 37, 195–204. [Google Scholar] [CrossRef] [Green Version]
- James, M.G.; Denyer, K.; Myers, A.M. Starch synthesis in the cereal endosperm. Curr. Opin. Plant Biol. 2003, 6, 215–222. [Google Scholar] [CrossRef]
- Nakamura, T.; Yamamori, M.; Hirano, H.; Hidaka, S. Identification of three Wx proteins in wheat (Triticum aestivum L.). Biochem. Genet. 1993, 31, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Urbano, M.; Margiotta, B.; Colaprico, G.; Lafiandra, D. Waxy proteins in diploid, tetraploid and hexaploid wheats. Plant Breed. 2002, 121, 465–469. [Google Scholar] [CrossRef]
- Caballero, L.; Bancel, E.; Debiton, C.; Branlard, G. Granule bound starch synthase (GBSS) diversity of ancient wheat and related species. Plant Breed. 2008, 127, 548–553. [Google Scholar] [CrossRef]
- Yamamori, M.; Yamamoto, K. Effects of two novel Wx-A1 alleles of common wheat (Triticum aestivum L.) on amylose and starch properties. J. Cereal Sci. 2011, 54, 229–235. [Google Scholar] [CrossRef]
- Rodríguez-Quijano, M.; Nieto-Taladriz, M.T.; Carrillo, J.M. Polymorphism of waxy proteins in Iberian hexaploid wheats. Plant Breed. 1998, 117, 341–344. [Google Scholar] [CrossRef]
- Guzmán, C.; Caballero, L.; Moral, A.; Alvarez, J. Genetic variation for waxy proteins and amylose content in Spanish spelt wheat (Triticum spelta L.). Genet. Resour. Crop Evol. 2010, 57, 721–725. [Google Scholar]
- McIntosh, R.A.; Yamazaki, Y.; Dubcovsky, J.; Rogers, W.J.; Morris, G.; Appels, R.; Xia, X.C. Catalogue of Gene Symbols for Wheat. 2013. Available online: http://www.shigen.nig.ac.jp/wheat/komugi/genes/macgene/2013/GeneSymbol.pdf (accessed on 30 April 2020).
- Guzmán, C.; Alvarez, J.B. Wheat waxy proteins: Polymorphism, molecular characterization and effects on starch properties. Theor. Appl. Genet. 2016, 129, 1–16. [Google Scholar] [CrossRef]
- Zi, Y.; Ding, J.; Song, J.; Humphreys, G.; Peng, Y.; Li, C.; Zhu, X.; Guo, W. Grain yield, starch content and activities of key enzymes of waxy and non-waxy wheat (Triticum aestivum L.). Sci. Rep. 2018, 8, 4548. [Google Scholar] [CrossRef]
- Nakamura, T.; Vrinten, P.; Saito, M.; Konda, M. Rapid classification of partial waxy wheats using PCR-based markers. Genome 2002, 45, 1150–1156. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Konda, M.; Vrinten, P.; Nakamura, K.; Nakamura, T. Molecular comparison of Waxy null alleles in common wheat and identification of a unique null allele. Theor. Appl. Genet. 2004, 108, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
- Guzmán, C.; Caballero, L.; Yamamori, M.; Alvarez, J. Molecular Characterization of a new Waxy allele with partial expression in spelt wheat. Planta 2012, 235, 1331–1339. [Google Scholar] [CrossRef] [PubMed]
- Nieto-Taladriz, M.T.; Rodriguez-Quijano, M.; Carrillo, J.M. Polymorphism of waxy proteins in Spanish durum wheats. Plant Breed. 2000, 119, 277–279. [Google Scholar] [CrossRef]
- Shariflou, M.R.; Sharp, P.J. A polymorphic microsatellite in the 3 ‘end of ‘waxy’ genes of wheat, Triticum aestivum. Plant Breed. 1999, 118, 275–277. [Google Scholar] [CrossRef]
- Ortega, R.; Guzmán, C.; Alvarez, J. Molecular characterization of several Wx alleles in durum wheat. Biol. Plant 2015, 59, 220–226. [Google Scholar] [CrossRef]
- Li, S.; Zhong, X.; Zhang, X.; Rahman, M.; Lan, J.; Tang, H.; Qi, P.; Ma, J.; Wang, J.; Chen, G.; et al. Production of waxy tetraploid wheat (Triticum turgidum durum L.) by EMS mutagenesis. Genet. Resour. Crop Evol. 2020, 67, 433–443. [Google Scholar] [CrossRef]
- Lan, J.; Li, Y.; Xu, K.; Zhang, X.; Tang, H.; Qi, P.; Ma, J.; Wang, J.; Chen, G.; Pu, Z.; et al. EMS induced SNP changes led to mutation of Wx protein in common wheat. Cereal Res. Commun. 2020, 48, 233–238. [Google Scholar] [CrossRef]
- Divashuk, M.; Klimushina, M.; Karlov, G. Molecular genetic characteristics of the Wx-B1e allele from common wheat and applicability of the DNA markers for its identification. Russ. J. Genet. 2011, 47, 1428–1432. [Google Scholar] [CrossRef]
- Yan, L.; Bhave, M. Characterization of waxy proteins and waxy genes of Triticum timopheevii and T. zhukovskyi and implications for evolution of wheat. Genome 2001, 44, 582–588. [Google Scholar] [CrossRef]
- Guzmán, C.; Ortega, R.; Yamamori, M.; Peña, R.; Alvarez, J. Molecular characterization of two novel null waxy alleles in Mexican bread wheat landraces. J. Cereal Sci. 2015, 62, 8–14. [Google Scholar] [CrossRef]
- Guzmán, C.; Caballero, L.; Alvarez, J. Molecular characterisation of the Wx-B1 allelic variants identified in cultivated emmer wheat and comparison with those of durum wheat. Mol. Breed. 2010, 28, 403–411. [Google Scholar] [CrossRef] [Green Version]
- Guzmán, C.; Caballero, L.; Gutierrez, M.; Alvarez, J. Polymorphism of waxy proteins in Spanish hulled wheats. Plant Genet. Resour. 2011, 9, 330–333. [Google Scholar] [CrossRef] [Green Version]
- Guzmán, C.; Caballero, L.; Alvarez, J.; Yamamori, M. Amylose content and starch properties in emmer and durum wheat lines with different waxy proteins composition. J. Sci. Food Agric. 2011, 91, 1625–1629. [Google Scholar] [CrossRef]
- Guzmán, C.; Alvarez, J. Molecular characterization of a novel waxy allele (Wx-A u 1a) from Triticum urartu Thum. ex Gandil. Genet. Resour. Crop Evol. 2012, 59, 971–979. [Google Scholar] [CrossRef]
- Guzmán, C.; Caballero, L.; Martín, L.; Alvarez, J. Waxy genes from spelt wheat: New alleles for modern wheat breeding and new phylogenetic inferences about the origin of this species. Ann. Bot. 2012, 110, 1161–1171. [Google Scholar] [CrossRef] [Green Version]
- Ortega, R.; Alvarez, J.; Guzmán, C. Characterization of the Wx gene in diploid Aegilops species and its potential use in wheat breeding. Genet. Resour. Crop Evol. 2013, 61, 369–382. [Google Scholar] [CrossRef]
- Ortega, R.; Guzmán, C.; Alvarez, J. Wx gene in diploid wheat: Molecular characterization of five novel alleles from einkorn (Triticum monococcum L. ssp. monococcum) and T. urartu. Mol. Breed. 2014, 34, 1137–1146. [Google Scholar] [CrossRef]
- Maryami, Z.; Fazeli, A. Molecular diversity and detection of Waxy genes in the Iranian wheat populations by multiplex PCR. Biotechnol. Biotechnol. Equip. 2015, 29, 869–875. [Google Scholar] [CrossRef]
- Ayala, M.; Alvarez, J.; Yamamori, M.; Guzmán, C. Molecular characterization of Waxy alleles in three subspecies of hexaploid wheat and identification of two novel Wx-B1 alleles. Theor. Appl. Genet. 2015, 128, 2427–2435. [Google Scholar] [CrossRef]
- Li, W.; Fu, B.; Li, Z.; Liu, Y.; Pu, Z.; Qi, P.; Jiang, Q.; Chen, G.; Wang, J.; Wei, Y.; et al. Characterization of the waxy gene in diploid Triticum L. and Aegilops L. species and its geographic distribution. Genet. Resour. Crop Evol. 2015, 63, 987–1002. [Google Scholar] [CrossRef]
- Alvarez, J.; Guzmán, C. Interspecific and intergeneric hybridization as a source of variation for wheat grain quality improvement. Theor. Appl. Genet. 2018, 131, 225–251. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Contreras, A.; Chaires-González, C.; Rosas-Burgos, E.; Borboa-Flores, J.; Wong-Corral, F.; Cortez-Rocha, M.; Cinco-Moroyoqui, F. Comparison of protein and starch content of substituted and complete triticales (× Triticosecale Wittmack): Contribution To Functional Properties. Int. J. Food Prop. 2014, 17, 421–432. [Google Scholar] [CrossRef] [Green Version]
- Cornejo-Ramírez, Y.; Ramírez-Reyes, F.; Cinco-Moroyoqui, F.; Rosas-Burgos, E.; Martínez-Cruz, O.; Carvajal-Millán, E.; Cárdenas-López, J.; Torres-Chavez, P.; Osuna-Amarillas, P.; Borboa-Flores, J.; et al. Starch debranching enzyme activity and its effects on some starch physicochemical characteristics in developing substituted and complete triticales (× Triticosecale Wittmack). Cereal Chem. J. 2016, 93, 64–70. [Google Scholar] [CrossRef]
- Mergoum, M.; Sapkota, S.; ElDoliefy, A.; Naraghi, S.; Pirseyedi, S.; Alamri, M.; AbuHammad, W. Triticale (× Triticosecale Wittmack) Breeding. In Advances in Plant Breeding Strategies: Cereals; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer: Cham, Switzerland, 2019; pp. 404–452. [Google Scholar]
- Dai, S.; Jiang, J.; Jia, Y.; Xue, X.; Liu, D.; Wei, Y.; Zheng, Y.; Yan, Z. Molecular characterization and phylogenetic analysis of Wx genes from three Taeniatherum diploid species. Biol. Plant 2016, 60, 505–512. [Google Scholar] [CrossRef]
- Alvarez, J.; Castellano, L.; Recio, R.; Cabrera, A. Wx gene in Hordeum chilense: Chromosomal location and characterisation of the allelic variation in the two main ecotypes of the species. Agronomy 2019, 9, 261. [Google Scholar] [CrossRef] [Green Version]
- Mason-Gamer, R. Phylogeny of a genomically diverse group of Elymus (Poaceae) allopolyploids reveals multiple levels of reticulation. PLoS ONE 2013, 8, e78449. [Google Scholar] [CrossRef] [Green Version]
- Sha, L.; Fan, X.; Li, J.; Liao, J.; Zeng, J.; Wang, Y.; Kang, H.; Zhang, H.; Zheng, Y.; Zhou, Y. Contrasting evolutionary patterns of multiple loci uncover new aspects in the genome origin and evolutionary history of Leymus (Triticeae; Poaceae). Mol. Phylogenet. Evol. 2017, 114, 175–188. [Google Scholar] [CrossRef]
- Gill, B.S.; Friebe, B.; Koo, D.H.; Li, W. Crop species origins, the impact of domestication and the potential of wide hybridisation for crop improvement. In Sustaining Global Food Security: The Nexus of Science and Policy; Zeigler, R., Ed.; CSIRO Publishing: Victoria, Australia, 2019; pp. 2–35. [Google Scholar]
- Dennett, A.; Schofield, P.; Roake, J.; Howes, N.; Chin, J. Starch swelling power and amylose content of triticale and Triticum timopheevii germplasm. J. Cereal Sci. 2009, 49, 393–397. [Google Scholar] [CrossRef]
- Cui, L.; Ren, Y.; Murray, T.; Yan, W.; Guo, Q.; Niu, Y.; Sun, Y.; Li, H. Development of perennial wheat through hybridization between wheat and wheatgrasses: A review. Engineering 2018, 4, 507–513. [Google Scholar] [CrossRef]
- Alvarez, J.; Ballesteros, J.; Sillero, J.; Martin, L. Tritordeum: A new crop of potential importance in the food industry. Hereditas 1992, 116, 193–197. [Google Scholar] [CrossRef]
- Martín, A.; Alvarez, J.; Martín, L.; Barro, F.; Ballesteros, J. The development of Tritordeum: A novel cereal for food processing. J. Cereal Sci. 1999, 30, 85–95. [Google Scholar] [CrossRef]
- Alvarez, J.; Campos, L.; Martin, A.; Martin, L. Influence of HMW and LMW glutenin subunits on gluten strength in hexaploid Tritordeum. Plant Breed. 1999, 118, 456–458. [Google Scholar] [CrossRef]
- Shapovalova, N. Two New Agricultural Crops will Appear on Russian Fields at Once (In Russian). Agro XXI (Agroindustrial Portal). 2019. Available online: https://www.agroxxi.ru/zhurnal-agromir-xxi/novosti/na-rossiiskih-poljah-pojavjatsja-srazu-dve-novye-agrokultury.html (accessed on 30 April 2020).
- Kroupin, P.; Divashuk, M.; Karlov, G. Gene resources of perennial wild cereals involved in breeding to improve wheat crop. Sel’skokhozyaistvennaya Biol. 2019, 54, 409–425. [Google Scholar] [CrossRef]
- Kocheshkova, A.; Kroupin, P.; Bazhenov, M.; Karlov, G.; Pochtovyy, A.; Upelniek, V.; Belov, V.; Divashuk, M. Pre-harvest sprouting resistance and haplotype variation of ThVp-1 gene in the collection of wheat-wheatgrass hybrids. PLoS ONE 2017, 12, e0188049. [Google Scholar] [CrossRef] [Green Version]
- Hayes, R.; Wang, S.; Newell, M.; Turner, K.; Larsen, J.; Gazza, L.; Anderson, J.; Bell, L.; Cattani, D.; Frels, K.; et al. The performance of early-generation perennial winter cereals at 21 sites across four continents. Sustainability 2018, 10, 1124. [Google Scholar] [CrossRef] [Green Version]
- Trifonova, A.; Boris, K.; Dedova, L.; Melnik, V.; Ivanova, L.; Kuzmina, N.; Zavgorodniy, S.; Upelniek, V. genome polymorphism of the synthetic species X Trititrigia cziczinii Tsvel. inferred from AFLP analysis. Vavilov J. Genet. Breed. 2018, 22, 648–653. [Google Scholar] [CrossRef]
- Larkin, P.; Newell, M.; Hayes, R.; Aktar, J.; Norton, M.; Moroni, S.; Wade, L. Progress in developing perennial wheats for grain and grazing. Crop Pasture Sci. 2014, 65, 1147. [Google Scholar] [CrossRef]
- Tsitsin, N.V. Mnogoletnyaya Pshenitsa; Nauka: Moscow, Russia, 1987. (In Russian) [Google Scholar]
- Gazza, L.; Galassi, E.; Ciccoritti, R.; Cacciatori, P.; Pogna, N. Qualitative traits of perennial wheat lines derived from different Thinopyrum species. Genet. Resour. Crop Evol. 2016, 63, 209–219. [Google Scholar] [CrossRef]
- Zhong, Y.; Mogoginta, J.; Gayin, J.; Annor, G. Structural characterization of intermediate wheatgrass (Thinopyrum intermedium) starch. Cereal Chem. 2019, 96, 927–936. [Google Scholar] [CrossRef]
- Marti, A.; Bock, J.; Pagani, M.; Ismail, B.; Seetharaman, K. Structural characterization of proteins in wheat flour doughs enriched with intermediate wheatgrass (Thinopyrum intermedium) flour. Food Chem. 2016, 194, 994–1002. [Google Scholar] [CrossRef]
- Li, H.; Wang, X. Thinopyrum ponticum and Th. intermedium: The promising source of resistance to fungal and viral diseases of wheat. J. Genet. Genom. 2009, 36, 557–565. [Google Scholar] [CrossRef]
- Mago, R.; Zhang, P.; Xia, X.; Zhang, J.; Hoxha, S.; Lagudah, E.; Graner, A.; Dundas, I. Transfer of stem rust resistance gene Srb from Thinopyrum ponticum into wheat and development of a closely linked PCR-based marker. Theor. Appl. Genet. 2018, 132, 371–382. [Google Scholar] [CrossRef]
- Wang, S.; Wang, C.; Wang, Y.; Wang, Y.; Chen, C.; Ji, W. Molecular cytogenetic identification of two wheat–Thinopyrum ponticum substitution lines conferring stripe rust resistance. Mol. Breed. 2019, 39, 143. [Google Scholar] [CrossRef]
- Zheng, X.; Tang, C.; Han, R.; Zhao, J.; Qiao, L.; Zhang, S.; Qiao, L.; Ge, C.; Zheng, J.; Liu, C. Identification, characterization, and evaluation of novel stripe rust-resistant wheat–Thinopyrum intermedium chromosome translocation lines. Plant Dis. 2020, 104, 875–881. [Google Scholar] [CrossRef]
- Ali, N. Wheat–Thinopyrum intermedium introgression lines enhancing wheat streak mosaic virus (WSMV) resistance. In Climate Change and Food Security with Emphasis on Wheat; Ozturk, M., Gul, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 243–255. [Google Scholar]
- Mahelka, V.; Kopecký, D.; Paštová, L. On the genome constitution and evolution of intermediate wheatgrass (Thinopyrum intermedium: Poaceae, Triticeae). BMC Evol. Biol. 2011, 11, 127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Larson, S.; Jensen, K.; Bushman, B.; DeHaan, L.; Wang, S.; Yan, X. genome evolution of intermediate wheatgrass as revealed by EST-SSR markers developed from its three progenitor diploid species. Genome 2015, 58, 63–70. [Google Scholar] [CrossRef]
- Guo, J.; Yu, X.; Yin, H.; Liu, G.; Li, A.; Wang, H.; Kong, L. Phylogenetic relationships of thinopyrum and triticum species revealed by Scot and CDDP markers. Plant Syst. Evol. 2016, 302, 1301–1309. [Google Scholar] [CrossRef]
- Divashuk, M.; Khuat, T.; Kroupin, P.; Kirov, I.; Romanov, D.; Kiseleva, A.; Khrustaleva, L.; Alexeev, D.; Zelenin, A.; Klimushina, M.; et al. Variation in copy number of Ty3/Gypsy centromeric retrotransposons in the genomes of Thinopyrum intermedium and its diploid progenitors. PLoS ONE 2016, 11, e0154241. [Google Scholar] [CrossRef] [Green Version]
- Linc, G.; Gaál, E.; Molnár, I.; Icsó, D.; Badaeva, E.; Molnár-Láng, M. Molecular cytogenetic (FISH) and genome analysis of diploid wheatgrasses and their phylogenetic relationship. PLoS ONE 2017, 12, e0173623. [Google Scholar] [CrossRef] [Green Version]
- Divashuk, M.; Karlov, G.; Kroupin, P. Copy number variation of transposable elements in Thinopyrum intermedium and its diploid relative species. Plants 2019, 9, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kroupin, P.; Kuznetsova, V.; Nikitina, E.; Martirosyan, Y.; Karlov, G.; Divashuk, M. Development of new cytogenetic markers for Thinopyrum ponticum (Podp.) Z.-W. Liu & R.-C. Wang. Comp. Cytogenet. 2019, 13, 231–243. [Google Scholar] [PubMed]
- Adebiyi, J.; Schmitt Olabisi, L.; Snapp, S. Understanding perennial wheat adoption as a transformative technology: Evidence from the literature and farmers. Renew. Agric. Food Syst. 2016, 31, 101–110. [Google Scholar] [CrossRef] [Green Version]
- Marquardt, K.; Vico, G.; Glynn, C.; Weih, M.; Eksvärd, K.; Dalin, P.; Björkman, C. Farmer perspectives on introducing perennial cereal in Swedish farming systems: A sustainability analysis of plant traits, farm management, and ecological implications. Agroecol. Sustain. Food Syst. 2016, 40, 432–450. [Google Scholar] [CrossRef]
- Snapp, S.; Rogé, P.; Okori, P.; Chikowo, R.; Peter, B.; Messina, J. Perennial grains for Africa: Possibility or pipedream? Exp. Agric. 2018, 55, 251–272. [Google Scholar] [CrossRef] [Green Version]
- Kroupin, P.; Divashuk, M.; Belov, V.; Glukhova, L.; Aleksandrov, O.; Karlov, G. Comparative Molecular Cytogenetic Characterization of Partial Wheat-Wheatgrass Hybrids. Russ. J. Genet. 2011, 47, 432–437. [Google Scholar] [CrossRef]
- Litvinov, D.; Chernook, A.; Kroupin, P.; Bazhenov, M.; Karlov, G.; Avdeev, S.; Divashuk, M. A Convenient co-dominant marker for height-reducing Ddw1 allele useful for marker-assisted selection. Agriculture 2020, 10, 110. [Google Scholar] [CrossRef] [Green Version]
- Monari, A.; Simeone, M.; Urbano, M.; Margiotta, B.; Lafiandra, D. Molecular characterization of new waxy mutants identified in bread and durum wheat. Theor. Appl. Genet. 2005, 110, 1481–1489. [Google Scholar] [CrossRef]
- Nicholas, K.B.; Nikolas, H.B., Jr. GeneDoc: A Tool for Editing and Annotating Multiple Sequence Alignments. 1997. Available online: https://genedoc.software.informer.com/download/ (accessed on 30 April 2020).
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Sharp, P. An improved 1-D SDS–PAGE method for the identification of three bread wheat «Waxy» proteins. J. Cereal Sci. 1996, 23, 191–193. [Google Scholar] [CrossRef]
- Gibson, T.; Solah, V.; McCleary, B. A procedure to measure amylose in cereal starches and flours with Concanavalin A. J. Cereal Sci. 1997, 25, 111–119. [Google Scholar] [CrossRef]
- Leterrier, M.; Holappa, L.; Broglie, K.; Beckles, D. Cloning, characterisation and comparative analysis of a starch synthase IV gene in wheat: Functional and evolutionary implications. BMC Plant Biol. 2008, 8, 98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Divashuk., M.; Krupin, P.; Bazhenov., M.; Klimushina, M.; Belov, V.; Semyonova, E.; Karlov, I. Molecular-genetic characterization of seed storage protein composition in partial wheat-wheatgrass hybrids. Izv. Tskha 2012, 5, 29–37. (In Russian) [Google Scholar]
- Lubimova, V.; Belov, V. Grain-Forage Wheat Cultivars Zernokormovaya 169 and Zernokormovaya 26 (Recommendations for Cultivating Grain-Forage Wheat); Tsitsin Main Botanical Garden, Academy of Sciences of USSR: Moscow, Russia, 1990. [Google Scholar]
- Upelniek, V.; Belov, V.; Ivanova, l.; Dolgova, S.; Demidov, A. Heritage of academician N.V. Tsitsin: State-of-the-art and potential of the collection of intermediate wheat × couch-grass hybrids. Vavilov J. Genet. Breed. 2020, 6, 85–89. (In Russian) [Google Scholar]
- Schoenfuss, T.; Seetharaman, K.; Peterson, D. Incorporation of intermediate wheat grass in food products. (Abstr.). Cereal Foods World 2014, 59, A12. [Google Scholar]
- Marti, A.; Qiu, X.; Schoenfuss, T.; Seetharaman, K. Characteristics of perennial wheatgrass (Thinopyrum intermedium) and refined wheat flour blends: Impact on rheological properties. Cereal Chem. J. 2015, 92, 434–440. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Dundas, I.; Xu, S.; Friebe, B.; McIntosh, R.; Raupp, W. Chromosome engineering techniques for targeted introgression of rust resistance from wild wheat relatives. Methods Mol. Biol. 2017, 1659, 163–172. [Google Scholar]




| Primer | Sequence | Annealing Temperature, °C |
|---|---|---|
| WxF3 | 5′-TCT GGT CAC GTC CCA GCT CGC CAC CT-3′ | 62 |
| WxVT1R | 5′-ACC CCG CGC TTG TAG CAG TGG AAG T-3′ | |
| WxBAF | 5′-ACT TCC ACT GCT ACA AGC GCG GGG T-3′ | 62 |
| WxBAR | 5′-GCT GAC GTC CAT GCC GTT GAC GAT G-3′ | |
| WxVT1F | 5′-CAT CGT CAA CGG CAT GGA CGT CAG C-3′ | 64 |
| WxVTR | 5′-CCA GAA GCA CGT CCT CCC AGT TCT TG-3′ | |
| WXTH F | 5’-AGG ATC CTG AAC CTC AAC AA-3’ | 64 |
| WXTH R | 5’-GAA GTC GTC GAA GGA GAA GC-3’ |
| № | Accession | Mean Amylose Content (% in Starch) |
|---|---|---|
| 1 | 1865 | 27.4 ± 4.4 |
| 2 | 1432 | 28.4 ± 17.2 |
| 3 | 1779 | 28.7 ± 1.9 |
| 4 | 150 | 28.8 ± 7.6 |
| 5 | 166 | 29.3 ± 6.4 |
| 6 | 1512 | 29.5 ± 6.4 |
| 7 | 548 | 29.6 ± 2.5 |
| 8 | 1416 | 29.9 ± 4.4 |
| 9 | 1783 | 30.4 ± 5.1 |
| 10 | 4044/4 | 30.5 ± 2.5 |
| 11 | ZP26/1 | 30.5 ± 1.3 |
| 12 | 1765 | 30.8 ± 10.8 |
| 13 | Wx-B1b | 24.0 ± 1.2 |
| 14 | Wx-B1a | 27.0 ± 1.2 |
| 15 | Wx-B1e | 29.4 ± 1.1 |
| Exon/Intron | Wx-A1 | Wx-B1 | Wx-D1 | Wx-Jb1 | Wx-Psstip1 | Wx-Thinter1 | Wx-Thpon1 |
|---|---|---|---|---|---|---|---|
| Exon 2 ** | 321 | 324 | 321 | 321 | 318/321 | 321/321 | 321 |
| Exon 3 | 81 | 81 | 81 | 81 | 81/81 | 81/81 | 81 |
| Exon 4 | 99 | 99 | 99 | 99 | 99/99 | 99/99 | 99 |
| Exon 5 | 154 | 154 | 154 | 154/154 | 154/154/154 | 154/154/154 | 154/154/154 |
| Exon 6 | 101 | 101 | 101 | 101/101 | 80 */80 */80 * | 80 */80 */80 * | 80 */80 */101 |
| Exon 7 | 354 | 354 | 354 | 354/354 | 354/354 | 354/348/354 | 354/354/354 |
| Exon 8 | 180 | 180 | 180 | 180 | 180/180 | 180 | 180/180/180/180/180 |
| Exon 9 | 192 | 192 | 192 | 192 | 192/192 | 192 | 192/192/192/192/192 |
| Exon 10 | 87 | 87 | 87 | 87 | 87/87 | 87 | 87/87/87/87/87 |
| Exon 11 | 129 | 129 | 129 | 129 | 129/129 | 129 | 129/129/129/129/129 |
| Exon 12 ** | 117 | 117 | 117 | 117 | 117/117 | 117 | 117/117/117/117/117 |
| Intron 2 | 82 | 99 | 90 | 81 | 81/84 | 74/81 | 81 |
| Intron 3 | 84 | 88 | 95 | 83 | 81/84 | 80/84 | 84 |
| Intron 4 | 109 | 113 | 104 | 105/108 | 106/106/106 | 106/106/106 | 106/106/105 |
| Intron 5 | 125 | 133 | 152 | 125/132 | 144 */144 */144 * | 144 */149 */144 * | 149 */145 */125 |
| Intron 6 | 99 | 69 | 141 | 99/99 | 103/103/103 | 103/103/103 | 103/103/96 |
| Intron 7 | 91 | 92 | 85 | 80 | 80/89 | 80 | 89/89/89/93/80 |
| Intron 8 | 95 | 86 | 82 | 82 | 82/99 | 82 | 82/82/82/86/82 |
| Intron 9 | 90 | 84 | 84 | 83 | 84/84 | 83 | 89/84/84/90/85 |
| Intron 10 | 98 | 97 | 98 | 98 | 98/96 | 98 | 99/98/94/96/98 |
| Intron 11 | 93 | 115 | 116 | 110 | 110/111 | 110 | 115/115/115/115/115 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klimushina, M.V.; Kroupin, P.Y.; Bazhenov, M.S.; Karlov, G.I.; Divashuk, M.G. Waxy Gene-Orthologs in Wheat × Thinopyrum Amphidiploids. Agronomy 2020, 10, 963. https://doi.org/10.3390/agronomy10070963
Klimushina MV, Kroupin PY, Bazhenov MS, Karlov GI, Divashuk MG. Waxy Gene-Orthologs in Wheat × Thinopyrum Amphidiploids. Agronomy. 2020; 10(7):963. https://doi.org/10.3390/agronomy10070963
Chicago/Turabian StyleKlimushina, Marina V., Pavel Yu. Kroupin, Mikhail S. Bazhenov, Gennady I. Karlov, and Mikhail G. Divashuk. 2020. "Waxy Gene-Orthologs in Wheat × Thinopyrum Amphidiploids" Agronomy 10, no. 7: 963. https://doi.org/10.3390/agronomy10070963
APA StyleKlimushina, M. V., Kroupin, P. Y., Bazhenov, M. S., Karlov, G. I., & Divashuk, M. G. (2020). Waxy Gene-Orthologs in Wheat × Thinopyrum Amphidiploids. Agronomy, 10(7), 963. https://doi.org/10.3390/agronomy10070963





