Improvement of a RD6 Rice Variety for Blast Resistance and Salt Tolerance through Marker-Assisted Backcrossing
Abstract
1. Introduction
2. Materials and Methods
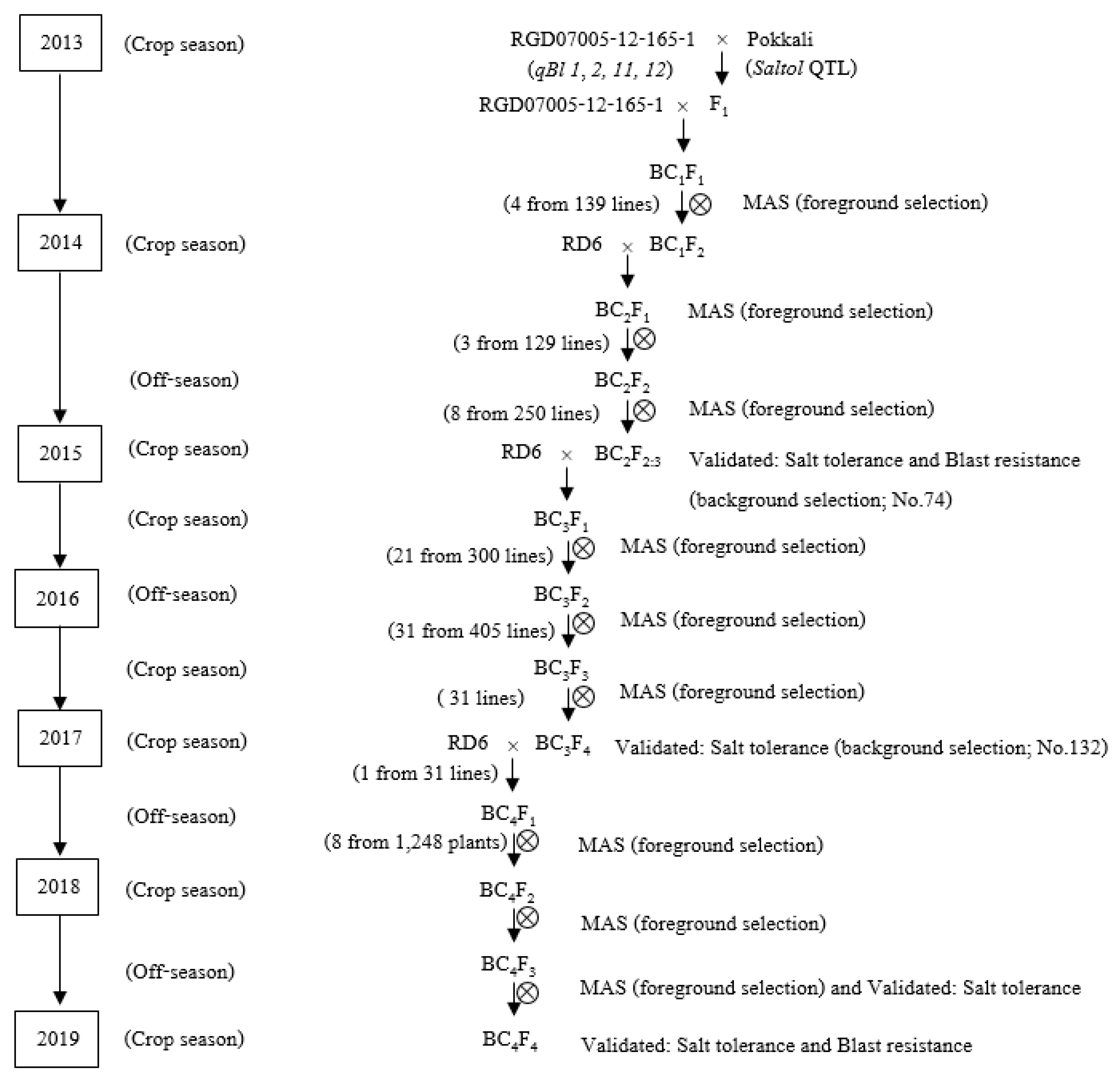
2.1. Plant Materials and Marker-Assisted Backcrossing Selection (MABS)
2.2. Evaluation of Salt Tolerance and Blast Resistance in the BC2F2:3 Populations (Exp. 1)
2.3. Evaluation of Salt Tolerance in the BC3F4 Populations (Exp. 2)
2.4. Evaluation of Salt Tolerance and Blast Resistance Evaluations in the BC4F3 Populations (Exp. 3)
2.5. Evaluation of Salt Tolerance in the BC4F4 Populations (Exp. 4)
2.6. Data Analysis
3. Results
3.1. Development of Populations Through MABS
3.2. Evaluation of Salt Tolerance and Blast Resistance in the BC2F2:3 Populations
3.3. Evaluation of Salt Tolerance in the BC3F4 Populations
3.4. Evaluation of Salt Tolerance and Blast Resistance in the BC4F3 Populations
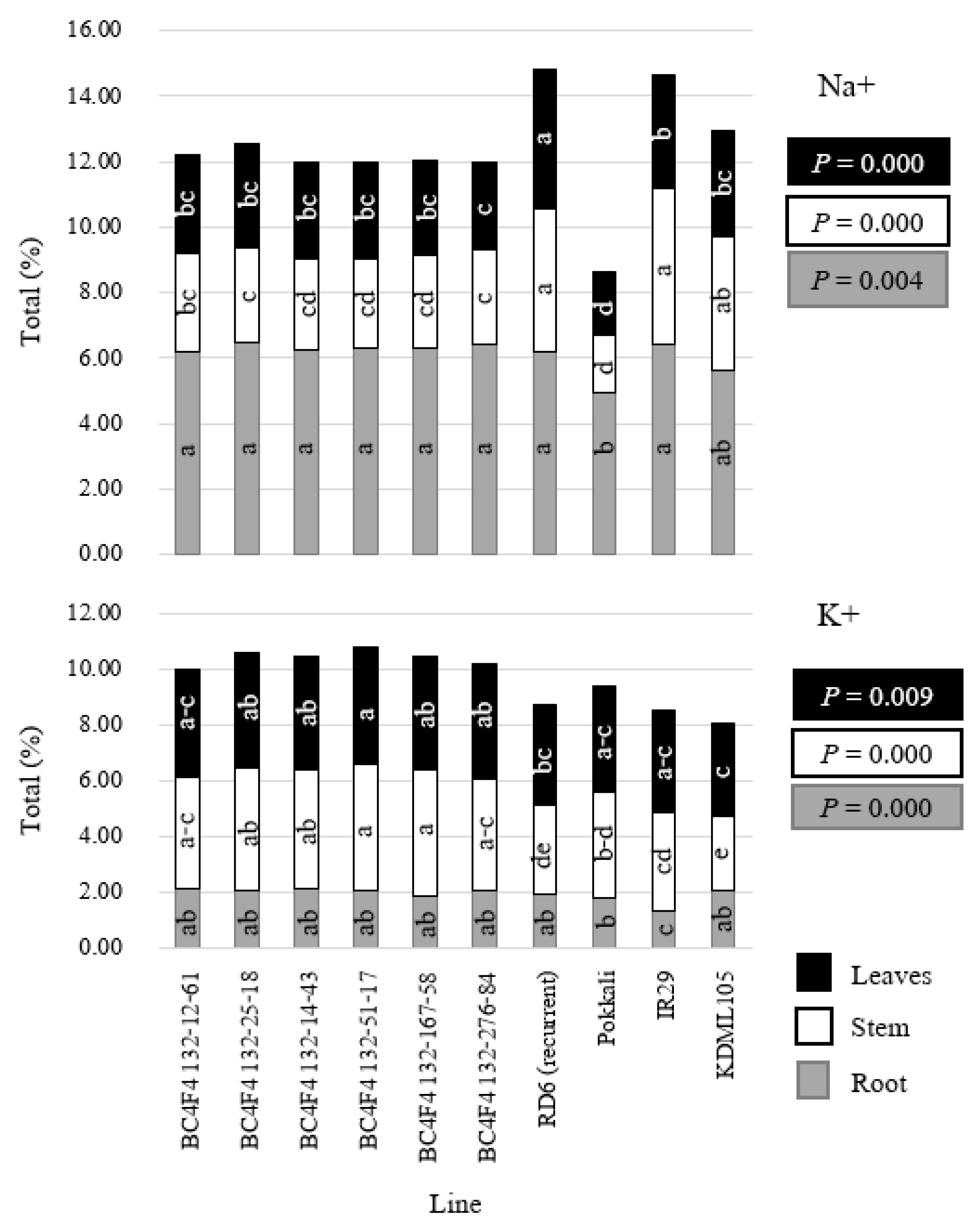
3.5. Evaluation of Salt Tolerance in the BC4F4 Population
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Keeratipibul, S.; Luangsakul, N.; Lertsatchayarn, T. The effect of Thai glutinous rice cultivars, grain length and cultivating locations on the quality of rice cracker (arare). Food Sci. Technol. 2008, 41, 1934–1943. [Google Scholar] [CrossRef]
- Chairote, E.; Jannoey, P.; Chairote, G. Improvement of Monacolin K and minimizing of citrinin content in Korkor 6 (RD 6) red yeast rice. World Acad. Sci. Eng. Technol. 2015, 9, 43–46. [Google Scholar]
- Raj, S.V.; Saranya, R.S.; Kumar, D.S.; Chinnadurai, M. Farm-level economic impact of rice blast: A Bayesian approach. Agric. Econ. Res. Rev. 2018, 31, 141. [Google Scholar] [CrossRef][Green Version]
- Mentlak, T.A.; Kombrink, A.; Shinya, T.; Ryder, L.S.; Otomo, I.; Saitoh, H.; Terauchi, R.; Nishizawa, Y.; Shibuya, N.; Thomma, B.P.H.J.; et al. Effector-mediated suppression of chitin-triggered immunity by magnaporthe oryzae is necessary for rice blast disease. Plant Cell 2012, 24, 322–335. [Google Scholar] [CrossRef]
- Chumpol, A.; Chankaew, S.; Saepaisan, S.; Monkham, T.; Sanitchon, J. New sources of rice blast resistance obtained from Thai indigenous upland rice germplasm. Euphytica 2018, 214, 183. [Google Scholar] [CrossRef]
- Greer, C.A.; Webster, R.K. Occurrence, distribution, epidemiology, cultivar reaction, and management of rice blast disease in California. Plant Dis. 2001, 85, 1096–1102. [Google Scholar] [CrossRef]
- Wang, D.; Guo, C.; Huang, J.; Yang, S.; Tian, D.; Zhang, X. Allele-mining of rice blast resistance genes at AC134922 locus. Biochem. Biophys. Res. Commun. 2014, 446, 1085–1090. [Google Scholar] [CrossRef]
- Noenplab, A.; Vanavichit, A.; Toojinda, T.; Sirithunya, P.; Tragoonrung, S.; Sriprakhon, S.; Vongsaprom, C. QTL mapping for leaf and neck blast resistance in Khao Dawk Mali105 and Jao Hom Nin recombinant inbred lines. Sci. Asia 2006, 32, 133–142. [Google Scholar] [CrossRef]
- Suwannual, T.; Chankaew, S.; Monkham, T.; Saksirirat, W.; Sanitchon, J. Pyramiding of four blast resistance QTLs into Thai rice cultivar RD6 through marker-assisted selection. Czech J. Genet. Plant Breed. 2017, 53, 1–8. [Google Scholar] [CrossRef]
- Arunin, S.; Pongwichian, P. Salt-affected soils and management in Thailand. Bull. Soc. Sea Water Sci. 2015, 69, 319–325. [Google Scholar]
- Wongsomsak, S. Salinization in Northeast Thailand. Southeast Asian Stud. 1986, 24, 133–153. [Google Scholar]
- Arunin, S. Salt effect soil in Southeast Asia. In Proceedings of the International Symposium on Salt Affected Lagoon Ecosystem, Valencia, Spain, 18–25 September 1995. [Google Scholar]
- Akbar, M. Breeding for salinity resistance in rice. In Prospects for Bio-Saline Research; Ahmed, R., Pietro, A.S., Eds.; Department of Botany, University of Karachi: Karachi, Sindh, Pakistan, 1986; pp. 37–55. [Google Scholar]
- Summart, J.; Thanonkeo, P.; Panichajakul, S.; Prathepha, P.; McManus, M.T. Effect of salt stress on growth, inorganic ion and proline accumulation in Thai aromatic rice, KhaoDawk Mali 105, callus culture. Afr. J. Biotechnol. 2010, 9, 145–152. [Google Scholar]
- Kumar, K.; Kumar, M.; Kim, S.-R.; Ryu, H.; Cho, Y.-G. Insights into genomics of salt stress response in rice. Rice 2013, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.I.; Yamaji, N.; Yamamoto, H.; Okubo, K.; Ueno, H.; Costa, A.; Tanoi, K.; Matsumura, H.; Fujii-Kashino, M.; Horiuchi, T.; et al. OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J. 2017, 91, 657–670. [Google Scholar] [CrossRef] [PubMed]
- Gregorio, G.B.; Senadhira, D.; Mendoza, R.D. Screening rice for salinity tolerance. IRRI Discuss. Pap. Ser. 1997, 22, 2–23. [Google Scholar]
- Bhowmik, S.K.; Islam, M.M.; Emon, R.M.; Begum, S.N.; Siddika, A.; Sultana, S. Identification of salt tolerant rice cultivars via phenotypic and marker-assisted procedures. Pak. J. Boil. Sci. 2007, 10, 4449–4454. [Google Scholar]
- Kavitha, P.G.; Miller, A.J.; Mathew, M.K.; Maathuis, F.J.M. Rice cultivars with differing salt tolerance contain similar cation channels in their root cells. J. Exp. Bot. 2012, 63, 3289–3296. [Google Scholar] [CrossRef]
- Ferreira, L.J.; Azevedo, V.S.; Marôco, J.P.; Oliveira, M.M.; Santos, A.P. Salt tolerant and sensitive rice varieties display differential methylome flexibility under salt stress. PLoS ONE 2015, 10, e0124060. [Google Scholar] [CrossRef]
- Mohammadi-Nejad, G.; Arzani, A.; Rezai, A.M.; Singh, R.K.; Gregorio, G.B. Assessment of rice genotypes for salt tolerance using microsatellite markers associated with the saltol QTL. Afr. J. Biotechnol. 2008, 7, 730–736. [Google Scholar]
- Huyen, L.T.N.; Cuc, L.M.; Ham, L.H.; Khanh, T.D. Introgression the Saltol QTL into Q5BD, the elite variety of Vietnam using marker assisted selection (MAS). Am. J. Biosci. 2013, 1, 80–84. [Google Scholar] [CrossRef]
- Ali, S.; Gautam, R.K.; Mahajan, R.; Krishnamurthy, S.L.; Sharma, S.K.; Singh, R.K. Stress indices and selectable traits in Saltol QTL introgressed rice genotypes for reproductive stage tolerance to sodicity and salinity stresses. Field Crops Res. 2013, 154, 65–73. [Google Scholar] [CrossRef]
- Waziri, A.; Kumar, P.; Purty, R.S. Saltol QTL and their role in salinity tolerance in rice. Austin. J. Biotechnol. Bioeng. 2016, 3, 1067. [Google Scholar]
- Ganie, S.A.; Borgohain, M.J.; Kritika, K.; Talukdar, A.; Pani, D.R.; Mondal, T.K. Assessment of genetic diversity of Saltol QTL among the rice (Oryza sativa L.) genotypes. Physiol. Mol. Boil. Plants 2016, 22, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Aala, W.F.; Gregorio, G.B. Morphological and molecular characterization of novel salt-tolerant rice germplasms from the philippines and bangladesh. Rice Sci. 2019, 26, 178–188. [Google Scholar] [CrossRef]
- Tanksley, S.; Nelson, J. Advanced backcross QTL analysis: A method for simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor. Appl. Genet. 1996, 92, 191–203. [Google Scholar] [CrossRef]
- Steele, K.; Price, A.H.; Shashidhar, H.E.; Witcombe, J.R. Marker-assisted selection to introgress rice QTLs controlling root traits into an Indian upland rice variety. Theor. Appl. Genet. 2005, 112, 208–221. [Google Scholar] [CrossRef]
- Neeraja, C.N.; Maghirang-Rodriguez, R.; Pamplona, A.; Heuer, S.; Collard, B.C.; Septiningsih, E.M.; Vergara, G.; Sanchez, D.; Xu, K.; Ismail, A.M.; et al. A marker-assisted backcross approach for developing submergence-tolerant rice cultivars. Theor. Appl. Genet. 2007, 115, 767–776. [Google Scholar] [CrossRef]
- Sundaram, R.M.; Vishnupriya, M.R.; Biradar, S.K.; Laha, G.S.; Reddy, G.A.; Rani, N.S.; Sarma, N.P.; Sonti, R.V. Marker assisted introgression of bacterial blight resistance in Samba Mahsuri, an elite indica rice variety. Euphytica 2007, 160, 411–422. [Google Scholar] [CrossRef]
- Iftekharuddaula, K.M.; Newaz, M.A.; Salam, M.A.; Ahmed, H.U.; Mahbub, M.A.A.; Septiningsih, E.M.; Collard, B.C.; Sanchez, D.L.; Pamplona, A.M.; Mackill, D. Rapid and high-precision marker assisted backcrossing to introgress the SUB1 QTL into BR11, the rainfed lowland rice mega variety of Bangladesh. Euphytica 2010, 178, 83–97. [Google Scholar] [CrossRef]
- Srichant, N.; Chankaew, S.; Monkham, T.; Thammabenjapone, P.; Sanitchon, J. Development of Sakon Nakhon rice variety for blast resistance through marker assisted backcross breeding. Agronomy 2019, 9, 67. [Google Scholar] [CrossRef]
- Kranto, S.; Chankaew, S.; Monkham, T.; Theerakulpisuta, P.; Sanitchon, J. Evaluation for salt tolerance in rice using multiple screening methods. J. Agric. Sci. Technol. 2016, 18, 1921–1931. [Google Scholar]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA minipreparation: Version II. Plant Mol. Boil. Rep. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Yoshida, S.; Forno, D.A.; Cock, J.H.; Gomez, K.A. Routine procedure for growing rice plants in cultures solution. Lab. Man. Physiol. Stud. Rice 1976, 17, 60–65. [Google Scholar]
- International Rice Research Institute. Standard Evaluation System Manual; International Rice Research Institute: Manila, Philippines, 1996. [Google Scholar]
- International Rice Research Institute. Standard Evaluation System for Rice (SES). Leaf blast; International Rice Research Institute: Manila, Philippines, 2002; p. 15. [Google Scholar]
- Sriseadka, T.; Wongpornchai, S.; Kitsawatpaiboon, P. Rapid method for quantitative analysis of the aroma impact compound, 2-Acetyl-1-pyrroline, in fragrant rice using automated headspace Gas chromatography. J. Agric. Food Chem. 2006, 54, 8183–8189. [Google Scholar] [CrossRef] [PubMed]
- Havre, G.N. The flame photometric determination of sodium, potassium and calcium in plant extracts with special reference to interference effects. Anal. Chim. Acta 1961, 25, 557. [Google Scholar] [CrossRef]
- Wongsaprom, C.; Sirithunya, P.; Vanavichit, A.; Pantuwan, G.; Jongdee, B.; Sidhiwong, N.; Lanceras-Siangliw, J.; Toojinda, T. Two introgressed quantitative trait loci confer a broad-spectrum resistance to blast disease in the genetic background of the cultivar RD6 a Thai glutinous jasmine rice. Field Crop. Res. 2010, 119, 245–251. [Google Scholar] [CrossRef]
- Pinta, W.; Toojinda, T.; Thummabenjapone, P.; Sanitchon, J. Pyramiding of blast and bacterial leaf blight resistance genes into rice cultivar RD6 using marker assisted selection. Afr. J. Biotechnol. 2013, 12, 4432–4438. [Google Scholar] [CrossRef]
- Nan, M.S.A.; Janto, J.; Sribunrueang, A.; Monkham, T.; Sanitchon, J.; Chankaew, S. Field evaluation of RD6 introgression lines for yield performance, blast, bacterial blight resistance, and cooking and eating Qualities. Agronomy 2019, 9, 825. [Google Scholar] [CrossRef]
- Suwanich, K. Geology and geological structure of potash and rock salt deposits in Chalerm Phrakiat District, Nakhon Ratchasima Province in Northeastern Thailand. Kasetsart J. (Nat. Sci.) 2010, 44, 1058–1068. [Google Scholar]
- Chunthaburee, S.; Dongsansuk, A.; Sanitchon, J.; Pattanagul, W.; Theerakulpisut, P. Physiological and biochemical parameters for evaluation and clustering of rice cultivars differing in salt tolerance at seedling stage. Saudi J. Boil. Sci. 2015, 23, 467–477. [Google Scholar] [CrossRef]
- Suriya-arunroj, D.; Supapoj, N.; Vanavichit, A.; Toojinda, T. Screening and selection for physiological characters contributing to salinity tolerance in rice. Kasetsart J. (Nat. Sci.) 2005, 39, 174–185. [Google Scholar]
- Kao, C.H. Mechanisms of salt tolerance in rice plants: Na+ transporters. Crop Environ. Bioinform. 2015, 12, 113–119. [Google Scholar]
- Miyakawa, S. Expansion of an improved variety into rain-fed rice cultivation in Northeast Thailand. Southeast Asian Stud. 1995, 33, 187–203. [Google Scholar]
- Jongdee, B.; Pantuwan, G.; Fukai, S.; Fischer, K. Improving drought tolerance in rainfed lowland rice: An example from Thailand. Agric. Water Manag. 2006, 80, 225–240. [Google Scholar] [CrossRef]
- Vasudevan, K.; Cruz, C.M.V.; Gruissem, W.; Bhullar, N.K. Large scale germplasm screening for identification of novel rice blast resistance sources. Front. Plant Sci. 2014, 5, 505. [Google Scholar] [CrossRef]
- Wang, R.; Fang, N.; Guan, C.; He, W.; Bao, Y.-M.; Zhang, H. Characterization and fine mapping of a blast resistant gene Pi-jnw1 from the japonica rice landrace Jiangnanwan. PLoS ONE 2016, 11, e0169417. [Google Scholar] [CrossRef][Green Version]
- Goto, K. Estimating losses from rice blast in Japan. In The Rice Blast Disease; Johns Hopkins Press: Baltimore, MD, USA, 1965; pp. 195–202. [Google Scholar]
| BC2F2:3 Populations | Salt Solution | Artificial Soil Salinity | Blast Resistance | |||
|---|---|---|---|---|---|---|
| Score | Reaction | Score | Reaction | Score | Reaction | |
| No. 74 | 4.3 c | T | 4.5 cd | T | 2.0 c | R |
| No. 42 | 5.5 b | MT | 4.8 cd | T | 1.0 c | R |
| No. 44 | 5.5 b | MT | 4.5 cd | T | 2.0 c | R |
| No. 23 | 4.8 bc | T | 4.5 cd | T | 1.0 c | R |
| No. 56 | 5.0 bc | MT | 4.5 cd | T | 2.0 c | R |
| No. 67 | 4.0 c | T | 4.0 d | T | 2.0 c | R |
| No. 33 | 5.5 b | MT | 5.0 cd | MT | 2.0 c | R |
| No. 7 | 4.0 c | T | 5.8 bc | MT | 2.0 c | R |
| Pokkali (tolerant check) | 4.0 c | T | 4.0 d | T | 6.0 a | MS |
| IR29 (susceptible check) | 9.0 a | HS | 9.0 a | HS | 4.0 b | MS |
| RD6 (recurrent parent) | 9.0 a | HS | 9.0 a | HS | 7.0 a | S |
| RGD07005-12-165-1 | 8.0 a | S | 7.0 b | S | 2.0 c | R |
| Jao Hom Nil (resistant check) | - | - | - | - | 1.0 c | R |
| P0489 (resistant check) | - | - | - | - | 2.0 c | R |
| F-Test | ** | ** | ** | |||
| C.V. (%) | 11.22 | 13.23 | 24.12 | |||
| Line | STS | 1000SW (g) | SL (cm) | SW (cm) | SS | Line | STS | 1000SW (g) | SL (cm) | SW (cm) | SS |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BC3F4 no.2 | 4.33 b–d | 26.80 a–g | 9.95 h–l | 2.62 c–h | 3.81 b–g | BC3F4 no.124 | 3.67 cd | 27.83 a–d | 10.64 a–d | 2.70 b–h | 3.97 a–e |
| BC3F4 no.14 | 4.33 b–d | 23.93 hi | 10.06 f–l | 2.64 c–h | 3.83 b–g | BC3F4 no.126 | 3.67 cd | 27.60 a–d | 10.36 b–i | 2.64 b–h | 3.93 a–f |
| BC3F4 no.22 | 3.67 cd | 24.40 f–i | 9.86 i–m | 2.58 f–h | 3.83 b–g | BC3F4 no.129 | 3.00 d | 26.23 b–h | 10.18 d–k | 2.68 b–h | 3.81 b–g |
| BC3F4 no.26 | 5.00 a–c | 24.17 g–i | 9.79 j–m | 2.59 f–h | 3.79 b–g | BC3F4 no.132 | 3.67 cd | 26.47 a–h | 10.39 b–i | 2.73 b–g | 3.81 b–g |
| BC3F4 no.35 | 4.33 b–d | 24.47 e–i | 10.14 d–k | 2.72 b–h | 3.74 c–h | BC3F4 no.136 | 4.33 b–d | 24.23 g–i | 9.88 h–m | 2.65 b–h | 3.73 c–h |
| BC3F4 no.36 | 3.67 cd | 26.00 c–h | 9.76 k–m | 2.56 gh | 3.82 b–g | BC3F4 no.151 | 6.33 a | 25.97 c–h | 10.02 g–l | 2.75 b–f | 3.68 d–h |
| BC3F4 no.40 | 5.00 a–c | 24.30 f–i | 9.93 h–l | 2.64 b–h | 3.47 gh | BC3F4 no.152 | 5.67 ab | 27.13 a–f | 9.92 h–l | 2.79 bc | 3.56 f–h |
| BC3F4 no.55 | 5.67 ab | 24.40 f–i | 9.35 mn | 2.76 b–e | 3.40 h | BC3F4 no.153 | 5.00 a–c | 25.07 d–i | 10.05 g–l | 2.73 b–g | 3.69 c–h |
| BC3F4 no.58 | 5.00 a–c | 24.50 e–i | 9.57 l–n | 2.62 c–h | 3.66 d–h | BC3F4 no.195 | 5.00 a–c | 27.40 a–d | 10.59 a–f | 2.55 h | 4.15 ab |
| BC3F4 no.67 | 6.33 a | 22.90 i | 9.04 n | 2.68 b–h | 3.38 h | BC3F4 no.235 | 3.67 cd | 25.80 d–h | 10.32 b–j | 2.61 d–h | 4.00 a–d |
| BC3F4 no.69 | 5.67 ab | 27.67 a–d | 10.76 a–c | 2.73 b–g | 3.97 a–e | BC3F4 no.241 | 3.67 cd | 27.27 a–e | 10.37 b–i | 2.74 b–g | 3.81 b–g |
| BC3F4 no.89 | 4.33 b–d | 27.57 a–d | 10.40 b–h | 2.72 b–h | 3.84 b–g | BC3F4 no.254 | 3.00 d | 29.07 ab | 10.71 a–c | 2.64 b–h | 4.06 a–c |
| BC3F4 no.93 | 3.67 cd | 26.40 a–h | 10.53 a–g | 2.73 b–g | 3.86 b–f | BC3F4 no.265 | 3.67 cd | 27.50 a–d | 10.83 ab | 2.58 f–h | 4.25 a |
| BC3F4 no.109 | 4.33 b–d | 29.13 a | 10.97 a | 2.62 c–h | 3.91 a–f | BC3F4 no.298 | 3.67 cd | 26.47 a–h | 10.28 c–k | 2.64 b–h | 3.91 a–f |
| BC3F4 no.113 | 4.33 b–d | 27.63 a–d | 10.73 a–c | 2.68 b–h | 4.01 a–d | Pokkali | 4.33 b–d | 13.77 j | 8.47 o | 3.42 a | 2.48 i |
| BC3F4 no.115 | 3.00 d | 28.80 a–c | 10.64 a–e | 2.81 b | 3.79 b–g | RD6 | 6.33 a | 24.20 g–i | 9.98 h–l | 2.78 b–d | 3.61 e–h |
| BC3F4 no.118 | 3.67 cd | 25.63 d–i | 10.10 e–k | 2.59 e–h | 3.92 a–f | BC2F5 no.74 | 5.67 ab | 24.53 e–i | 9.88 h–m | 2.68 b–h | 3.70 c–h |
| F-test | ** | ** | ** | ** | ** | F-test | ** | ** | ** | ** | ** |
| C.V. (%) | 25.4 | 6.8 | 3.96 | 3.2 | 6.18 | C.V. (%) | 25.4 | 6.8 | 3.96 | 3.2 | 6.18 |
| Line | QTLs | Leaf Blast | Neck Blast | Upland Short Row Method | Salt Score | ||||
|---|---|---|---|---|---|---|---|---|---|
| Resistance | Mean | Resistance | Mean | Resistance | Mean | Tolerance | Mean | ||
| BC4F3 132-12-61 | Saltol, qBl (1 2 11 12) | HR | 0.00 c | R | 1.53 b | HR | 0.00 c | T | 3.67 b |
| BC4F3 132-25-18 | Saltol, qBl (1 11 12) | HR | 0.00 c | R | 1.27 b | HR | 0.00 c | T | 3.45 b |
| BC4F3 132-14-43 | Saltol, qBl (1 11 12) | HR | 0.00 c | R | 1.67 b | HR | 0.00 c | T | 3.67 b |
| BC4F3 132-98-87 | Saltol, qBl (11 12) | HR | 0.00 c | R | 1.67 b | HR | 0.00 c | T | 3.67 b |
| BC4F3 132-174-54 | Saltol, qBl (1 11) | HR | 0.00 c | R | 1.93 b | - | - | T | 3.89 b |
| BC4F3 132-51-17 | Saltol, qBl (12) | HR | 0.87 c | R | 1.00 b | R | 1.00 c | T | 3.67 b |
| BC4F3 132-167-58 | Saltol, qBl (11) | HR | 0.23 c | R | 2.53 b | HR | 0.33 c | T | 3.67 b |
| BC4F3 132-276-84 | Saltol, qBl (2) | HR | 0.43 c | R | 1.13 b | HR | 0.67 c | T | 3.89 b |
| RD6(recurrent) | - | MS | 6.77 a | MS | 5.80 a | MR | 4.33 b | MS | 6.56 a |
| JHN(resistant check) | qBl (1 11) | MR | 3.67 b | R | 1.00 b | HR | 0.00 c | - | - |
| P0489(resistant check) | qBl (2 12) | HR | 0.00 c | R | 1.00 b | HR | 0.00 c | - | - |
| KDML105(susceptible check) | - | - | - | - | - | S | 7.67 a | - | - |
| IR 29(susceptible check) | - | - | - | - | - | - | - | S | 7.45 a |
| F-test | ** | ** | ** | ** | |||||
| C.V. (%) | 48.2 | 60.73 | 31.03 | 17.20 | |||||
| Line | PH (cm) | PL (cm) | SW4P (g) | 1000SW (g) | TDW (g) | TSW (g) | HI | SL (cm) | SW (cm) | SS | 2AP Content (ppm) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BC4F3 132-12-61 | 161.33 ab | 27.10 a–c | 20.98 a–c | 27.90 c–e | 354.66 ab | 146.05 a–c | 0.42 a | 1.09 b | 0.26 bc | 4.11 cd | 3.63 b–d |
| BC4F3 132-25-18 | 165.90 a | 27.67 a | 20.33 a–c | 29.07 ab | 367.31 a | 157.76 ab | 0.43 a | 1.09 b | 0.26 c | 4.20 bc | 3.15 d–f |
| BC4F3 132-14-43 | 163.90 ab | 27.30 ab | 22.03 ab | 29.10 ab | 330.40 ab | 146.54 a–c | 0.44 a | 1.07 b | 0.27 ab | 4.06 c–e | 4.05 b |
| BC4F3 132-98-87 | 163.00 ab | 25.60 de | 20.81 a–c | 26.93 e | 325.53 ab | 147.19 a–c | 0.45 a | 1.02 c | 0.26 bc | 3.88 ef | 4.68 a |
| BC4F3 132-174-54 | 164.43 ab | 25.70 c–e | 20.80 a–c | 28.50 bc | 345.89 ab | 151.41 a–c | 0.44 a | 1.05 bc | 0.26 bc | 4.06 c–f | 3.44 c–e |
| BC4F3 132-51-17 | 166.10 a | 27.00 a–d | 19.82 a–c | 28.17 bc | 291.88 bc | 122.30 bc | 0.42 a | 1.08 b | 0.26 bc | 4.08 cd | 3.73 bc |
| BC4F3 132-167-58 | 162.90 ab | 27.93 a | 23.27 a | 27.97 cd | 334.45 ab | 146.77 a–c | 0.44 a | 1.09 b | 0.26 c | 4.22 bc | 3.84 bc |
| BC4F3 132-276-84 | 165.10 a | 26.73 a–d | 20.91 a–c | 28.53 bc | 368.80 a | 164.07 a | 0.44 a | 1.05 bc | 0.27 a | 3.86 f | 3.06 ef |
| RD6 (recurrent) | 167.57 a | 25.87 b–e | 18.46 bc | 25.10 f | 295.79 a–c | 115.33 cd | 0.39 ab | 1.02 c | 0.26 bc | 3.88 ef | 4.62 a |
| JHN (resistant check) | 109.80 d | 25.03 e | 12.99 d | 21.80 h | 224.05 c | 60.30 e | 0.26 c | 1.07 b | 0.24 d | 4.43 a | - |
| P0489 (resistant check) | 137.70 c | 25.53 de | 17.36 c | 23.30 g | 232.65 c | 82.13 de | 0.34 b | 0.95 d | 0.23 d | 4.05 c–f | - |
| F-test | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** |
| C.V. (%) | 2.46 | 3.33 | 11.28 | 2.16 | 13.95 | 16.74 | 10.15 | 2.79 | 1.70 | 2.85 | 3.72 |
| Line | LDW (g) | SDW (g) | RDW (g) | TDW (g) | Salt Score | ||||
|---|---|---|---|---|---|---|---|---|---|
| BC4F4 132-12-61 | 0.56 | bc | 0.47 | b | 0.27 | b | 1.3 | bc | 5.2 c–e |
| BC4F4 132-25-18 | 0.47 | b–d | 0.33 | bc | 0.22 | b | 1.02 | bc | 5.9 b–d |
| BC4F4 132-14-43 | 0.55 | bc | 0.41 | bc | 0.25 | b | 1.21 | bc | 6.0 b–e |
| BC4F4 132-51-17 | 0.5 | bc | 0.35 | bc | 0.22 | b | 1.07 | bc | 5.2 c–e |
| BC4F4 132-167-58 | 0.49 | bc | 0.42 | bc | 0.21 | bc | 1.12 | bc | 4.9 de |
| BC4F4 132-276-84 | 0.44 | b–d | 0.39 | bc | 0.23 | b | 1.06 | bc | 5.5 b–e |
| RD6 (recurrent) | 0.39 | cd | 0.27 | cd | 0.2 | bc | 0.85 | cd | 6.2 a–c |
| Pokkali (tolerant check) | 1.21 | a | 1.18 | a | 0.47 | a | 2.86 | a | 3.6 f |
| IR29 (susceptible check) | 0.23 | d | 0.11 | d | 0.13 | c | 0.47 | d | 7.4 a |
| KDML105 | 0.63 | b | 0.51 | b | 0.25 | b | 1.39 | b | 6.5 ab |
| F-test | ** | ** | ** | ** | ** | ||||
| CV (%) | 22.24 | 21.68 | 17.1 | 18.88 | 11.35 | ||||
| LDW | SDW | RDW | TDW | Leaves Na+ | Leaves K+ | Stem Na+ | Stem K+ | Root Na+ | Root K+ | |
|---|---|---|---|---|---|---|---|---|---|---|
| SDW | 0.9926 ** | |||||||||
| RDW | 0.9863 ** | 0.9864 ** | ||||||||
| TDW | 0.9978 ** | 0.9981 ** | 0.9911 ** | |||||||
| Leaves Na+ | −0.7490 * | −0.7762 ** | −0.7513 * | −0.7665 ** | ||||||
| Leaves K+ | −0.0157 | −0.0096 | 0.0313 | −0.0044 | −0.4369 | |||||
| Stem Na+ | −0.6957 * | −0.6999 * | −0.7268 * | −0.7056 * | 0.8532 ** | −0.6924 * | ||||
| Stem K+ | −0.0578 | −0.0566 | −0.0188 | −0.0500 | −0.3733 | 0.9680 ** | −0.6326 * | |||
| Root Na+ | −0.9140 ** | −0.9037 ** | −0.8716 ** | −0.9054 ** | 0.5259 | 0.3967 | 0.3537 | 0.4257 | ||
| Root K+ | 0.1762 | 0.1411 | 0.2168 | 0.1666 | −0.1362 | 0.3735 | −0.4801 | 0.2748 | 0.0495 | |
| Salt score | −0.7899 ** | −0.8136 ** | −0.8140 ** | −0.8065 ** | 0.7670 ** | −0.4516 | 0.8917 ** | −0.4183 | 0.5386 | −0.3405 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thanasilungura, K.; Kranto, S.; Monkham, T.; Chankaew, S.; Sanitchon, J. Improvement of a RD6 Rice Variety for Blast Resistance and Salt Tolerance through Marker-Assisted Backcrossing. Agronomy 2020, 10, 1118. https://doi.org/10.3390/agronomy10081118
Thanasilungura K, Kranto S, Monkham T, Chankaew S, Sanitchon J. Improvement of a RD6 Rice Variety for Blast Resistance and Salt Tolerance through Marker-Assisted Backcrossing. Agronomy. 2020; 10(8):1118. https://doi.org/10.3390/agronomy10081118
Chicago/Turabian StyleThanasilungura, Korachan, Sukanya Kranto, Tidarat Monkham, Sompong Chankaew, and Jirawat Sanitchon. 2020. "Improvement of a RD6 Rice Variety for Blast Resistance and Salt Tolerance through Marker-Assisted Backcrossing" Agronomy 10, no. 8: 1118. https://doi.org/10.3390/agronomy10081118
APA StyleThanasilungura, K., Kranto, S., Monkham, T., Chankaew, S., & Sanitchon, J. (2020). Improvement of a RD6 Rice Variety for Blast Resistance and Salt Tolerance through Marker-Assisted Backcrossing. Agronomy, 10(8), 1118. https://doi.org/10.3390/agronomy10081118





