High-Efficiency Agrobacterium rhizogenes-Mediated Transgenic Hairy Root Induction of Lens culinaris
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
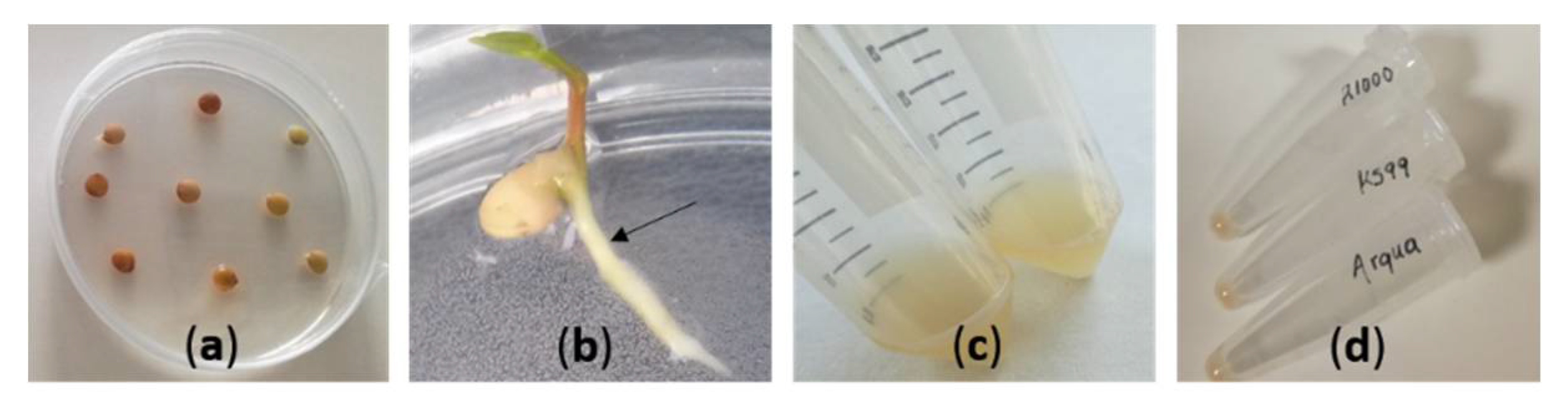
2.2. Preparation of Inoculum for Transformation
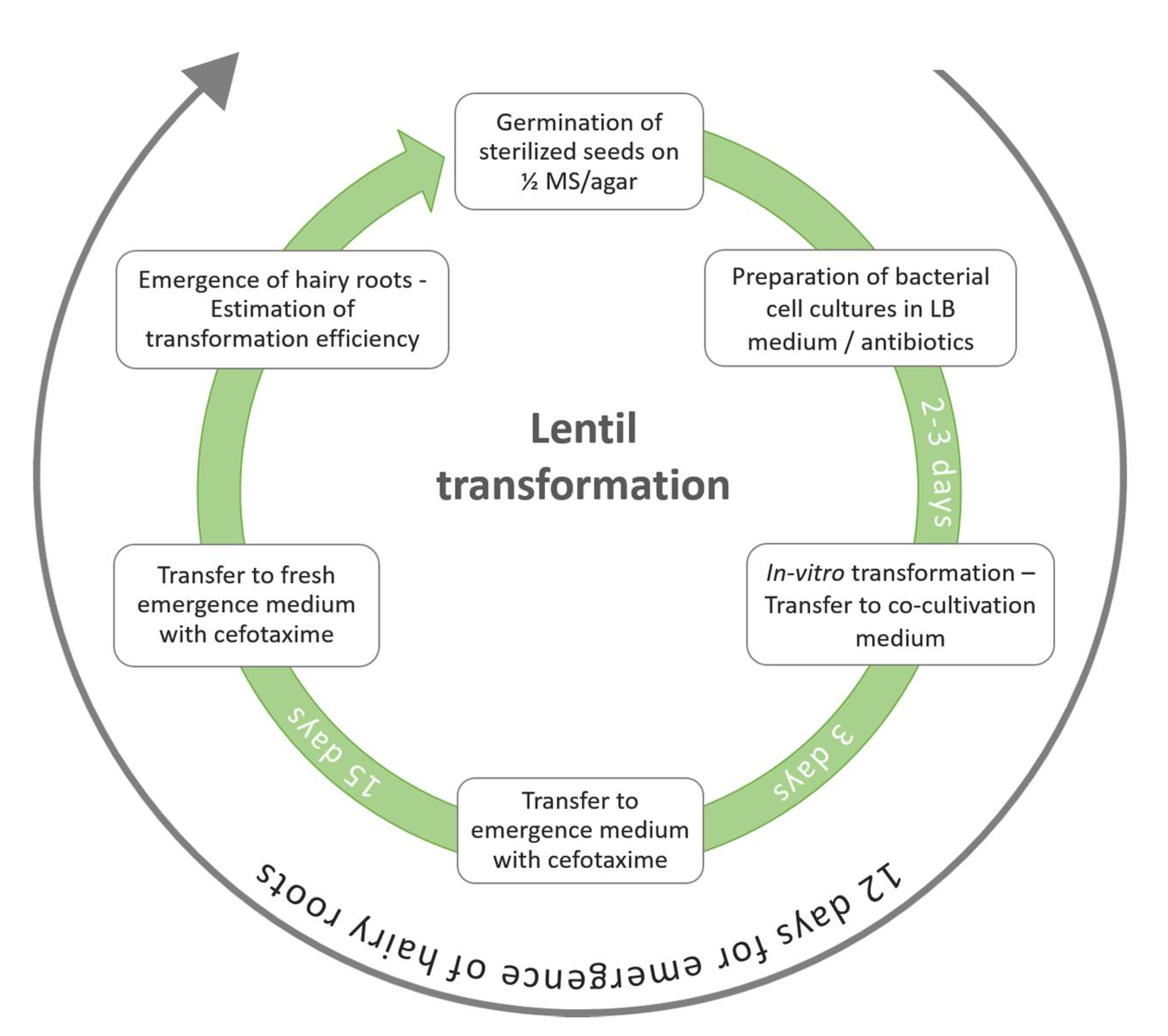
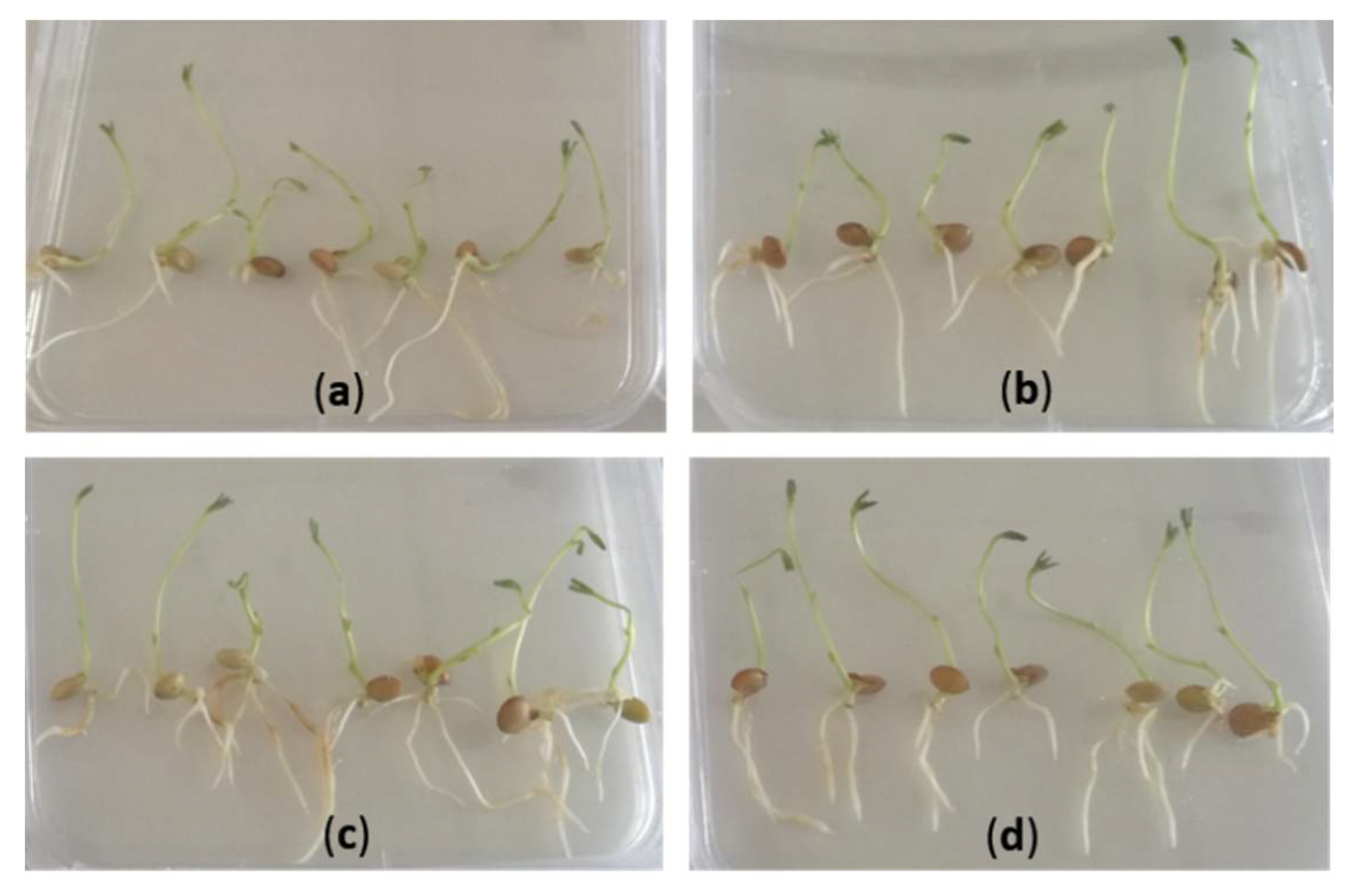
2.3. Hairy Root Induction
2.4. Evaluation of Transformed Roots
2.5. Statistical Analysis
3. Results
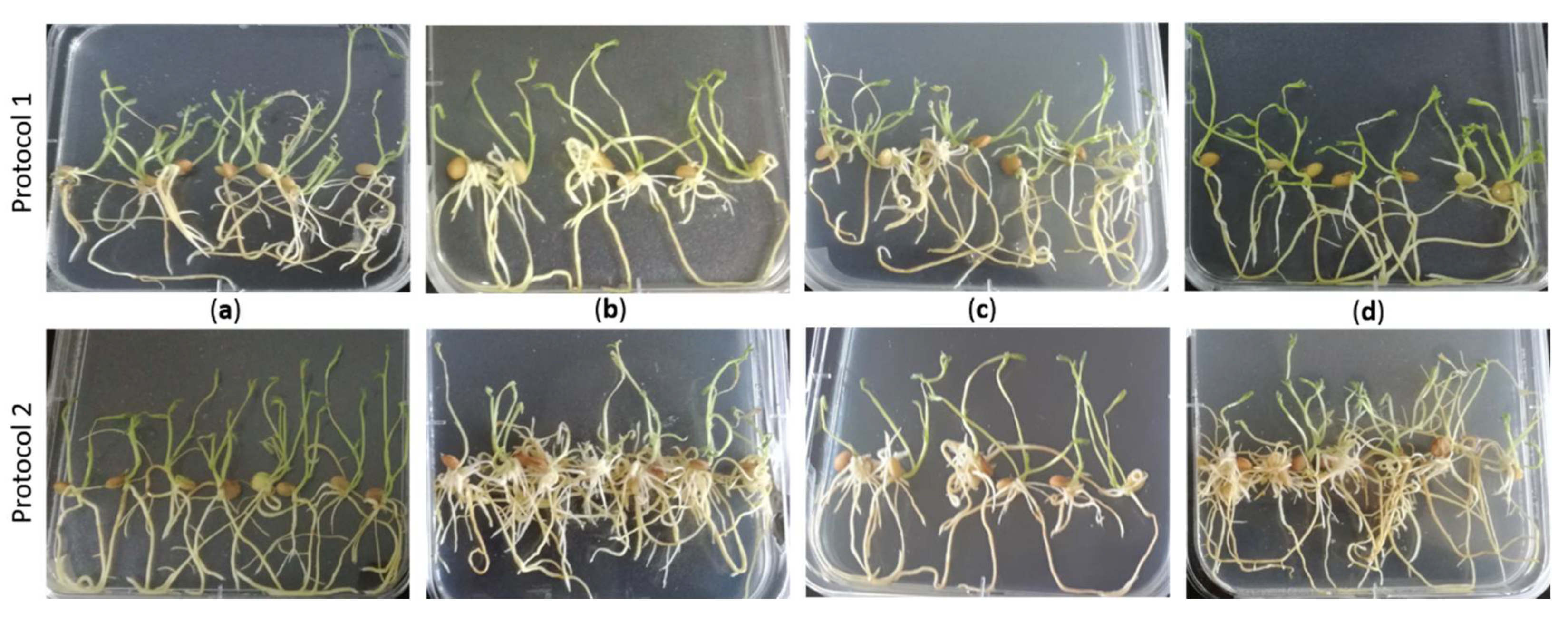
3.1. Protocol Optimization for the Generation of Composite Plants in Lentil
3.2. Verification of the Transgenic Nature of Hairy Roots
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Christou, P. Biotechnology applied to grain legumes. Field Crop Res. 1997, 53, 83–97. [Google Scholar] [CrossRef]
- Khazaei, H.; Subedi, M.; Nickerson, M.; Martínez-Villaluenga, C.; Frias, J.; Vandenberg, A. Seed Protein of Lentils: Current status, progress and food applications. Foods 2019, 9, 391. [Google Scholar] [CrossRef] [PubMed]
- Sarker, A.; Erskine, W.; Singh, M. Regression models for lentil seed and straw yields in Near East. Agri. Meteorol. 2003, 116, 61–72. [Google Scholar] [CrossRef]
- Somers, A.D.; Samac, D.A.; Olhoft, R.M. Recent Advances in Legume Transformation. Plant Physiol. 2003, 131, 892–899. [Google Scholar] [CrossRef] [PubMed]
- Khatib, F.; Makris, A.; Yamaguchi-Shinozakic, K.; Kumar, S.; Sarker, A.; Erskine, W.; Baum, M. Expression of DREB1A gene in lentil (Lens culinaris Medik. culinaris) transformed with the Agrobacterium system. Crop Pasture Sci. 2011, 62, 488–495. [Google Scholar] [CrossRef]
- Barton, J.; Klyne, A.; Tennakon, D.; Francis, C.; Hamblin, J. Development of a system for gene transfer to lentils. In Proceedings of the International Food Legume Research Conference III, Adelaide, Australia, 22–26 September 1997; p. 85. [Google Scholar]
- Lurquin, P.F.; Cai, Z.; Stiff, C.M.; Fuerst, E.P. Half-embryo cocultivation technique for estimating the susceptibility of pea (Pisum sativum L.) and lentil (Lens culinaris Medik.) cultivars to Agrobacterium tumefaciens. Mol. Biotechnol. 1998, 9, 175–179. [Google Scholar] [CrossRef]
- Gulati, A.; Schryer, P.; McHughen, A. Production of fertile transgenic lentil (Lens culinaris Medik) plants using particle bombardment. Vitr. Cell Dev. Biol-Plant 2002, 38, 316–324. [Google Scholar] [CrossRef]
- Mahmoudian, M.; Yucel, M.; Oktem, H.A. Transformation of lentil (Lens culinaris M.) cotyledonary nodes by vacuum in filtration of Agrobacterium tumefaciens. Plant Mol. Biol. Rep. 2002, 20, 251–257. [Google Scholar] [CrossRef]
- Chowrira, G.M.; Akella, V.; Fuerst, P.E.; Lurquin, P.F. Transgenic grain legumes obtained by in planta electroporation-mediated gene transfer. Mol. Biotechnol. 1996, 5, 85–96. [Google Scholar] [CrossRef]
- Khatib, F.; Koudsieh, S.; Ghazal, B.; Barton, J.; Tsujimoto, H.; Baum, M. Developing herbicide resistant lentil (Lens culinaris Medikus subsp. culinaris) through Agrobacterium-mediated transformation. Arab. J. Plant Prot. 2007, 25, 185–192. [Google Scholar]
- Akcay, U.C.; Mahmoudian, M.; Kamci, H.; Yucel, M.; Oktem, H.A. Agrobacterium tumefaciens-mediated genetic transformation of a recalcitrant grain legume, lentil (Lens culinaris Medik). Plant Cell Rep. 2009, 28, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Das, S.K.; Shethi, K.J.; Hoque, M.I.; Sarker, R.H. Agrobacterium-mediated genetic transformation of lentil (Lens culinaris Medik.) with chitinase gene followed by in vitro flower and pod formation. Plant Tissue Cult. Biotechnol. 2012, 29, 99–109. [Google Scholar] [CrossRef][Green Version]
- Christey, M.C.; Braun, R.H. Production of hairy root cultures and transgenic plants by Agrobacterium rhizogenes-mediated transformation. In Transgenic Plants: Methods and Protocols; Peña, L., Ed.; Humana Press: Totowa, NJ, USA, 2005; Volume 286, pp. 47–60. [Google Scholar]
- Chilton, M.D.; Tepfer, D.A.; Petit, A.; David, C.; Casse-Delbert, F.; Tempe, J. Agrobacterium rhizogenes inserts T-DNA into the genomes of the host plant root cells. Nature 1982, 295, 432–434. [Google Scholar] [CrossRef]
- White, F.F.; Taylor, B.H.; Huffman, G.A.; Gordon, M.P.; Nester, E.W. Molecular and genetic analysis of the transferred DNA regions of the root-inducing plasmid of Agrobacterium rhizogenes. J. Bacteriol. 1985, 164, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Jensen, J.S.; Marcker, K.A.; Otten, L.; Schell, J. Nodule-specific expression of a chiamaeric soybean leghaemoglobin gene in transgenic Lotus corniculatus. Nature 1986, 321, 669–674. [Google Scholar] [CrossRef]
- Constantino, P.; Spano, L.; Pomponi, M.; Benvenuto, E.; Ancora, G. The T-DNA of Agrobacterium rhizogenes is transmitted through meiosis to the progeny of hairy root plants. J. Mol. Appl. Genet. 1984, 2, 465–470. [Google Scholar]
- Collier, R.; Fuchs, B.; Walter, N.; Lutke, W.K.; Taylor, C.G. Ex vitro composite plants: An inexpensive, rapid method for root biology. Plant J. 2005, 43, 449–457. [Google Scholar] [CrossRef]
- Tόth, K.; Batek, J.; Stacey, G. Generation of soybean (Glycine max) transient transgenic roots. Curr. Protoc. Plant Biol. 2016, 5, 1–13. [Google Scholar] [CrossRef]
- Thilip, C.; Raju, C.S.; Varutharaju, K.; Aslam, A.; Shajahan, A. Improved Agrobacterium rhizogenes-mediated hairy root culture system of Withania omnifera (L.) Dunal using sonication and heat treatment. Biotechnology 2015, 5, 949–956. [Google Scholar] [CrossRef]
- Stiller, J.; Martirani, L.; Tuppale, S.; Chian, R.J.; Chiurazzi, M.; Gresshoff1, P.M. High frequency transformation and regeneration of transgenic plants in the model legume Lotus japonicus. J. Exp. Bot. 1997, 48, 1357–1365. [Google Scholar] [CrossRef]
- Boisson-Dernier, A.; Chabaud, M.; Garcia, F.; Bécard, G.; Rosenberg, C.; Barker, D.G. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant-Microbe Interact. 2001, 14, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Hou, W.S.; Liu, W.; Yao, W.W.; Wu, C.X.; Liu, X.B.; Han, T.F. Overexpression of TaNHX2 enhances salt tolerance of ‘composite’ and whole transgenic soybean plants. Plant Cell Tissue Organ Cult. 2011, 107, 541–552. [Google Scholar] [CrossRef]
- Chen, L.; Caia, Y.; Liua, X.; Guoa, C.; Sunb, S.; Wub, C.; Jiangb, B.; Hanb, T.; Houa, W. Soybean hairy roots produced in vitro by Agrobacterium rhizogenes-mediated transformation. Crop J. 2018, 6, 162–171. [Google Scholar] [CrossRef]
- Grant, J.E.; Cooper, P.A.; Gilpin, B.J.; Hoglund, S.J.; Reader, J.K.; Pither-Joyce, M.D.; Timmerman-Vaughan, G.M. Kanamycin is effective for selecting transformed peas. Plant Sci. 1998, 139, 159–164. [Google Scholar] [CrossRef]
- Clemow, S.R.; Clairmont, L.; Madsen, L.H.; Guinel, F. Reproducible hairy root transformation and spot-inoculation methods to study root symbioses of pea. Plant Methods 2011, 7, 46. [Google Scholar] [CrossRef]
- Leppyanen, I.V.; Kirienko, A.N.; Dolgikh, E.A. Agrobacterium rhizogenes-mediated transformation of Pisum sativum L. roots as a tool for studying the mycorrhizal and root nodule symbioses. PeerJ 2019, 7, e6552. [Google Scholar] [CrossRef]
- Bajrovic, K.; Gozukirmizi, N. Regeneration and hairy root formation of chickpea using callus derived plantlets and seedlings. Int. Chickpea Pigeonpea Newsl. 1997, 4, 30–31. [Google Scholar]
- Krishnamurthy, K.V.; Suhasini, K.; Sagare, A.P.; Meixner, M.; de Kathen, A.; Pickardt, T.; Schieder, O. Agrobacterium mediated transformation of chickpea (Cicer arietinum L.) embryo axes. Plant Cell Rep. 2000, 19, 235–240. [Google Scholar] [CrossRef]
- Khawar, K.M.; Ozcan, S. Hairy root transformation in turkish chickpea (Cicer arietinum L.) cultivars. Biotechnol. Biotechnol. Equip. 2004, 3, 51–54. [Google Scholar] [CrossRef]
- Aggarwal, P.R.; Nag, P.; Choudhary, P.; Chakraborty, N.; Chakraborty, S. Genotype-independent Agrobacterium rhizogenes-mediated root transformation of chickpea: A rapid and efficient method for reverse genetics studies. Plant Methods 2018, 14, 55. [Google Scholar] [CrossRef]
- Estrada-Navarrete, G.; Alvarado-Affantranger, X.; Olivares, J.E.; Diaz-Camino, C.; Santana, O.; Murillo, E.; Guillén, G.; Sánchez-Guevara, N.; Acosta, J.; Quinto, C.; et al. Agrobacterium rhizogenes-transformation of the Phaseolus spp.: A tool for functional genomics. Mol. Plant Microbe Interact. 2006, 19, 1385–1393. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dogan, D.; Khawar, K.M.; Ozcan, S. Agrobacterium mediated tumor and hairy root formation from different explants of lentils derived from young seedlings. Int. J. Agric. Biol. 2005, 7, 1019–1025. [Google Scholar]
- Pavli, O.I.; Skaracis, G.N. Fast and efficient genetic transformation of sugar beet by Agrobacterium rhizogenes. Sci. Protoc. 2010. [Google Scholar] [CrossRef]
- Hansen, J.; Jørgensen, J.E.; Stougaard, J.; Marcker, K.A. Hairy roots: A short cut to transgenic root nodules. Plant Cell Rep. 1989, 8, 12–15. [Google Scholar] [CrossRef]
- Mankin, S.L.; Hill, D.S.; Olhoft, P.M.; Toren, E.; Wenck, A.R.; Nea, L.; Xing, L.; Brown, J.A.; Fu, H.; Ireland, L.; et al. Disarming and sequencing of Agrobacterium rhizogenes strain K599 (NCPPB2659) plasmid pRi2659. Vitr. Cell Dev. Biol. Plant 2007, 6, 521–535. [Google Scholar] [CrossRef]
- Mrosk, C.; Forner, S.; Hause, G.; Küster, H.; Kopka, J.; Hause, B. Composite Medicago truncatula plants harbouring Agrobacterium rhizogenes-transformed roots reveal normal mycorrhization by Glomus intraradices. J. Exp. Bot. 2009, 13, 3797–3807. [Google Scholar] [CrossRef]
- Quandt, H.J.; Pühler, A.; Broer, I. Transgenic root nodules of Vicia hirsuta: A fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol. Plant Microbe Interact. 1993, 6, 699–706. [Google Scholar] [CrossRef]
- Tao, J.; Li, L. Genetic transformation of Torenia fournieri L. mediated by Agrobacterium rhizogenes. South Afr. J. Bot. 2006, 72, 211–216. [Google Scholar] [CrossRef][Green Version]
- Thwe, A.; Valan Arasu, M.; Li, X.; Park, C.H.; Kim, S.J.; Al-Dhabi, N.A.; Park, S.U. Effect of different Agrobacterium rhizogenes strains on hairy root induction and phenylpropanoid biosynthesis in Tartary buckwheat (Fagopyrum tataricum Gaertn). Front Microbiol. 2016, 7, 318. [Google Scholar] [CrossRef]
- Mariashibu, T.S.; Subramanyam, K.; Arun, M.; Mayavan, S.; Rajesh, M.; Theboral, J.; Manickavasagam, M.; Ganapathi, A. Vacuum infiltration enhances the Agrobacterium-mediated genetic transformation in Indian soybean cultivars. Acta Physiol. Plant 2013, 35, 41–54. [Google Scholar] [CrossRef]
- Tiwari, R.K.; Trivedi, M.; Guang, Z.C.; Guo, G.Q.; Zheng, G.C. Genetic transformation of Gentiana macrophylla with Agrobacterium rhizogenes: Growth and production of secoiridoid glucoside gentiopicroside in transformed hairy root cultures. Plant Cell Rep. 2007, 26, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Sujatha, G.; Zdravković-Korać, S.; Ćalić, D.; Flamini, G.; Ranjitha Kumaria, B.D. High-efficiency Agrobacterium rhizogenes-mediated genetic transformation in Artemisia vulgaris: Hairy root production and essential oil analysis. Ind. Crop. Prod 2013, 44, 643–652. [Google Scholar] [CrossRef]
- Zhou, M.L.; Zhu, X.M.; Shao, J.R.; Tang, Y.X.; Wu, Y.M. Production and metabolic engineering of bioactive substances in plant hairy root culture. Appl. Microbiol. Biotechnol. 2011, 90, 1229–1239. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Sharma, A.; Prasad, B.; Gururaj, H.B.; Gokare, R. Agrobacterium rhizogenes mediated genetic transformation resulting in hairy root formation is enhanced by ultrasonication and acetosyringone. Electron. J. Biotechnol. 2006, 9, 4. [Google Scholar] [CrossRef]
- Ishida, Y.; Hiei, Y.; Komari, T. Agrobacterium-mediated transformation of maize. Nat. Protoc. 2007, 2, 1614–1621. [Google Scholar] [CrossRef]
- Beigmohammadi, M.; Sharafi, A.; Jafari, S. An optimized protocol for Agrobacterium rhizogenes-mediated genetic transformation of Citrullus Colocynthis. J. Appl. Biotechnol. Rep. 2019, 6, 113–117. [Google Scholar] [CrossRef][Green Version]
- Ogaki, M.; Furuichi, Y.; Kuroda, K.; Chin, D.P.; Ogawa, Y.; Mi, M. Importance of co-cultivation medium pH for successful Agrobacterium-mediated transformation of Lilium x formolongi. Plant Cell Rep. 2008, 27, 699–705. [Google Scholar] [CrossRef]
- Mano, H.; Fujii, T.; Sumikawa, N.; Hiwatashi, Y.; Hasebe, M. Development of an Agrobacterium-mediated stable transformation method for the sensitive plant Mimosa pudica. PLoS ONE 2014, 9, e88611. [Google Scholar] [CrossRef]
- Ankenbauer, R.G.; Nester, E.W. Sugar-mediated induction of Agrobacterium tumefaciens virulence genes: Structural specificity and activities of monosaccharides. J. Bacteriol. 1990, 172, 6442–6446. [Google Scholar] [CrossRef]
- Nilsson, O.; Olsson, O. Getting to the root: The role of the Agrobacterium rhizogenes rol-genes in the formation of hairy roots. Physiol. Plant 1997, 100, 463–473. [Google Scholar] [CrossRef]
- Georgiev, M.I.; Agostini, E.; Ludwig-Muller, J.; Xu, J. Genetically transformed roots: From plant disease to biotechnological resource. Trends Biotechnol. 2012, 30, 528–537. [Google Scholar] [CrossRef] [PubMed]

| Culture Medium | Protocol 1 | Protocol 2 |
|---|---|---|
| Inoculation (MS) | MS: 4.4 g/L | MS/MES/vitamins: 4.9 g/L |
| Sucrose: 30 g/L | Sucrose: 30 g/L | |
| - | Acetosyringone: 150 μM | |
| Co-cultivation | MS: 2.2 g/L | MS/MES/vitamins: 2.2 g/L |
| Sucrose: 20 g/L | Sucrose: 20 g/L | |
| Agar: 9.0 g/L | Agar: 9.0 g/L | |
| - | Acetosyringone: 100 μM | |
| Emergence | MS: 2.2 g/L | MS/MES/vitamins: 2.46 g/L |
| Sucrose: 30 g/L | Sucrose: 30 g/L | |
| Agar: 9.0 g/L | Agar: 9.0 g/L |
| A. rhizogenes Strain | N. Inoculated Explants | Time to Hairy Root Emergence (dpt) | Seedlings That Formed Hairy Roots (%) 1 | MEAN (S) | |||
|---|---|---|---|---|---|---|---|
| 1 2 | 2 | 1 | 2 | 1 | 2 | ||
| R1000 | 60 | 60 | 12 ± 1 | 10 ± 1 | 76.7a 3 | 95.0a | 85.8a |
| K599 | 60 | 60 | 16 ± 1 | 15 ± 1 | 10.0b | 38.3c | 24.15b |
| Arqua | 60 | 60 | - | 15 ± 1 | 0.0b | 56.7b | 28.3b |
| MEAN (P) | 28.9a | 63.3b | |||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Foti, C.; Pavli, O.I. High-Efficiency Agrobacterium rhizogenes-Mediated Transgenic Hairy Root Induction of Lens culinaris. Agronomy 2020, 10, 1170. https://doi.org/10.3390/agronomy10081170
Foti C, Pavli OI. High-Efficiency Agrobacterium rhizogenes-Mediated Transgenic Hairy Root Induction of Lens culinaris. Agronomy. 2020; 10(8):1170. https://doi.org/10.3390/agronomy10081170
Chicago/Turabian StyleFoti, Chrysanthi, and Ourania I. Pavli. 2020. "High-Efficiency Agrobacterium rhizogenes-Mediated Transgenic Hairy Root Induction of Lens culinaris" Agronomy 10, no. 8: 1170. https://doi.org/10.3390/agronomy10081170
APA StyleFoti, C., & Pavli, O. I. (2020). High-Efficiency Agrobacterium rhizogenes-Mediated Transgenic Hairy Root Induction of Lens culinaris. Agronomy, 10(8), 1170. https://doi.org/10.3390/agronomy10081170





