Genetics of Height and Branching in Faba Bean (Vicia faba)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Hybridization
2.3. Growing Conditions
2.3.1. Growth Chamber
2.3.2. Field
2.4. Phenotyping
2.5. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hedden, P. The gene of the Green Revolution. Trends Genet. 2003, 19, 5–9. [Google Scholar] [CrossRef]
- Huyghe, C. Genetics and genetic modifications of plant architecture in grain legumes: A review. Agronomie 1998, 18, 383–411. [Google Scholar] [CrossRef]
- Bond, D.A.; Fyfe, J.L. Breeding field beans. Plant Breed. 1962, 4–26. [Google Scholar]
- Sjödin, J. Induced morphological variation in Vicia faba L. Hereditas 1971, 67, 155–180. [Google Scholar] [CrossRef]
- Ward, S.; Chapman, G.P. (Eds.) ICARDA (International Center for Agricultural Research in the Dry Areas). In Third Conspectus of Genetic Variation within Vicia faba; Faba Bean Information Service: Aleppo, Syria, 1986; p. 54. [Google Scholar]
- Filippetti, A. Inheritance of dwarf growth habit, induced in Vicia faba L. var major by ethyl methane sulfonate (EMS). FABIS Newsl. 1988, 20, 15–18. [Google Scholar]
- Fukuta, N.; Fujiok, S.; Takatsuto, S.; Yoshida, S.; Fukuta, Y.; Nakayama, M. ‘Rinrei’, a brassinosteroid-deficient dwarf mutant of faba bean (Vicia faba L.). Physiol. Plant. 2004, 121, 506–512. [Google Scholar] [CrossRef]
- Fukuta, N.; Fukuzono, K.; Kawaide, H.; Abe, H.; Nakayama, M. Physical restriction of pods causes seed size reduction of a brassinosteroid-deficient faba bean (Vicia faba). Ann. Bot. 2006, 97, 65–69. [Google Scholar] [CrossRef] [Green Version]
- van Norel, A.; Hoogendoorn, J. Faba bean dwarf selections outyield Dutch top varieties. FABIS Newsl. 1989, 25, 10–13. [Google Scholar]
- Webb, A.; Cottage, A.; Wood, T.; Khamassi, K.; Hobbs, D.; Gostkiewicz, K.; White, M.; Khazaei, H.; Ali, M.; Street, D.; et al. A SNP-based consensus genetic map for synteny-based trait targeting in faba bean (Vicia faba L.). Plant. Biotech. J. 2016, 14, 177–185. [Google Scholar] [CrossRef]
- Ogas, J. Plant hormones: Dissecting the gibberellin response pathway. Curr. Biol. 1998, 8, 165–167. [Google Scholar] [CrossRef] [Green Version]
- Peng, J.R.; Richards, D.E.; Hartley, N.M.; Murphy, G.P.; Devos, K.M.; Flintham, J.E.; Beales, J.; Fish, L.J.; Worland, A.J.; Pelica, F.; et al. ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 1999, 400, 256–261. [Google Scholar] [CrossRef]
- Bajguz, A.; Tretyn, A. The chemical characteristic and distribution of brassinosteroids in plants. Phytochemistry 2003, 62, 1027–1046. [Google Scholar] [CrossRef]
- Nomura, T.; Nakayama, M.; Reid, J.B.; Takeuchi, Y.; Yokota, T. Blockage of brassinosteroid biosynthesis and sensitivity cause dwarfism in garden pea. Plant. Physiol. 1997, 113, 31–37. [Google Scholar] [CrossRef] [Green Version]
- Kwon, M.; Choe, S. Brassinosteroid biosynthesis and dwarf mutants. J. Plant. Biol. 2005, 48, 1. [Google Scholar] [CrossRef]
- Castorina, G.; Consonni, G. The role of brassinosteroids in controlling plant height in Poaceae: A genetic perspective. Int. J. Mol. Sci. 2020, 21, 1191. [Google Scholar] [CrossRef] [Green Version]
- Pilbeam, C.J.; Hebblethwaite, P.D.; Clark, A.S. Effect of different inter-row spacings on faba beans of different form. Field Crops Res. 1989, 21, 203–214. [Google Scholar] [CrossRef]
- Spies, J.M.; Warkentin, T.D.; Shirtliffe, S.J. Variation in field pea (Pisum sativum) cultivars for basal branching and weed competition. Weed Sci. 2011, 59, 218–223. [Google Scholar] [CrossRef]
- Franklin, K.A. Shade avoidance. New Phytol. 2008, 179, 930–944. [Google Scholar] [CrossRef]
- Duc, G. Faba bean (Vicia faba L.). Field Crops Res. 1997, 53, 99–109. [Google Scholar] [CrossRef]
- Zong, X.; Cheng, X.; Wang, S. Food legume crops. In Crops and Its Relative Species in China Grain Crops; Yuchen, D., Diansheng, Z., Eds.; China Agriculture: Beijing, China, 2006; pp. 406–479. [Google Scholar]
- Khazaei, H.; Stoddard, F.L.; Purves, R.W.; Vandenberg, A. A multi-parent faba bean (Vicia faba L.) population for future genomic studies. Plant Genet. Resour. 2018, 16, 419–423. [Google Scholar] [CrossRef] [Green Version]
- Khazaei, H.; Wach, D.; Pecio, A.; Vandenberg, A.; Stoddard, F.L. Genetic analysis of photosynthesis-related traits in faba bean (Vicia faba L.) for crop improvement. Plant. Breed. 2019, 138, 761–769. [Google Scholar] [CrossRef]
- R Core TeamR. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: http://www.R-project.org/ (accessed on 20 June 2020).
- Knott, C.M. A key for stages of development of the faba bean (Vicia faba). Ann. Appl. Biol. 1990, 116, 391–404. [Google Scholar] [CrossRef]
- Foundation for Arable Research. Faba Beans—A Growers’ Guide. FAR Focus, 2012. Available online: https://www.far.org.nz/assets/files/uploads/26313_FAR_focus_8_-_faba_beans.pdf (accessed on 28 June 2020).
- Milach, S.C.K.; Federizzi, L.C. Dwarfing genes in plant improvement. Adv. Agron. 2001, 73, 35–63. [Google Scholar]
- Heath, M.C.; Pilbeam, C.J.; McKenzie, B.A.; Hebblethwaite, P.D. Plant architecture, competitive ability and crop productivity in food legumes with particular emphasis on pea (Pisum sativum L.) and faba bean (Vicia faba L.). In Expanding the Production and Use of Cool Season Food Legumes. Current Plant Science and Biotechnology in Agriculture; Muehlbauer, F.J., Kaiser, W.J., Eds.; Springer: Dordrecht, The Netherlands, 1994; Volume 19, pp. 771–790. [Google Scholar]
- Mathan, J.; Bhattacharya, J.; Ranjan, A. Enhancing crop yield by optimizing plant developmental features. Development 2016, 143, 3283–3294. [Google Scholar] [CrossRef] [Green Version]
- Clark, D.; Klee, H.; Dandekar, A. Despite benefits, commercialization of transgenic horticultural crops lags. Calif. Agric. 2004, 58, 89–98. [Google Scholar] [CrossRef] [Green Version]
- Filippetti, A. Inheritance of determinate growth habit induced in Vicia faba major by ethyl methane sulphonate (EMS). FABIS Newsl. 1986, 15, 12–14. [Google Scholar]
- Avila, C.; Atienza, S.; Moreno, M.; Torres, A. Development of a new diagnostic marker for growth habit selection in faba bean (Vicia faba L.) breeding. Theor. Appl. Genet. 2007, 115, 1075–1082. [Google Scholar] [CrossRef]
- Cottage, A.; Gostkiewicz, K.; Thomas, J.; Borrows, R.; Torres, A.M.; O’Sullivan, D.M. Heterozygosity and diversity analysis using mapped single nucleotide polymorphisms in a faba bean inbreeding programme. Mol. Breed. 2012, 30, 1799–1809. [Google Scholar] [CrossRef]
- Abu-Amer, J.H.; Saoub, H.M.; Akash, M.W.; Al-Abdallat, A.M. Genetic and phenotypic variation among faba bean landraces and cultivars. Int. J. Veg. Sci. 2010, 17, 45–59. [Google Scholar] [CrossRef]
- Busov, V.B.; Brunner, A.M.; Strauss, S.H. Genes for control of plant stature and form. New Phytol. 2008, 177, 589–607. [Google Scholar] [CrossRef]
- Wang, Y.; Li, J. Molecular basis of plant architecture. Ann. Rev. Plant. Biol. 2008, 59, 253–279. [Google Scholar] [CrossRef]
- Sun, Z.; Su, C.; Yun, J.; Jiang, Q.; Wang, L.; Wang, Y.; Cao, D.; Zhao, F.; Zhao, Q.; Zhang, M.; et al. Genetic improvement of the shoot architecture and yield in soya bean plants via the manipulation of GmmiR156b. Plant. Biotech. J. 2019, 17, 50–62. [Google Scholar] [CrossRef] [Green Version]
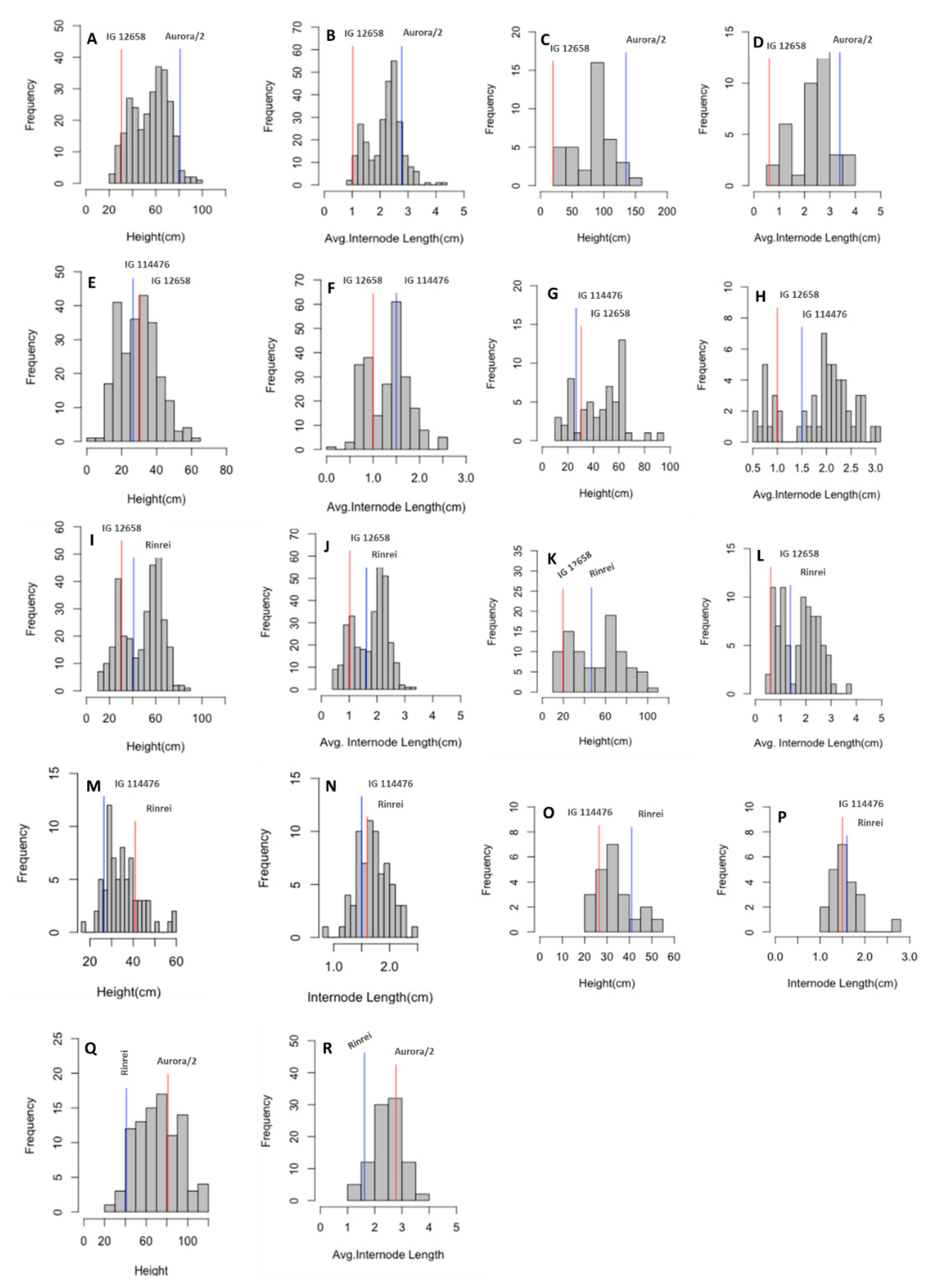
| Parental Line/F2 Cross | Height (cm) | Internode Length (cm) | |||
|---|---|---|---|---|---|
| Mean ± SD | Range | Mean ± SD | Range | ||
| Aurora/2 | Field | 81.00 ± 1.19 | 71–96 | 2.79 ± 0.55 | 2.29–3.69 |
| Growth chamber | 135.00 ± 16.06 | 112–147 | 3.39 ± 0.30 | 3.06–3.73 | |
| IG 114476 | Field | 26.57 ± 2.51 | 23–30 | 1.52 ± 0.30 | 0.91–1.86 |
| Growth chamber | 33.17 ± 8.08 | 23–45 | 1.75 ± 0.11 | 1.53–1.86 | |
| IG 12658 | Field | 30.57 ± 2.37 | 27–34 | 1.02 ± 0.09 | 0.94–1.15 |
| Growth chamber | 19.71 ± 0.95 | 19–21 | 0.62 ± 0.07 | 0.51–0.72 | |
| Rinrei | Field | 41.00 ± 6.08 | 37–48 | 1.62 ± 0.13 | 1.48–1.73 |
| Growth chamber | 46.75 ± 5.50 | 39–53 | 1.40 ± 0.13 | 1.28–1.68 | |
| Aurora/2 × IG 12658 | Field | 56.14 ± 15.54 | 20–99 | 2.20 ± 0.59 | 0.97–4.30 |
| Growth chamber | 85.55 ± 30.67 | 31–146 | 2.39 ± 0.81 | 0.84–3.76 | |
| IG 114476 × IG 12658 | Field | 30.07 ± 10.91 | 9–64 | 1.33 ± 0.44 | 0.56–2.56 |
| Growth chamber | 45.72 ± 18.20 | 12–94 | 1.86 ± 0.68 | 0.58–3.03 | |
| Rinrei × IG 12658 | Field | 48.60 ± 17.30 | 11–90 | 1.77 ± 0.61 | 0.44–3.29 |
| Growth chamber | 51.67 ± 24.33 | 14–105 | 1.72 ± 0.77 | 0.45–3.75 | |
| IG 114476 × Rinrei | Field | 35.99 ± 8.35 | 17–70 | 1.75 ± 0.31 | 0.90–3.11 |
| Growth chamber | 33.87 ± 7.83 | 20–52 | 1.60 ± 0.29 | 1.14–2.69 | |
| Rinrei × Aurora/2 | Field | 72.60 ± 20.33 | 29–117 | 2.50 ± 0.53 | 1.19–4.00 |
| S.E.M. 1 | 1.30 | 0.20 | |||
| Cross | F1 | F2 | ||||
|---|---|---|---|---|---|---|
| Normal | Dwarf | χ2(3:1), df = 1 | p | |||
| dwf1 gene | ||||||
| Aurora/2 × IG 12658 | Field | Normal | 194 | 79 | 2.258 | 0.133 |
| Growth chamber | Normal | 30 | 8 | 0.316 | 0.574 | |
| IG 114476 × IG 12658 | Field | Normal | 168 | 71 | 2.824 | 0.093 |
| Growth chamber | Normal | 44 | 17 | 0.268 | 0.605 | |
| Rinrei × IG 12658 | Field | Normal | 232 | 82 | 0.208 | 0.648 |
| Growth chamber | Normal | 68 | 20 | 0.242 | 0.622 | |
| Pooled | 736 | 277 | 2.970 | 0.085 | ||
| dwarf1 gene | Normal | Semi-dwarf | χ2(3:1), df = 1 | p | ||
| Rinrei × IG 12658 | Field | Normal | 244 | 70 | 1.227 | 0.268 |
| Growth chamber | Normal | 59 | 29 | 2.970 | 0.085 | |
| IG 114476 × Rinrei 1 | Field | Normal | 94 | 50 | 7.259 | 0.007 |
| Growth chamber | Normal | 12 | 2 | 0.857 | 0.355 | |
| Rinrei × Aurora/2 | Field | Normal | 75 | 18 | 1.581 | 0.209 |
| Pooled | 484 | 169 | 0.270 | 0.603 |
| F2 | ||||||
|---|---|---|---|---|---|---|
| Normal | Rinrei Semi-Dwf 1 | IG 12658 Dwf | Double Dwf | |||
| χ2(9:7), df = 1 | P | |||||
| Field | 182 | 132 | - | 0.374 | 0.541 | |
| Growth chamber | 46 | 42 | - | 0.566 | 0.452 | |
| χ2(9:3:3:1), df = 3 | ||||||
| Field | 182 | 50 | 62 | 20 | 1.674 | 0.643 |
| Growth chamber | 46 | 22 | 13 | 7 | 3.232 | 0.357 |
| χ2(15:1), df = 1 | ||||||
| Field | 294 | - | - | 20 | 0.008 | 0.930 |
| Growth chamber | 81 | - | - | 7 | 0.436 | 0.509 |
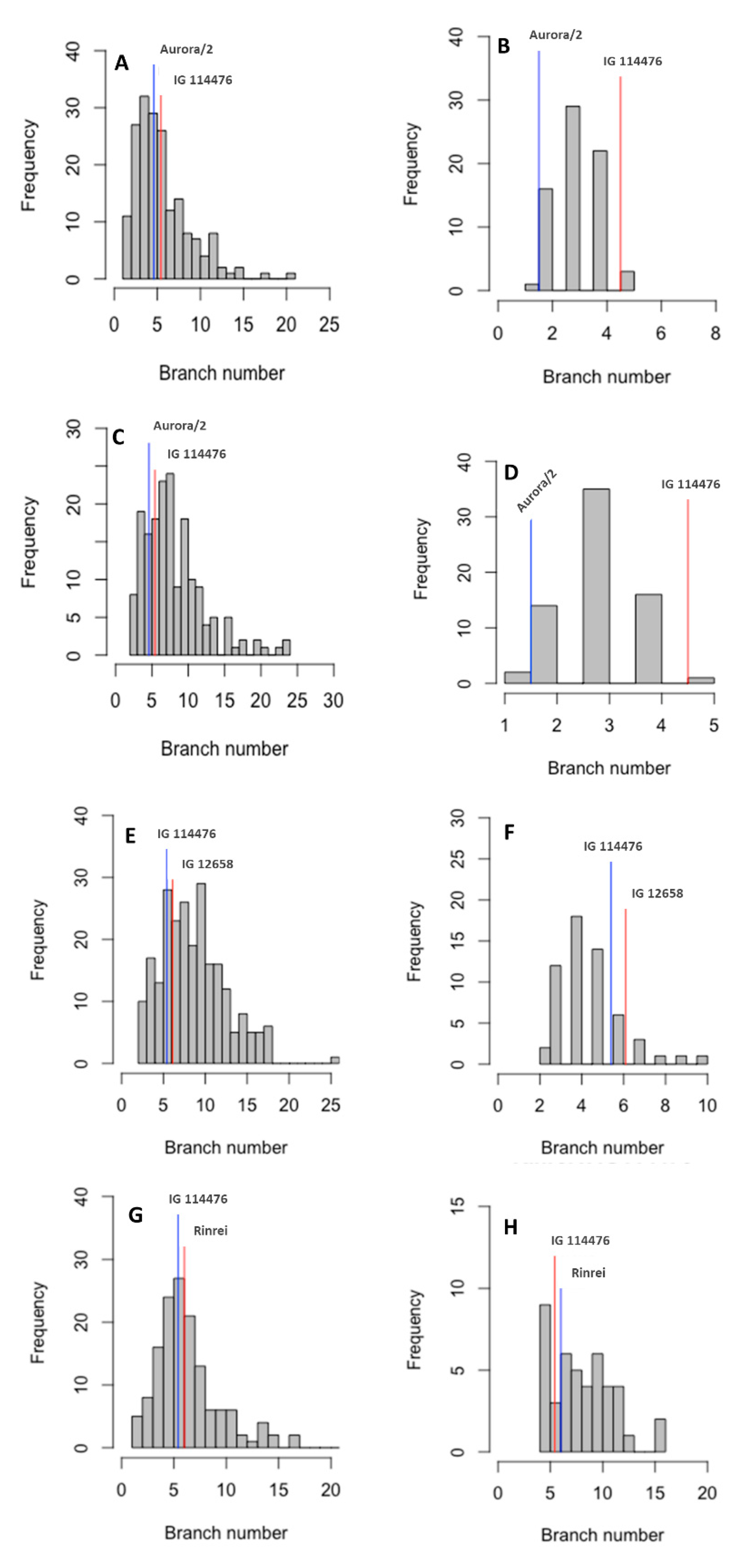
| Parental Line/F2 Cross | Number of Branches | ||
|---|---|---|---|
| Mean ± SD | Range | ||
| Aurora/2 | Field | 5.43 ± 1.81 | 3–6 |
| Growth chamber | 1.50 ± 0.58 | 1–2 | |
| IG 114476 | Field | 4.60 ± 1.52 | 3–8 |
| Growth chamber | 4.50 ± 0.84 | 3–6 | |
| IG 12658 | Field | 6.14 ± 1.07 | 5–8 |
| Growth chamber | 6.14 ± 1.00 | 5–8 | |
| Rinrei | Field | 6.00 ± 1.0 | 5–7 |
| Growth chamber | - | - | |
| Aurora/2 × IG 114476 | Field | 8.50 ± 4.20 | 2–24 |
| Growth chamber | 3.00 ± 0.79 | 1–5 | |
| IG 114476 × Aurora/2 | Field | 6.04 ± 3.24 | 1–21 |
| Growth chamber | 3.14 ± 0.87 | 1–5 | |
| IG 114476 × IG 12658 | Field | 9.07 ± 3.68 | 2–26 |
| Growth chamber | 4.59 ± 1.58 | 2–10 | |
| Rinrei × IG 114476 | Field | 8.52 ± 3.00 | 4–16 |
| IG 114476 × Rinrei | Field | 6.86 ± 3.30 | 1–22 |
| S.E.M. 1 | 0.38 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hughes, J.; Khazaei, H.; Vandenberg, A. Genetics of Height and Branching in Faba Bean (Vicia faba). Agronomy 2020, 10, 1191. https://doi.org/10.3390/agronomy10081191
Hughes J, Khazaei H, Vandenberg A. Genetics of Height and Branching in Faba Bean (Vicia faba). Agronomy. 2020; 10(8):1191. https://doi.org/10.3390/agronomy10081191
Chicago/Turabian StyleHughes, Jessa, Hamid Khazaei, and Albert Vandenberg. 2020. "Genetics of Height and Branching in Faba Bean (Vicia faba)" Agronomy 10, no. 8: 1191. https://doi.org/10.3390/agronomy10081191
APA StyleHughes, J., Khazaei, H., & Vandenberg, A. (2020). Genetics of Height and Branching in Faba Bean (Vicia faba). Agronomy, 10(8), 1191. https://doi.org/10.3390/agronomy10081191







