Adaptation of Grass Pea (Lathyrus sativus) to Mediterranean Environments
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Experimental Design
2.2. Statistical Analysis
2.3. AMMI Analysis
3. Results
3.1. Model Diagnosis
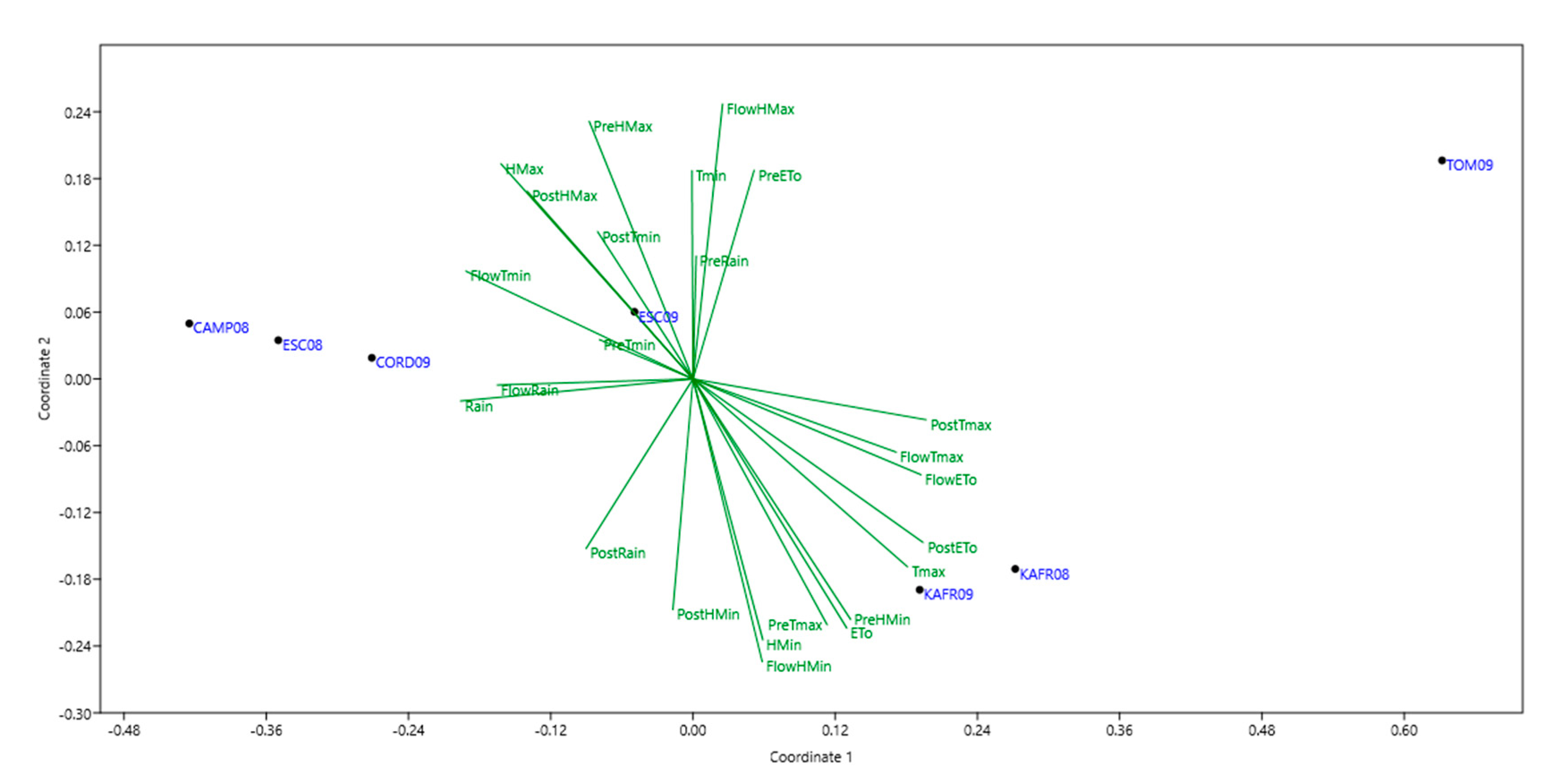
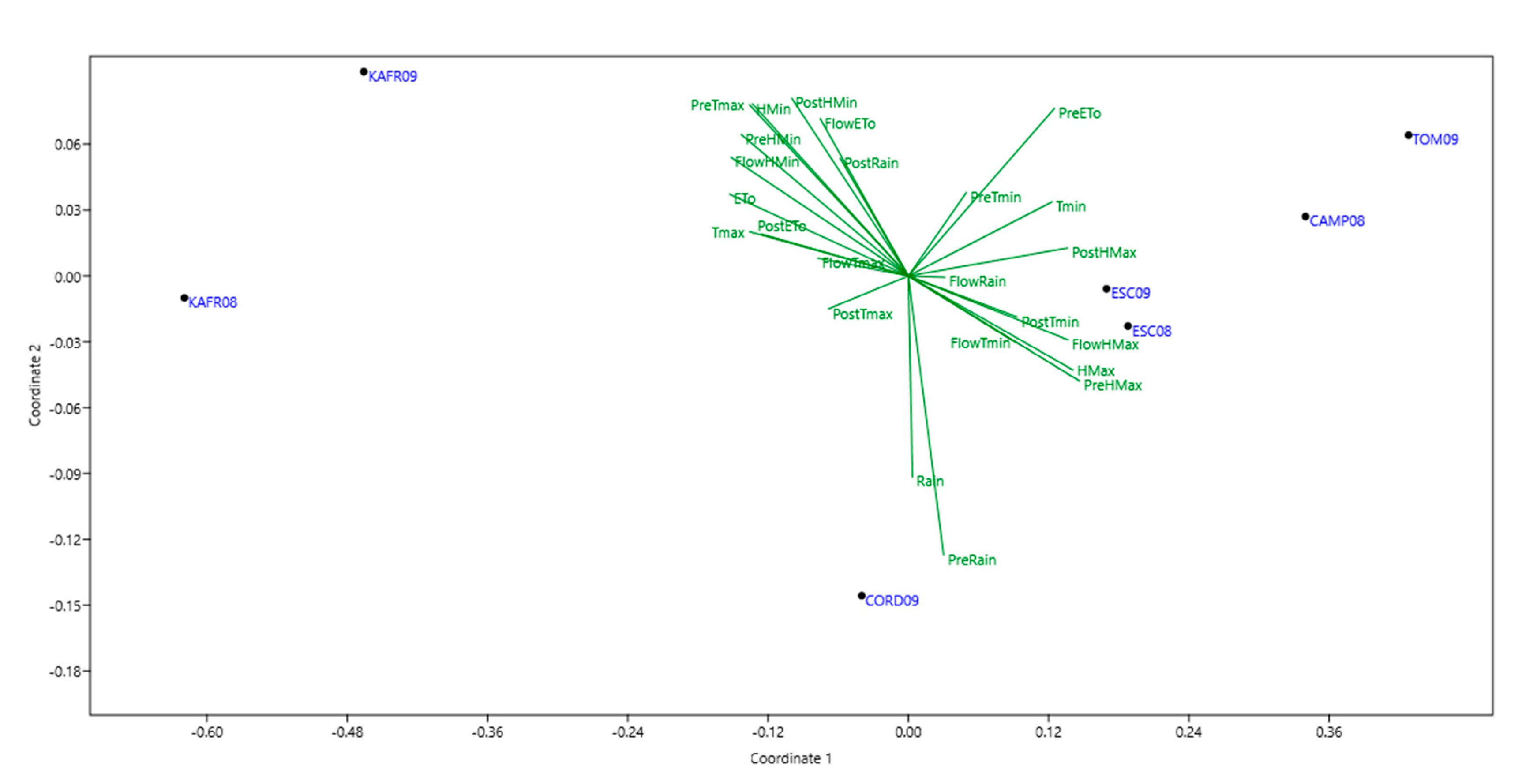
3.2. Mega-Environments (MEs) Delineation and Description
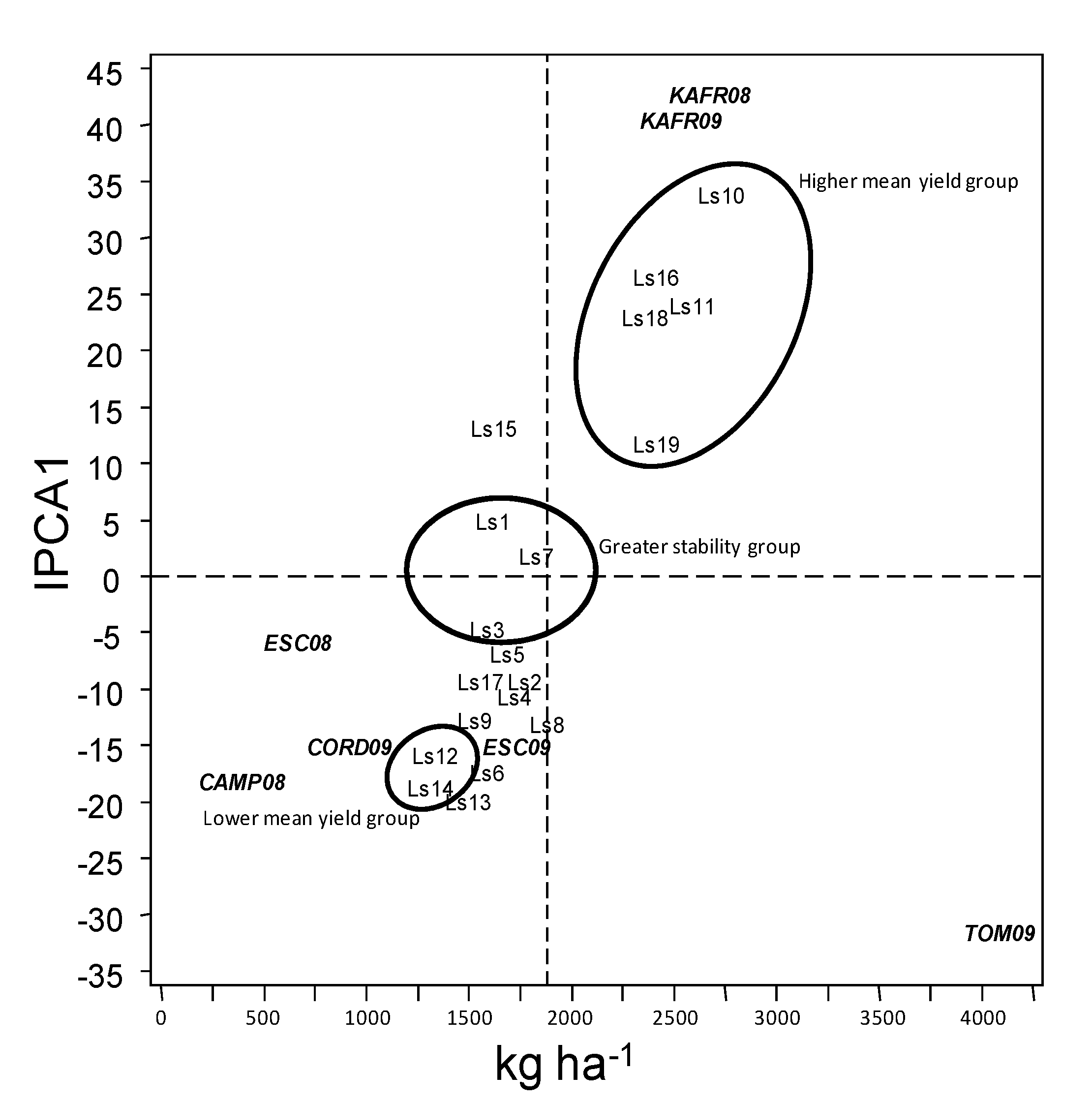
3.3. Selection or Recommendation of the Best Accessions
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Almeida, N.F.; Leitão, S.T.; Krezdorn, N.; Rotter, B.; Winter, P.; Rubiales, D.; Vaz Patto, M.C. Allelic diversity in the transcriptomes of contrasting rust-infected genotypes of Lathyrus sativus, a lasting resource for smart breeding. BMC Plant Biol. 2014, 14, 376. [Google Scholar] [CrossRef] [PubMed]
- Vaz Patto, M.C.; Skiba, B.; Pang, E.C.K.; Ochatt, S.J.; Lambein, F.; Rubiales, D. Lathyrus improvement for resistance against biotic and abiotic stresses: From classical breeding to marker assisted selection. Euphytica 2006, 147, 133–147. [Google Scholar] [CrossRef]
- Girma, D.; Korbu, L. Genetic improvement of grass pea (Lathyrus sativus) in Ethiopia: An unfulfilled promise. Plant Breed. 2012, 131, 231–236. [Google Scholar] [CrossRef]
- Kumar, S.; Bejiga, G.; Ahmed, S.; Nakkoul, H.; Sarker, A. Genetic improvement of grass pea for low neurotoxin (B-ODAP) content. Food Chem. Toxicol. 2011, 3, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Gonςalves, L.; Rubiales, D.; Vaz Patto, M.C. Grass pea prospective at the Mediterranean Basin. Legume Perspect. 2015, 10, 8–9. [Google Scholar]
- Lambein, F.; Travella, S.; Kuo, Y.; Van Montagu, M.; Heidje, M. Grass pea (Lathyrus sativus L.): Orphan crop, nutraceutical or just plain food? Planta 2019, 250, 821–838. [Google Scholar] [CrossRef]
- De-la-Rosa, L.; Martí, I. Morphological characterization of Spanish genetic resources of Lathyrus sativus L. Lathyrus Lathyrism Newsl. 2001, 2, 31–34. [Google Scholar]
- Tavoletti, S.; Iommarini, L.; Crinò, P.; Granati, E. Collection and evaluation of grass pea (Lathyrus sativus L.) germplasm of central Italy. Plant Breed. 2005, 124, 388–391. [Google Scholar] [CrossRef]
- Piergiovanni, A.R.; Lupo, F.; Zaccardelli, M. Environmental effect on yield, composition and technological seed traits of some Italian ecotypes of grass pea (Lathyrus sativus L.). J. Sci. Food Agric. 2011, 91, 122–129. [Google Scholar] [CrossRef]
- Grela, E.R.; Rybiński, W.; Matras, J.; Sobolewska, S. Variability of phenotypic and morphological characteristics of some Lathyrus sativus L. and Lathyrus cicera L. accessions and nutritional traits of their seeds. Genet. Resour. Crop Evol. 2012, 59, 1687–1703. [Google Scholar] [CrossRef]
- Aci, M.M.; Lupini, A.; Badagliacca, G.; Mauceri, A.; Lo Presti, E.; Preiti, G. Genetic diversity among Lathyrus ssp. based on agronomic traits and molecular markers. Agronomy 2020, 10, 1182. [Google Scholar] [CrossRef]
- Dixit, G.P.; Parihar, A.K.; Bohra, A.; Singh, N.P. Achievements and prospects of grass pea (Lathyrus sativus L.) improvement for sustainable food production. Crop J. 2016, 4, 407–416. [Google Scholar] [CrossRef]
- Hillocks, R.J.; Maruthi, M.N. Grass pea (Lathyrus sativus): Is there a case for further crop improvement? Euphytica 2012, 186, 647–654. [Google Scholar] [CrossRef]
- Fikre, A.; Negwo, T.; Kuo, Y.H.; Lambein, F.; Ahmed, S. Climatic, edaphic and altitudinal factors affecting yield and toxicity of Lathyrus sativus grown at five locations in Ethiopia. Food Chem. Toxicol. 2011, 49, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Jiao, C.J.; Jiang, J.L.; Ke, L.M.; Cheng, W.; Li, F.M.; Li, Z.X.; Wang, C.Y. Factors affecting beta-ODAP content in Lathyrus sativus and their possible physiological mechanisms. Food Chem. Toxicol. 2011, 49, 543–549. [Google Scholar] [CrossRef]
- Getahun, H.; Lambein, F.; Vanhoorne, M.; Van der Stuyft, P. Food-aid cereals to reduce neurolathyrism related to grass-pea preparations during famine. Lancet 2003, 362, 1808–1810. [Google Scholar] [CrossRef]
- Buta, M.B.; Emire, S.A.; Posten, C.; André, S.; Greiner, R. Reduction of β-ODAP and IP6 contents in Lathyrus sativus L. seed by high hydrostatic pressure. Food Res. Int. 2019, 120, 73–82. [Google Scholar] [CrossRef]
- Gauch, H.G. Model selection and validation for yield trials with interaction. Biometrics 1988, 88, 705–715. [Google Scholar] [CrossRef]
- Flores, F.; Moreno, M.T.; Cubero, J.I. A comparison of univariate and multivariate methods to analyze G*E interaction. Field Crops Res. 1998, 56, 271–286. [Google Scholar] [CrossRef]
- McIntosh, M.S. Analysis of combined experiments. Agron. J. 1983, 75, 153–155. [Google Scholar] [CrossRef]
- Zobel, R.W.; Wright, M.J.; Gauch, H.G. Statistical analysis of a yield trial. Agron. J. 1988, 80, 388–393. [Google Scholar] [CrossRef]
- Gauch, H.G. A simple protocol for AMMI analysis of yield trials. Crop Sci. 2013, 53, 1860–1869. [Google Scholar] [CrossRef]
- Iglesias-García, R.; Prats, E.; Flores, F.; Amri, M.; Mikic, A.; Rubiales, D. Assessment of field pea (Pisum sativum L.) grain yield, aerial biomass and flowering date stability in Mediterranean environments. Crop Past. Sci. 2017, 68, 915–923. [Google Scholar] [CrossRef]
- Burgueño, J.; Crossa, J.; Vargas, M. SAS Programs for Graphing GE and GGE Biplots; Biometrics and Statistics Unit, CIMMYT, Int.: México City, Mexico, 2003. [Google Scholar]
- Malik, W.A.; Forkman, J.; Piepho, H.P. Testing multiplicative terms in AMMI and GGE models for multienvironment trials with replicates. Theor. App. Gen. 2019, 132, 2087. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 2001, 26, 32–46. [Google Scholar]
- Gollob, H.F. A statistical model which combines features of factor analytic and analysis of variance techniques. Psychometrika 1968, 33, 73–115. [Google Scholar] [CrossRef]
- Hanbury, C.D.; Siddique, K.H.M.; Galwey, N.W.; Cocks, P.S. Genotype-environment interaction for seed yield and ODAP concentration of Lathyrus sativus L. and L. cicera L. in Mediterranean type environments. Euphytica 1999, 110, 45–60. [Google Scholar] [CrossRef]
- Vaz Patto, M.C.; Rubiales, D. Lathyrus diversity: Available resources with relevance to crop improvement. Ann. Bot. 2014, 113, 895–908. [Google Scholar] [CrossRef]
- Fernández-Aparicio, M.; Flores, F.; Rubiales, D. Escape and true resistance to crenate broomrape (Orobanche crenata Forsk.) in grass pea (Lathyrus sativus L.) germplasm. Field Crops Res. 2011, 125, 92–97. [Google Scholar] [CrossRef]
- Fernández-Aparicio, M.; Flores, F.; Rubiales, D. The effect of Orobanche crenata infection severity in faba bean, field pea, and grass pea productivity. Front. Plant Sci. 2016, 7, 1409. [Google Scholar] [CrossRef]
- Rubiales, D.; Alcántara, C.; Pérez-de-Luque, A.; Gil, J.; Sillero, J.C. Infection of chickpea (Cicer arietinum) by crenate broomrape (Orobanche crenata) as influenced by sowing date and weather conditions. Agronomie 2003, 23, 359–362. [Google Scholar] [CrossRef]
- Rubiales, D.; Pérez-de-Luque, A.; Cubero, J.I.; Sillero, J.C. Crenate broomrape (Orobanche crenata) infection in field pea cultivars. Crop Prot. 2003, 22, 865–872. [Google Scholar] [CrossRef]
- Pérez-de-Luque, A.; Flores, F.; Rubiales, D. Differences in crenate broomrape parasitism dynamics on three legume crops using a Thermal Time Model. Front. Plant Sci. 2016, 7, 1910. [Google Scholar] [CrossRef] [PubMed]
- Pérez-de-Luque, A.; Sillero, J.C.; Moral, A.; Cubero, J.I.; Rubiales, D. Effect of sowing date and host resistance on the establishment of Orobanche crenata in faba bean and common vetch. Weed Res. 2004, 44, 282–288. [Google Scholar] [CrossRef]
- Rubiales, D. Parasitic plants, wild relatives and the nature of resistance. New Phytol. 2003, 160, 459–461. [Google Scholar] [CrossRef]

| Environm | Location | Soil Type | Soil pH | Latitude | Longit. | Altit. (m) | Season | AvTmax (°C) | AvTmin (°C) | Rain (mm) |
|---|---|---|---|---|---|---|---|---|---|---|
| CAMP08 | Campillo, Spain | Vertisol | 7.5–8 | 37°20′ N | 4°51′ W | 461 | 2007–08 | 18.8 | 7.8 | 264 |
| TOM09 | Tomejil, Spain | Vertisol | 7–7.5 | 37°30′ N | 5°57′ W | 12 | 2008–09 | 22.6 | 7.5 | 219 |
| CORD09 | Córdoba, Spain | Fluvisol | 6.5–7 | 37°50′ N | 4°50′ W | 90 | 2008–09 | 21.7 | 7.9 | 280 |
| ESC08 | Escacena, Spain | Fluvisol | 7–7.5 | 37°25′ N | 6°15′ W | 88 | 2007–08 | 20.7 | 10.1 | 391 |
| ESC09 | Escacena, Spain | Fluvisol | 7–7.5 | 37°25′ N | 6°15′ W | 88 | 2008–09 | 21.4 | 9.4 | 252 |
| KAFR08 | Kafr El-Sheik, Egypt | Entisol | 7.5–8 | 30°47′ N | 30°59′ E | 0 | 2007–08 | 23.9 | 5.4 | 276 B |
| KAFR09 | Kafr El-Sheik, Egypt | Entisol | 7.5–8 | 30°47′ N | 30°59′ E | 0 | 2008–09 | 23.4 | 8.3 | 193 B |
| Grain Yield (kg ha−1) in Different Environments | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| SPAIN | EGYPT | ||||||||
| Accession | Synom. | CAMP08 | CORD09 | ESC08 | ESC09 | TOM09 | KAFR08 | KAFR09 | Mean |
| Ls1 | Sel190 | 533 | 926 | 667 | 1567 | 2903 | 2599 | 2343 | 1648 |
| Ls2 | Sel288 | 340 | 1021 | 667 | 1747 | 4468 | 2189 | 2069 | 1786 |
| Ls3 | Sel289 | 340 | 1370 | 645 | 1614 | 3631 | 2302 | 1889 | 1685 |
| Ls4 | Sel290 | 507 | 978 | 440 | 1786 | 4398 | 1903 | 2267 | 1754 |
| Ls5 | Sel299 | 355 | 900 | 520 | 1638 | 4238 | 2465 | 1908 | 1718 |
| Ls6 | Sel387 | 467 | 934 | 300 | 1692 | 4402 | 1459 | 1569 | 1546 |
| Ls7 | Sel390 | 393 | 873 | 457 | 1620 | 4220 | 2737 | 2717 | 1860 |
| Ls8 | Sel449 | 320 | 1238 | 889 | 1984 | 4724 | 2237 | 1995 | 1912 |
| Ls9 | SelB111 | 311 | 942 | 600 | 1708 | 4343 | 2076 | 1884 | 1695 |
| Ls10 | SelB222 | 140 | 1556 | 1534 | 2343 | 3963 | 4999 | 4800 | 2762 |
| Ls11 | Sel2177 | 607 | 2011 | 908 | 1619 | 4325 | 4789 | 4087 | 2621 |
| Ls12 | Sel2119 | 207 | 685 | 329 | 1436 | 4035 | 1373 | 1507 | 1368 |
| Ls13 | SelETH-7 | 307 | 985 | 400 | 1745 | 4374 | 1214 | 1674 | 1528 |
| Ls14 | SelETH-15 | 160 | 716 | 334 | 1472 | 4146 | 1604 | 978 | 1344 |
| Ls15 | Sel945 | 440 | 534 | 806 | 1637 | 2565 | 2800 | 2787 | 1653 |
| Ls16 | Sel1784 | 613 | 515 | 1045 | 2688 | 3892 | 3427 | 5092 | 2468 |
| Ls17 | Sel1942 | 338 | 840 | 480 | 1590 | 4256 | 2117 | 1861 | 1640 |
| Ls18 | Sel1959 | 547 | 1197 | 1227 | 1464 | 4083 | 4808 | 3399 | 2389 |
| Ls19 | Lisa | 810 | 753 | 703 | 1988 | 4985 | 3988 | 3567 | 2399 |
| Mean | 407 | 999 | 682 | 1755 | 4103 | 2689 | 2547 | 1883 | |
| SE | 30 | 57 | 52 | 50 | 117 | 208 | 178 | 76 | |
| Accession | CAMP08 | CORD09 | ESC08 | ESC09 | TOM09 | KAFR08 | KAFR09 | Mean |
|---|---|---|---|---|---|---|---|---|
| Ls1 | 0.13 | 0.48 | 0.37 | 0.34 | 0.0 | 0.50 | 0.56 | 0.40 |
| Ls2 | 0.23 | 1.15 | 0.63 | 0.66 | 0.0 | 2.18 | 0.86 | 0.95 |
| Ls3 | 0.10 | 0.40 | 0.30 | 0.47 | 0.0 | 0.38 | 0.19 | 0.31 |
| Ls4 | 0.37 | 1.12 | 0.50 | 0.49 | 0.0 | 2.52 | 1.47 | 1.08 |
| Ls5 | 0.10 | 1.02 | 0.70 | 0.79 | 0.0 | 2.07 | 1.94 | 1.10 |
| Ls6 | 0.33 | 1.58 | 0.63 | 0.68 | 0.0 | 3.61 | 3.30 | 1.69 |
| Ls7 | 0.17 | 0.98 | 0.50 | 0.51 | 0.0 | 0.99 | 0.69 | 0.64 |
| Ls8 | 0.00 | 0.80 | 0.57 | 0.40 | 0.0 | 1.02 | 1.18 | 0.66 |
| Ls9 | 0.03 | 0.98 | 0.60 | 0.59 | 0.0 | 1.54 | 1.51 | 0.88 |
| Ls10 | 0.10 | 1.63 | 0.50 | 0.53 | 0.0 | 0.99 | 1.00 | 0.79 |
| Ls11 | 0.10 | 0.83 | 0.40 | 0.39 | 0.03 | 2.40 | 1.64 | 0.96 |
| Ls12 | 0.27 | 1.18 | 0.63 | 0.68 | 0.0 | 3.20 | 3.30 | 1.54 |
| Ls13 | 0.13 | 0.97 | 0.50 | 0.45 | 0.0 | 1.26 | 1.32 | 0.77 |
| Ls14 | 0.27 | 1.17 | 0.60 | 0.67 | 0.0 | 2.59 | 2.47 | 1.29 |
| Ls15 | 0.17 | 0.93 | 0.63 | 0.38 | 0.0 | 2.21 | 1.72 | 1.01 |
| Ls16 | 0.20 | 1.15 | 0.53 | 0.55 | 0.0 | 1.65 | 1.49 | 0.93 |
| Ls17 | 0.40 | 1.22 | 0.50 | 0.74 | 0.0 | 3.28 | 2.47 | 1.43 |
| Ls18 | 0.03 | 0.28 | 0.57 | 0.46 | 0.0 | 0.85 | 0.70 | 0.48 |
| Ls19 | 0.50 | 0.98 | 0.83 | 1.40 | 0.65 | 3.09 | 3.00 | 1.63 |
| Mean | 0.19 | 0.99 | 0.55 | 0.59 | 0.04 | 1.91 | 1.62 | 0.98 |
| SE | 0.03 | 0.08 | 0.02 | 0.03 | 0.03 | 0.14 | 0.14 | 0.05 |
| Source | DF | SS | MS | % Variation |
|---|---|---|---|---|
| Environment (E) | 6 | 594,938,551 | 99,156,425 *** | 76 A |
| Replication/E | 14 | 10,326,066 | 737,576 | |
| Genotype (G) | 18 | 68,746,582 | 3,819,254 *** | 9 A |
| G*E | 108 | 113,679,415 | 1,052,587 *** | 15 A |
| IACP1 | 23 | 82,737,353 | 3,597,276 *** | 73 B |
| IACP2 | 21 | 14,543,665 | 692,555 | 13 B |
| Residual | 64 | 16,398,397 | 256,224 | |
| Error | 252 | 120,081,873 | 476,515 | |
| Total | 398 | 907,772,488 |
| Source | DF | SS | MS | % Variation |
|---|---|---|---|---|
| Environment (E) | 6 | 171 | 28.5 *** | 60 A |
| Replication/E | 14 | 3 | 0.19 | |
| Genotype (G) | 18 | 48 | 2.64 *** | 17 A |
| G*E | 108 | 64 | 0.58 *** | 23 A |
| IACP1 | 23 | 54 | 2.32 *** | 84 B |
| IACP2 | 21 | 5 | 0.23 | 8 B |
| Residual | 64 | 5 | 0.08 | |
| Error | 252 | 38 | 0.15 | |
| Total | 398 | 322 |
| AMMI1 Rank for Grain Yield | AMMI1 Rank for Number Broomrapes Per Plant | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Environment | 1 | 2 | 3 | 4 | 5 | 6 | 1 | 2 | 3 | 4 | 5 | 6 |
| CAMP08 | Ls11 | Ls19 | Ls10 | Ls8 | Ls16 | Ls18 | Ls3 | Ls1 | Ls18 | Ls11 | Ls8 | Ls5 |
| CORD09 | Ls11 | Ls10 | Ls19 | Ls8 | Ls16 | Ls18 | Ls3 | Ls1 | Ls18 | Ls8 | Ls7 | Ls11 |
| ESC08 | Ls10 | Ls11 | Ls19 | Ls8 | Ls16 | Ls18 | Ls18 | Ls3 | Ls11 | Ls1 | Ls8 | Ls15 |
| ESC09 | Ls10 | Ls11 | Ls19 | Ls16 | Ls8 | Ls18 | Ls3 | Ls1 | Ls18 | Ls11 | Ls8 | Ls13 |
| KAFR08 | Ls10 | Ls11 | Ls16 | Ls18 | Ls19 | Ls15 | Ls3 | Ls1 | Ls18 | Ls7 | Ls10 | Ls8 |
| KAFR09 | Ls10 | Ls11 | Ls16 | Ls18 | Ls19 | Ls15 | Ls3 | Ls1 | Ls18 | Ls7 | Ls10 | Ls8 |
| TOM09 | Ls8 | Ls13 | Ls6 | Ls2 | Ls4 | Ls9 | ||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rubiales, D.; Emeran, A.A.; Flores, F. Adaptation of Grass Pea (Lathyrus sativus) to Mediterranean Environments. Agronomy 2020, 10, 1295. https://doi.org/10.3390/agronomy10091295
Rubiales D, Emeran AA, Flores F. Adaptation of Grass Pea (Lathyrus sativus) to Mediterranean Environments. Agronomy. 2020; 10(9):1295. https://doi.org/10.3390/agronomy10091295
Chicago/Turabian StyleRubiales, Diego, Amero A. Emeran, and Fernando Flores. 2020. "Adaptation of Grass Pea (Lathyrus sativus) to Mediterranean Environments" Agronomy 10, no. 9: 1295. https://doi.org/10.3390/agronomy10091295
APA StyleRubiales, D., Emeran, A. A., & Flores, F. (2020). Adaptation of Grass Pea (Lathyrus sativus) to Mediterranean Environments. Agronomy, 10(9), 1295. https://doi.org/10.3390/agronomy10091295






