Salt Stress Induces Differentiated Nitrogen Uptake and Antioxidant Responses in Two Contrasting Barley Landraces from MENA Region
Abstract
1. Introduction
2. Materials and Methods
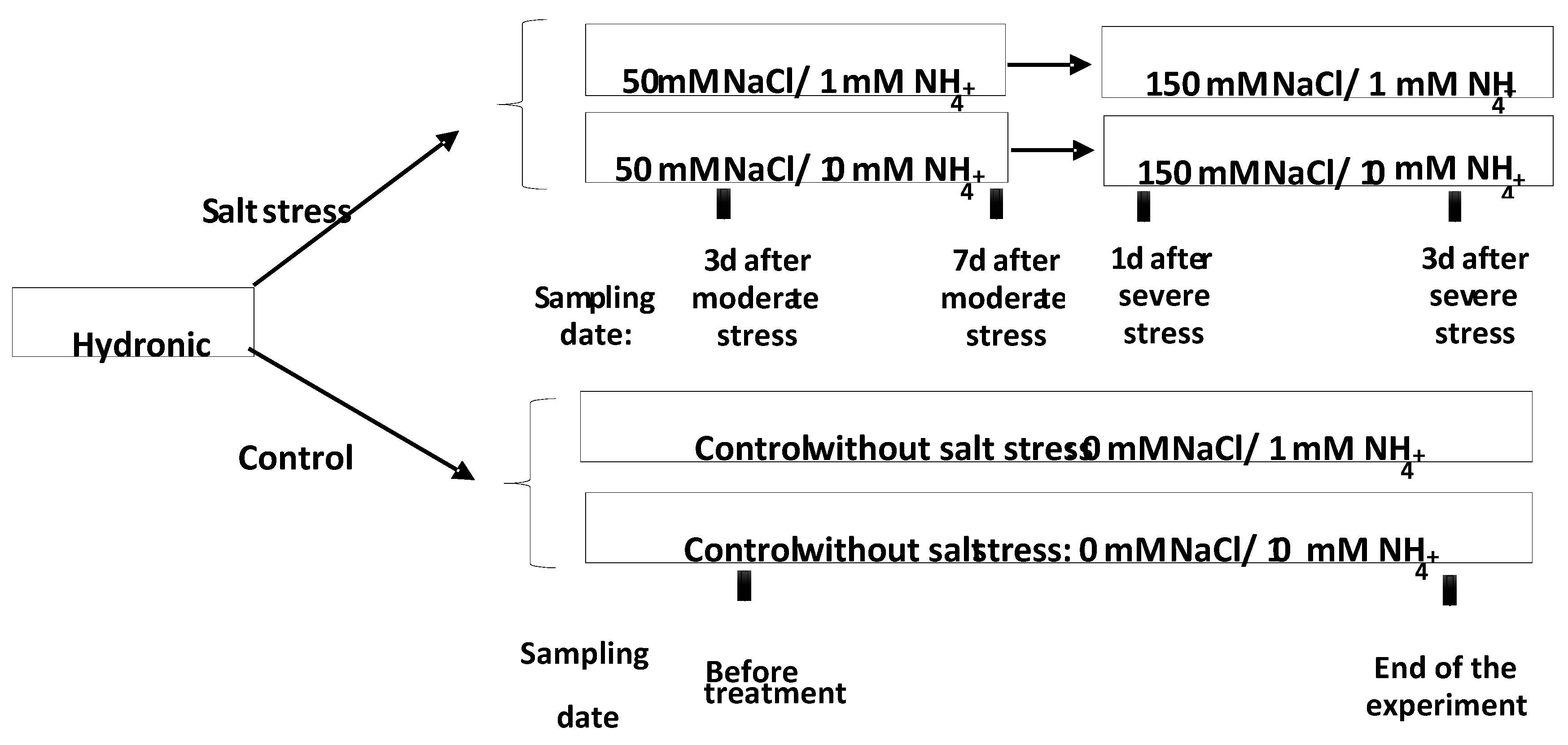
2.1. Plant Material and Growth Conditions
2.2. Nitrogen and Salt Treatments
2.3. Enzyme Extraction and Activities’ Determination of NADH-GOGAT and NADH-GDH
2.4. Enzyme Extraction and Activities Determination of G6PDH, APX, and PEPC
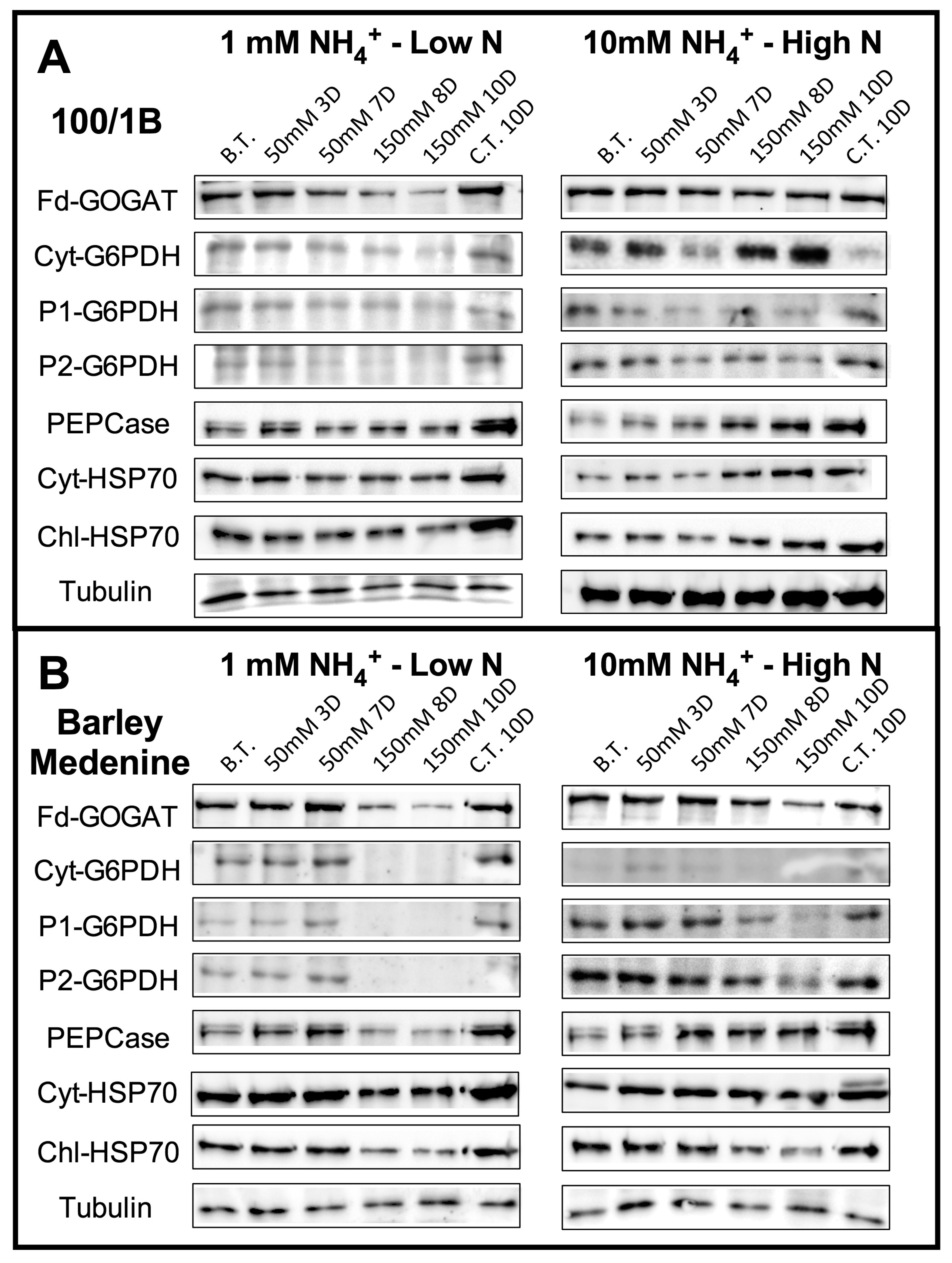
2.5. Western Blotting
2.6. Statistical Analyses
3. Results
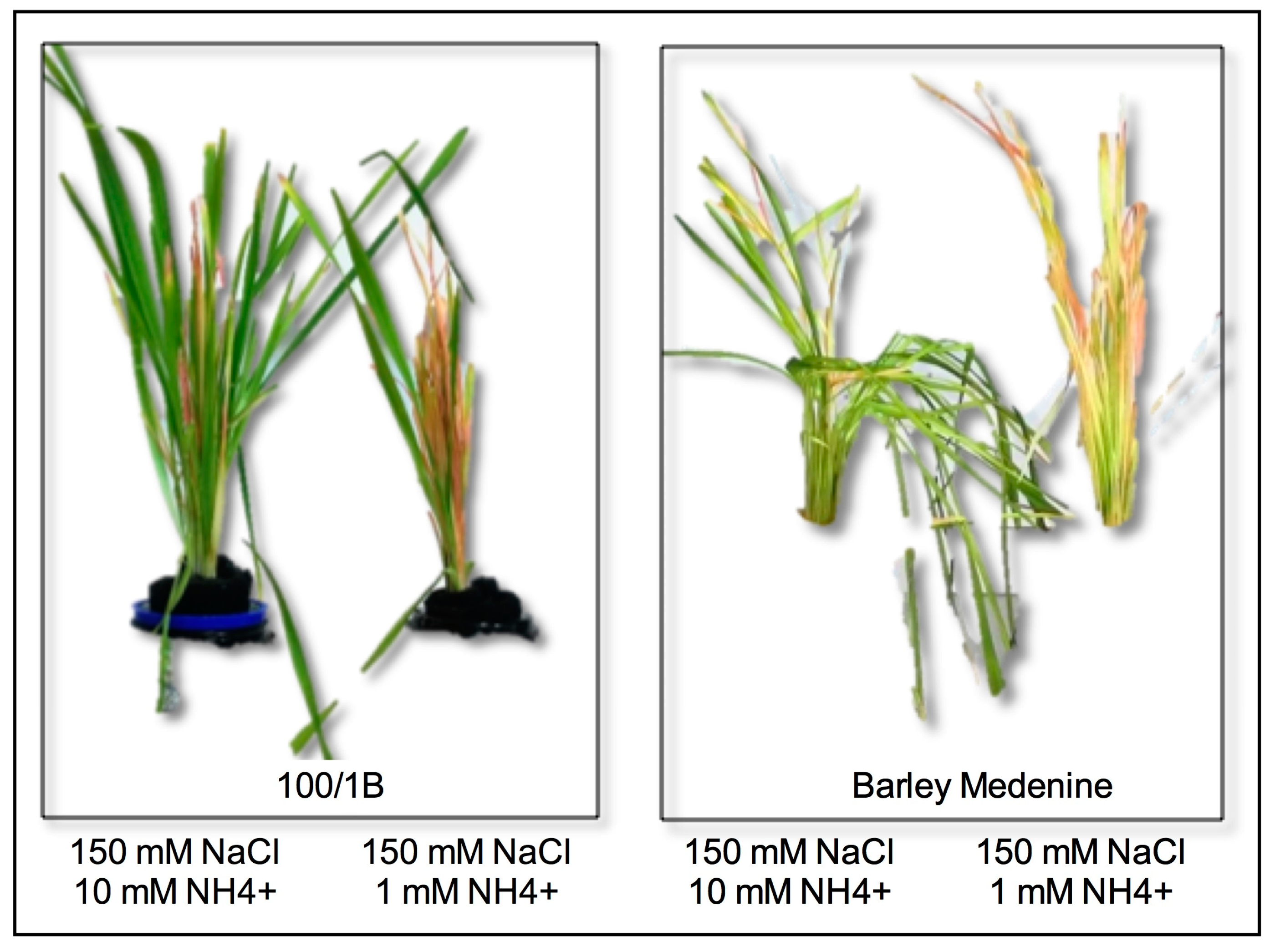
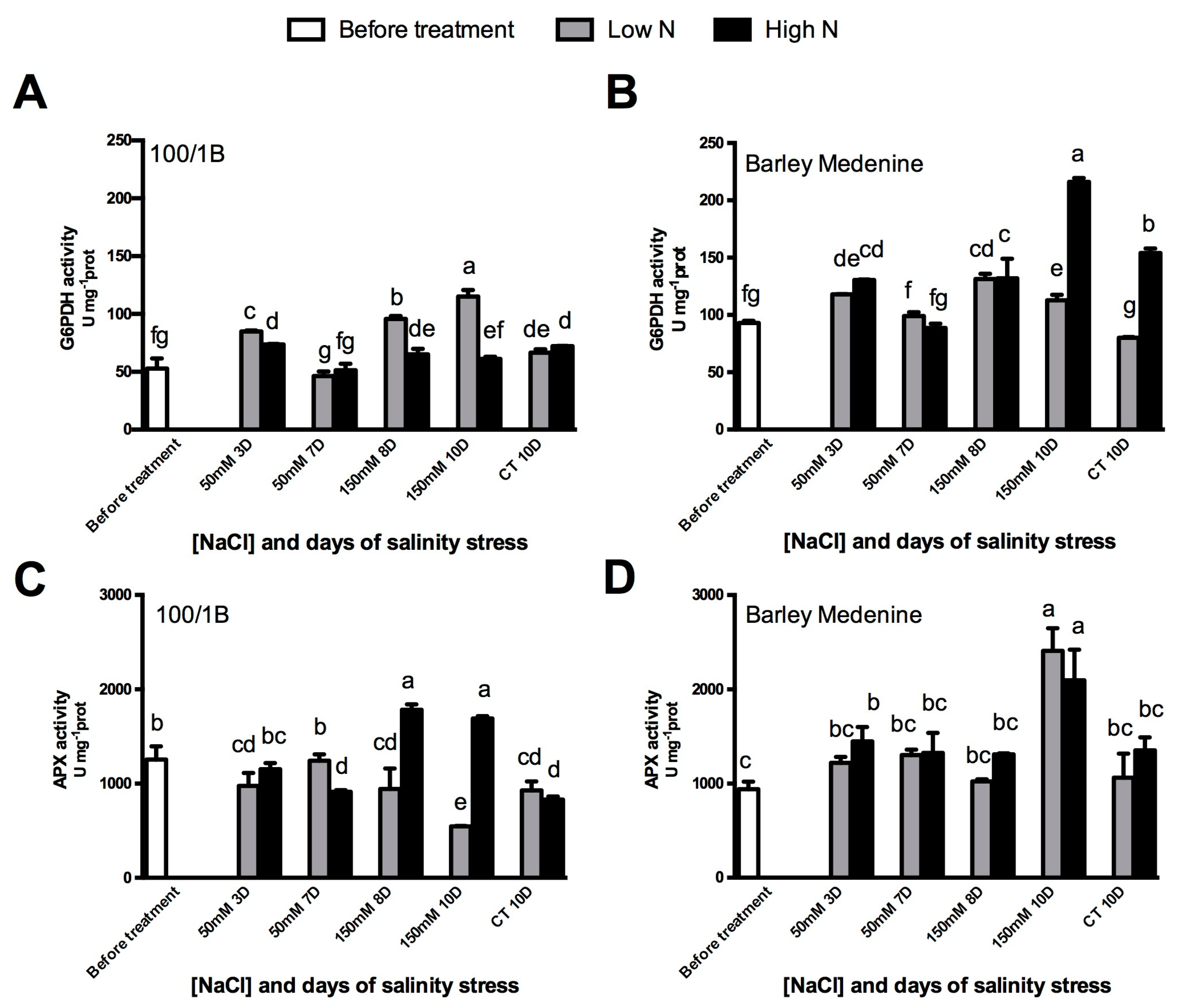
3.1. Contrasting Barley Landraces Showed Diversified Stress and Antioxidant Responses
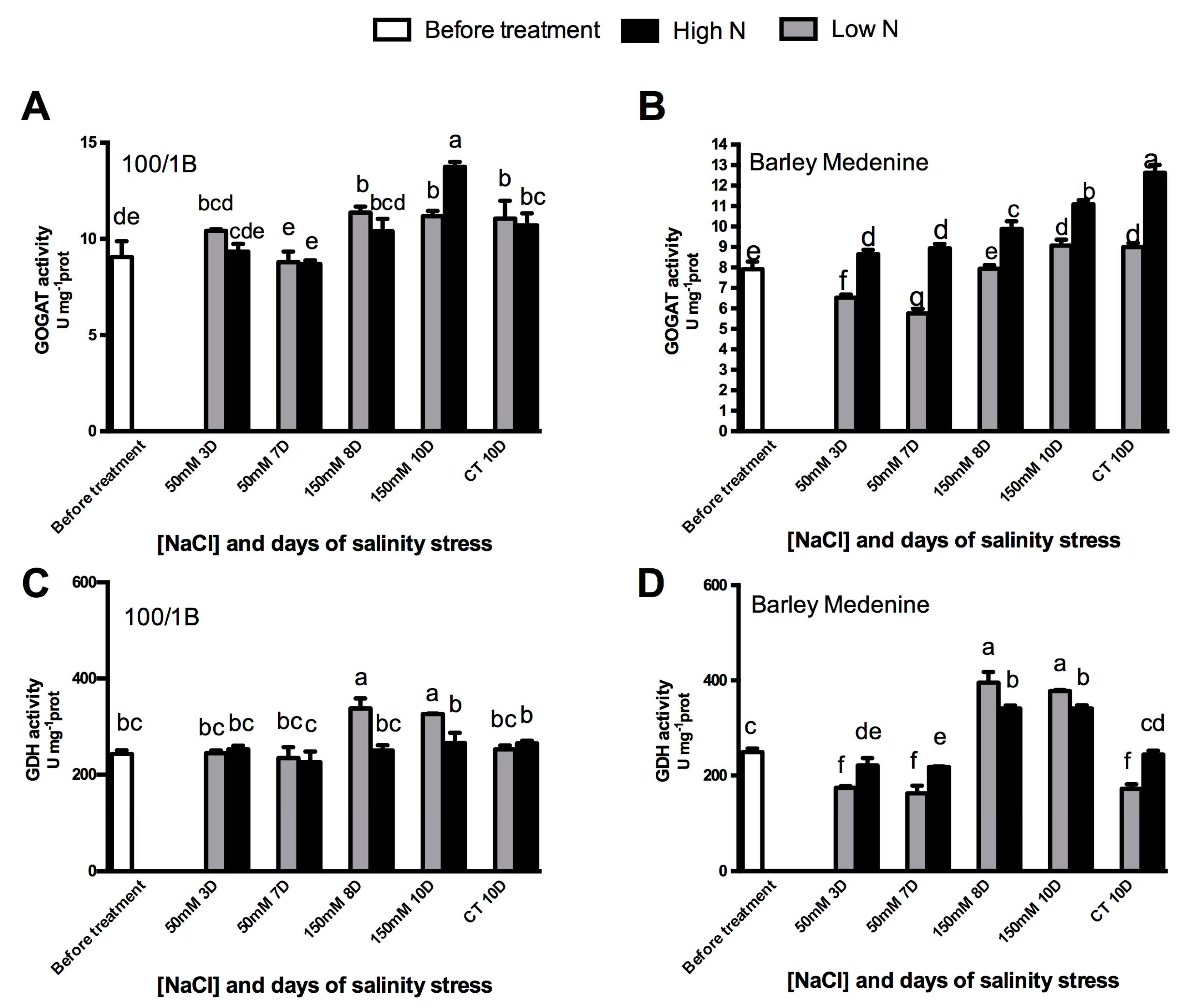
3.2. N Metabolism Modifications upon Salinity: The Role of GOGAT and GDH
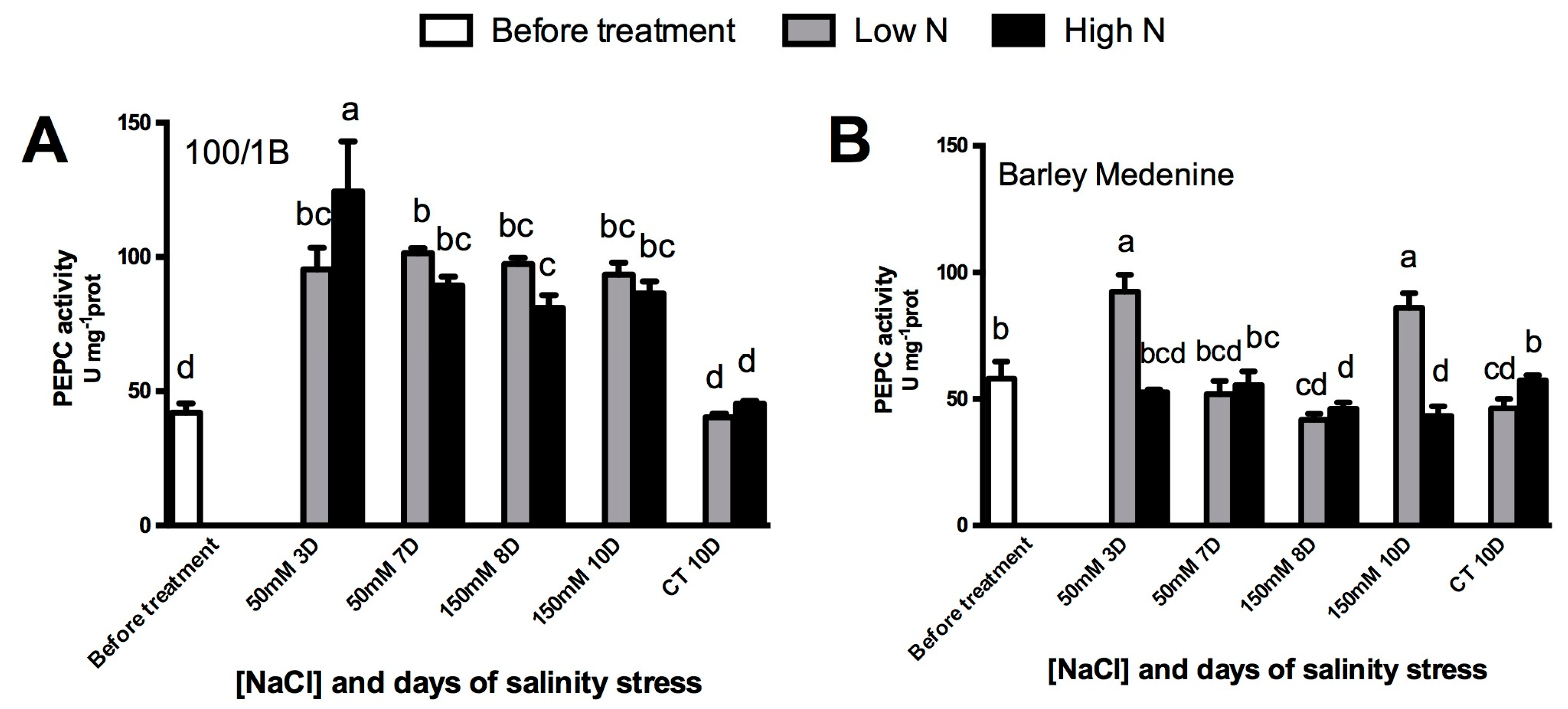
3.3. Change in the Activity of PEPcase
3.4. Correlation between Suppliers of Reducing Power (G6PDH and PEPC) and Different Enzymes Involved in N Metabolism or Defense against Stress
4. Discussions
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Martinez-Atienza, J.; Jiang, X.; Garciadeblas, B.; Mendoza, I.; Zhu, J.-K.; Pardo, J.M.; Quintero, F.J. Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 2007, 143, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, M.; Yang, C.; Shi, D.; Liu, B.; Lin, X.; Yang, C. Effects of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice (Oryza sativa L.). BMC Plant Biol. 2012, 12, 194. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, A.; Landi, S.; Punzo, P.; Possenti, M.; Van Oosten, M.J.; Costa, A.; Morelli, G.; Maggio, A.; Grillo, S.; Batelli, G. Salinity and ABA Seed Responses in Pepper: Expression and Interaction of ABA Core Signaling Components. Front. Plant Sci. 2019, 10, 304. [Google Scholar] [CrossRef]
- Lobell, D.B.; Schlenker, W.; Costa-Roberts, J. Climate trends and global crop production since 1980. Science 2011, 333, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Chen, T.; Hussain, N.; Zhang, G.; Jiang, L. Characterization of Salinity Tolerance of Transgenic Rice Lines Harboring HsCBL8 of Wild Barley (Hordeum spontanum) Line from Qinghai-Tibet Plateau. Front. Plant Sci. 2016, 7, 1678. [Google Scholar] [CrossRef]
- Halford, N.G.; Curtis, T.Y.; Chen, Z.; Huang, J. Effects of abiotic stress and crop management on cereal grain composition: Implications for food quality and safety. J. Exp. Bot. 2015, 66, 1145–1156. [Google Scholar] [CrossRef]
- Bartels, D.; Sunkar, R. Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 2005, 24, 23–58. [Google Scholar] [CrossRef]
- Ashraf, M.; Shahzad, S.M.; Imtiaz, M.; Rizwan, M.S. Salinity effects on nitrogen metabolism in plants–focusing on the activities of nitrogen metabolizing enzymes: A review. J. Plant Nutr. 2018, 41, 1065–1081. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Jallouli, S.; Ayadi, S.; Landi, S.; Capasso, G.; Santini, G.; Chamekh, Z.; Zouari, I.; Ben Azaiez, F.E.; Trifa, Y.; Esposito, S. Physiological and Molecular Osmotic Stress Responses in Three Durum Wheat (Triticum Turgidum ssp Durum) Genotypes. Agronomy 2019, 9, 550. [Google Scholar] [CrossRef]
- Caverzan, A.; Passaia, G.; Rosa, S.B.; Ribeiro, C.W.; Lazzarotto, F.; Margis-Pinheiro, M. Plant responses to stresses: Role of ascorbate peroxidase in the antioxidant protection. Genet. Mol. Biol. 2012, 35, 1011–1019. [Google Scholar] [CrossRef] [PubMed]
- Landi, S.; De Lillo, A.; Nurcato, R.; Grillo, S.; Esposito, S. In-field study on traditional Italian tomato landraces: The constitutive activation of the ROS scavenging machinery reduces effects of drought stress. Plant Physiol. Biochem. 2017, 118, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, L.; Li, Y.; Hou, J.; Huang, J.; Liang, W. Involvement of ABA- and H2O2-dependent cytosolic glucose-6-phosphate dehydrogenase in maintaining redox homeostasis in soybean roots under drought stress. Plant Physiol. Biochem. 2016, 107, 126–136. [Google Scholar] [CrossRef]
- Lentini, M.; De Lillo, A.; Paradisone, V.; Liberti, D.; Landi, S.; Esposito, S. Early responses to Cadmium exposure in barley plants: Effects on biometric and physiological parameters. Acta Physiol. Plant. 2018, 40, 178. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, X.; Wang, X.; Wu, K.; Li, P.; Chang, N.; Wang, J.; Wang, F.; Li, J.; Bi, Y. Glucose-6-phosphate dehydrogenase and alternative oxidase are involved in the cross tolerance of highland barley to salt stress and UV-B radiation. J. Plant Physiol. 2015, 181, 83–95. [Google Scholar] [CrossRef]
- Arghavani, M.; Zaeimzadeh, A.; Savadkoohi, S.; Samiei, L. Salinity tolerance of Kentucky bluegrass as affected by nitrogen fertilization. J. Agric. Sci. Technol. 2017, 19, 173–183. [Google Scholar]
- Singh, M.; Singh, V.P.; Prasad, S.M. Responses of photosynthesis, nitrogen and proline metabolism to salinity stress in Solanum lycopersicum under different levels of nitrogen supplementation. Plant Physiol. Biochem. 2016, 109, 72–83. [Google Scholar] [CrossRef]
- Esposito, S. Nitrogen assimilation, abiotic stress and glucose 6-phosphate dehydrogenase: The full circle of reductants. Plants 2016, 5, 24. [Google Scholar] [CrossRef]
- De Angeli, A.; Monachello, D.; Ephritikhine, G.; Frachisse, J.-M.; Thomine, S.; Gambale, F.; Barbier-Brygoo, H. The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature 2016, 442, 939–942. [Google Scholar] [CrossRef]
- Álvarez-Aragón, R.; Rodríguez-Navarro, A. Nitrate-dependent shoot sodium accumulation and osmotic functions of sodium in Arabidopsis under saline conditions. Plant J. 2017, 91, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Landi, S.; Esposito, S. Nitrate Uptake Affects Cell Wall Synthesis and Modeling. Front. Plant Sci. 2017, 8, 1376. [Google Scholar] [CrossRef] [PubMed]
- Landi, S.; Capasso, G.; Ben Azaiez, F.E.; Jallouli, S.; Ayadi, S.; Trifa, Y.; Esposito, S. Different roles of heat shock proteins (70 kDa) during abiotic stresses in barley (Hordeum vulgare) genotypes. Plants 2019, 8, 248. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, S.L.; Ceccarelli, S.; Blair, M.W.; Upadhyaya, H.D.; Are, A.K.; Ortiz, R. Landrace germplasm for improving yield and abiotic stress adaptation. Trends Plant Sci. 2016, 21, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Hammami, Z.; Gauffreteau, A.; BelhajFraj, M.; Sahli, A.; Jeuffroy, M.H.; Rezgui, S.; Bergaoui, K.; McDonnell, R.; Trifa, Y. Predicting yield reduction in improved barley (Hordeum vulgare L.) varieties and landraces under salinity using selected tolerance traits. Field Crop. Res. 2017, 211, 10–18. [Google Scholar] [CrossRef]
- FAOstat. Statistical Database 2017. Available online: http://www.fao.org/faostat/en/#home (accessed on 2 July 2020).
- Shen, Q.; Fu, L.; Dai, F.; Jiang, L.; Zhang, G.; Wu, D. Multi-omics analysis reveals molecular mechanisms of shoot adaption to salt stress in Tibetan wild barley. BMC Genom. 2016, 17, 889. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, T.; Yang, S.; Lee, Y. BarleyNet: A Network-Based Functional Omics Analysis Server for Cultivated Barley, Hordeum vulgare L. Front. Plant Sci. 2020, 11, 98. [Google Scholar] [CrossRef]
- Gürel, F.; Øztürk, Z.N.; Uçarlı, C.; Rosellini, D. Barley genes as tools to confer abiotic stress tolerance in crops. Front. Plant Sci. 2016, 7, 1137. [Google Scholar] [CrossRef]
- Tuberosa, R. Phenotyping for drought tolerance of crops in the genomics era. Front. Physiol. 2012, 3, 347. [Google Scholar] [CrossRef]
- Groat, R.G.; Vance, C.P. Root nodule enzymes of ammonia assimilation in alfalfa (Medicago sativa L). Plant Physiol. 1981, 67, 1198–1203. [Google Scholar] [CrossRef]
- Singh, R.P.; Srivastava, H.S. Increase in glutamate synthase (NADH) activity in maize seedlings in response to nitrate and ammonium nitrogen. Physiol. Plant. 1986, 66, 413–416. [Google Scholar] [CrossRef]
- Castiglia, D.; Cardi, M.; Landi, S.; Cafasso, D.; Esposito, S. Expression and characterization of a cytosolic glucose 6 phosphate dehydrogenase isoform from barley (Hordeum vulgare) roots. Protein Expr. Purif. 2015, 112, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Asada, K. Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 1981, 22, 867–880. [Google Scholar]
- Fontaine, V.; Pelloux, J.; Podor, M.; Afif, D.; Gérant, D.; Grieu, P.; Dizengremel, P. Carbon fixation in Pinus halepensis submitted to ozone. Opposite response of ribulose-1,5-bisphosphate carboxylase/oxygenase and phosphoenolpyruvate carboxylase. Physiol. Plant. 1999, 105, 187–192. [Google Scholar] [CrossRef]
- Wendt, U.K.; Wenderoth, I.; Tegeler, A.; Von Schaewen, A. Molecular characterization of a novel glucose-6-phosphate dehydrogenase from potato (Solanum tuberosum L.). Plant J. 2000, 23, 723–733. [Google Scholar] [CrossRef]
- Pajuelo, P.; Pajuelo, E.; Márquez, A.J. Proteolysis of barley (Hordeum vulgare L.) T. ferredoxin-glutamate synthase affects ferredoxin-and methyl viologen-dependent enzyme activities differently. J. Plant Physiol. 2000, 157, 575–579. [Google Scholar] [CrossRef]
- Li, Y.J.; Hai, R.L.; Du, X.H.; Jiang, X.N.; Lu, H. Over-expression of a Populus peroxisomal ascorbate peroxidase (PpAPX) gene in tobacco plants enhances stress tolerance. Plant Breed. 2009, 128, 404–410. [Google Scholar] [CrossRef]
- Maksimović, J.D.; Zhang, J.; Zeng, F.; Živanović, B.D.; Shabala, L.; Zhou, M.; Shabala, S. Linking oxidative and salinity stress tolerance in barley: Can root antioxidant enzyme activity be used as a measure of stress tolerance? Plant Soil 2013, 365, 141–155. [Google Scholar] [CrossRef]
- Chang, Z.; Liu, Y.; Dong, H.; Teng, K.; Han, L.; Zhang, X. Effects of cytokinin and nitrogen on drought tolerance of creeping bentgrass. PLoS ONE 2016, 11, e0154005. [Google Scholar] [CrossRef]
- Sagi, M.; Dovrat, A.; Kipnis, T.; Lips, H. Nitrate reductase, phosphoenolpyruvate carboxylase, and glutamine synthetase in annual ryegrass as affected by salinity and nitrogen. J. Plant Nutr. 1998, 21, 707–723. [Google Scholar] [CrossRef]
- Shi, W.M.; Muramoto, Y.; Ueda, A.; Takabe, T. Cloning of peroxisomal ascorbate peroxidase gene from barley and enhanced thermotolerance by overexpressing in Arabidopsis thaliana. Gene 2001, 273, 23–27. [Google Scholar] [CrossRef]
- Soussi, M.; Ocaña, A.; Lluch, C. Effects of salt stress on growth, photosynthesis and nitrogen fixation in chick-pea (Cicer arietinum L.). J. Exp. Bot. 1998, 49, 1329–1337. [Google Scholar] [CrossRef]
- Stewart, G.R.; Rhodes, D. Nitrogen metabolism of halophytes III. Enzymes of ammonia assimilation. New Phytol. 1978, 80, 307–316. [Google Scholar] [CrossRef]
- Abobatta, W.F. Plant Responses and Tolerance to Combined Salt and Drought Stress. In Salt and Drought Stress Tolerance in Plants; Hasanuzzaman, M., Tanveer, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2020; pp. 17–52. [Google Scholar] [CrossRef]
- Wang, Z.Q.; Yuan, Y.Z.; Ou, J.Q.; Lin, Q.H.; Zhang, C.F. Glutamine synthetase and glutamate dehydrogenase contribute differentially to proline accumulation in leaves of wheat (Triticum aestivum) seedlings exposed to different salinity. J. Plant Physiol. 2007, 164, 695–701. [Google Scholar] [CrossRef]
- Ullah, A.; Li, M.; Noor, J.; Tariq, A.; Liu, Y.; Shi, L. Effects of salinity on photosynthetic traits, ion homeostasis and nitrogen metabolism in wild and cultivated soybean. PeerJ 2019, 7, 8191. [Google Scholar] [CrossRef]
- Goel, P.; Singh, A.K. Abiotic stresses downregulate key genes involved in nitrogen uptake and assimilation in Brassica juncea L. PLoS ONE 2015, 10, e0143645. [Google Scholar] [CrossRef]
| “100/1B” | GOGAT | GDH | G6PDH | APX | PEPC |
|---|---|---|---|---|---|
| GOGAT | 0.53 (0.173) | 0.84 (0.011) | −0.28 (0.5) | −0.07 (0.875) | |
| GDH | −0.11 (0.799) | 0.4 (0.345) | 0.21 (0.622) | 0.54 (0.176) | |
| G6PDH | −0.35 (0.389) | 0.87 (0.005) | −0.24 (0.564) | 0.25 (0.558) | |
| APX | 0.26(0.539) | −0.85 (0.008) | −0.97 (0.001) | 0.52 (0.195) | |
| PEPC | 0.03 (0.942) | 0.90 (0.002) | 0.76 (0.030) | −0.78 (0.021) |
| “Barley Medenine” | GOGAT | GDH | G6PDH | APX | PEPC |
|---|---|---|---|---|---|
| GOGAT | 0.92 (0.001) | 0.04 (0.916) | 0.33 (0.430) | −0.35 (0.402) | |
| GDH | −0.84 (0.009) | 0.14 (0.737) | 0.55 (0.155) | −0.35 (0.389) | |
| G6PDH | 0.76 (0.027) | −0.57 (0.141) | 0.20 (0.642) | 0.26 (0.534) | |
| APX | 0.48 (0.229) | −0.17 (0.684) | 0.19 (0.645) | −0.39 (0.336) | |
| PEPC | −0.16 (0.705) | 0.23 (0.576) | −0.50 (0.206) | 0.7 (0.058) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ben Azaiez, F.E.; Ayadi, S.; Capasso, G.; Landi, S.; Paradisone, V.; Jallouli, S.; Hammami, Z.; Chamekh, Z.; Zouari, I.; Trifa, Y.; et al. Salt Stress Induces Differentiated Nitrogen Uptake and Antioxidant Responses in Two Contrasting Barley Landraces from MENA Region. Agronomy 2020, 10, 1426. https://doi.org/10.3390/agronomy10091426
Ben Azaiez FE, Ayadi S, Capasso G, Landi S, Paradisone V, Jallouli S, Hammami Z, Chamekh Z, Zouari I, Trifa Y, et al. Salt Stress Induces Differentiated Nitrogen Uptake and Antioxidant Responses in Two Contrasting Barley Landraces from MENA Region. Agronomy. 2020; 10(9):1426. https://doi.org/10.3390/agronomy10091426
Chicago/Turabian StyleBen Azaiez, Fatma Ezzahra, Sawsen Ayadi, Giorgia Capasso, Simone Landi, Valeria Paradisone, Salma Jallouli, Zied Hammami, Zoubeir Chamekh, Inès Zouari, Youssef Trifa, and et al. 2020. "Salt Stress Induces Differentiated Nitrogen Uptake and Antioxidant Responses in Two Contrasting Barley Landraces from MENA Region" Agronomy 10, no. 9: 1426. https://doi.org/10.3390/agronomy10091426
APA StyleBen Azaiez, F. E., Ayadi, S., Capasso, G., Landi, S., Paradisone, V., Jallouli, S., Hammami, Z., Chamekh, Z., Zouari, I., Trifa, Y., & Esposito, S. (2020). Salt Stress Induces Differentiated Nitrogen Uptake and Antioxidant Responses in Two Contrasting Barley Landraces from MENA Region. Agronomy, 10(9), 1426. https://doi.org/10.3390/agronomy10091426







