Genome-Wide Association Study (GWAS) Analysis of Camelina Seedling Germination under Salt Stress Condition
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Salt Stress Tolerance Assays
2.2. Phenotypic Analysis
2.3. Genotypic Data, Population Genetics, Linkage Disequilibrium (LD)
2.4. Genome Wide Association Study (GWAS) Analyses
2.5. Candidate Gene Identification
3. Results and Discussion
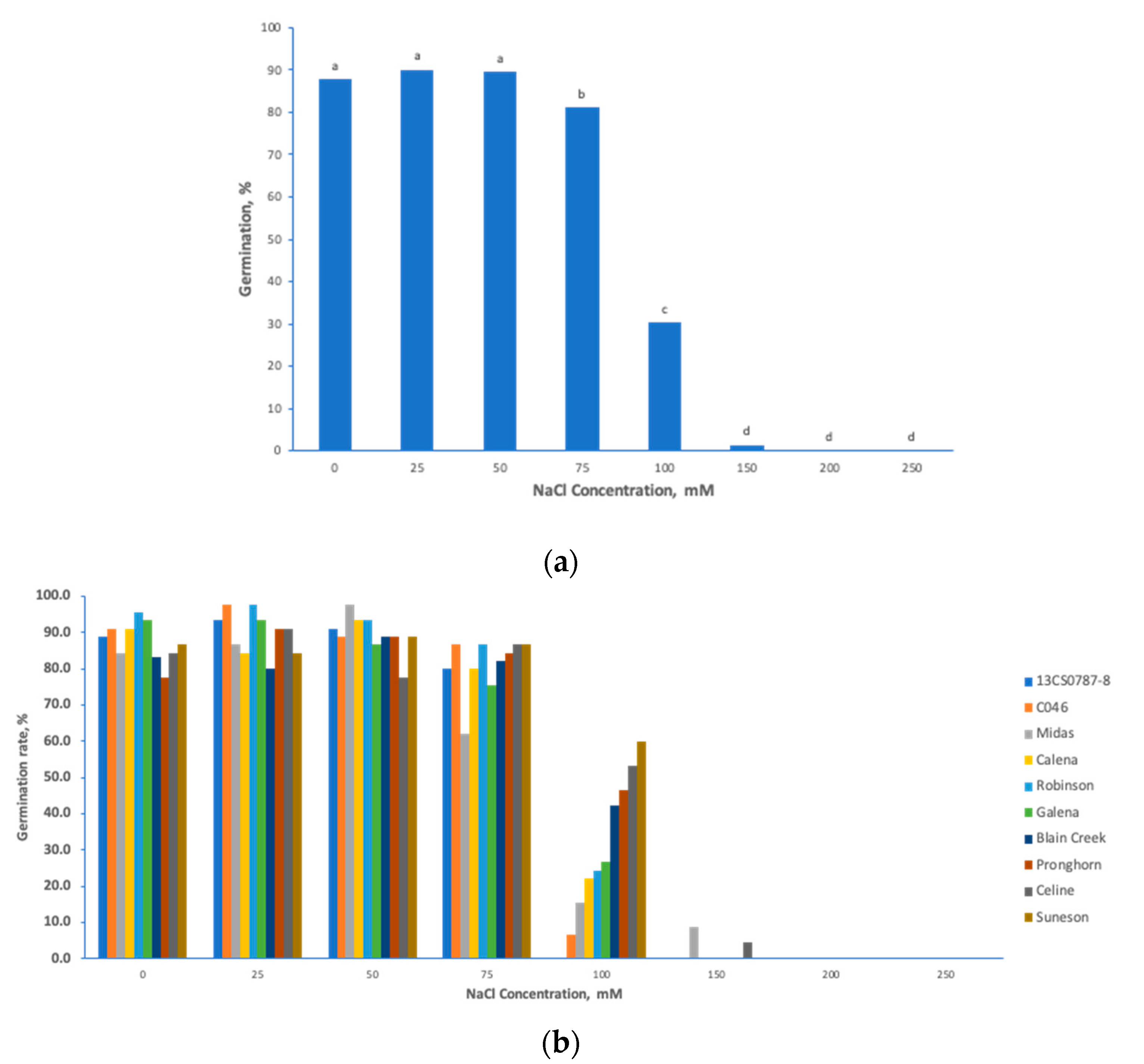
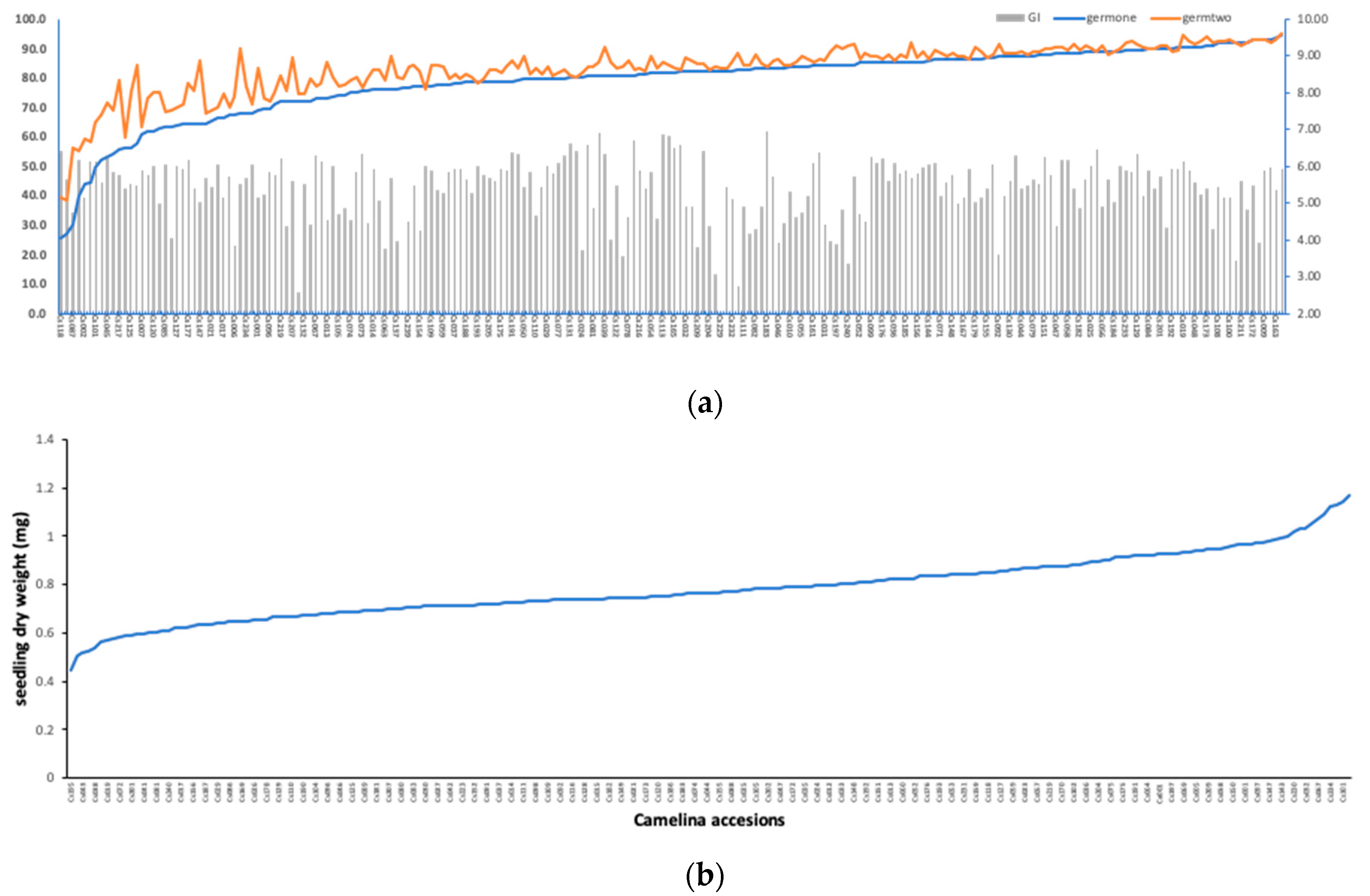
3.1. Phenotypic Diversity in Seedling Germination Traits under Salt Stress
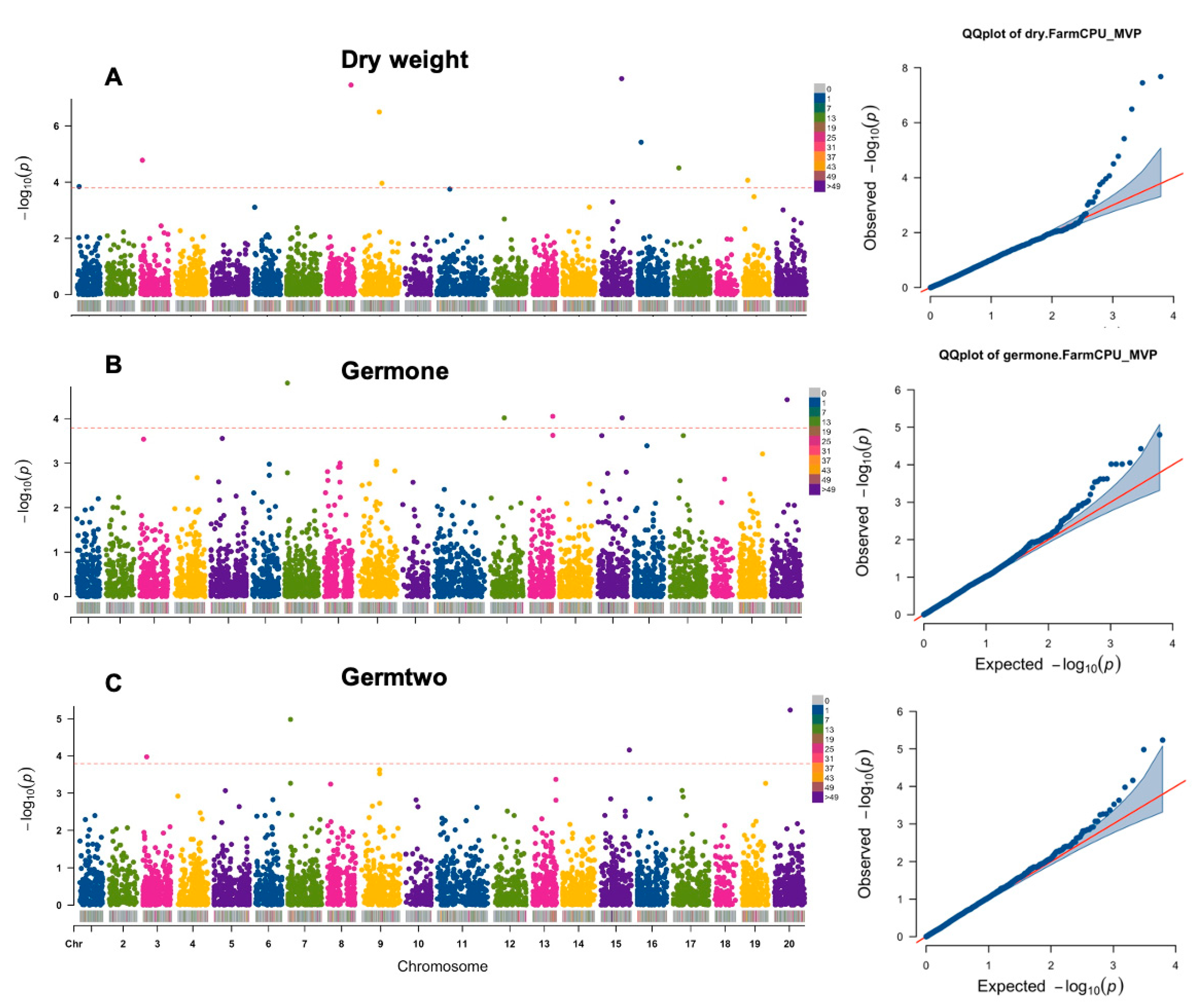
3.2. GWAS Analysis of Seedling Germination Traits under Salt Stress
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Singh, R.; Bollina, V.; Higgins, E.E.; Clarke, W.E.; Eynck, C.; Sidebottom, C.; Gugel, R.; Snowdon, R.; Parkin, I.A.P. Single-nucleotide polymorphism identification and genotyping in Camelina sativa. Mol. Breed. 2015, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kagale, S.; Koh, C.S.; Nixon, J.; Bollina, V.; Clarke, W.E.; Tuteja, R.; Spillane, C.; Robinson, S.J.; Links, M.G.; Clarke, C.; et al. The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Ghamkhar, K.; Croser, J.; Aryamanesh, N.; Campbell, M.; Kon’kova, N.; Francis, C. Camelina (Camelina sativa (L.) Crantz) as an alternative oilseed: Molecular and ecogeographic analyses. Genome 2010, 53, 558–567. [Google Scholar] [CrossRef] [PubMed]
- Seguin-Swartz, G.; Eynck, C.; Gugel, R.K.; Strelkov, S.E.; Olivier, C.Y.; Li, J.L.; Klein-Gebbinck, H.; Borhan, H.; Caldwell, C.D.; Falk, K.C. Diseases of Camelina sativa (false flax). Can. J. Plant Pathol. 2009, 31, 375–386. [Google Scholar] [CrossRef]
- Berti, M.; Gesch, R.; Eynck, C.; Anderson, J.; Cermak, S. Camelina uses, genetics, genomics, production, and management. Ind. Crop. Prod. 2016, 94, 690–710. [Google Scholar] [CrossRef]
- Shrivastava, P.; Kumar, R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.L.; Niu, L.; Wei, J.; Chen, X.Y.; Chen, Y.L. Phosphorus Limitation Improved Salt Tolerance in Maize Through Tissue Mass Density Increase, Osmolytes Accumulation, and Na+ Uptake Inhibition. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Heydarian, Z.; Yu, M.; Gruber, M.; Coutu, C.; Robinson, S.J.; Hegedus, D.D. Changes in gene expression in Camelina sativa roots and vegetative tissues in response to salinity stress. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [Green Version]
- Manchanda, G.; Garg, N. Salinity and its effects on the functional biology of legumes. Acta Physiol. Plant 2008, 30, 595–618. [Google Scholar] [CrossRef]
- Vicente, O.; Boscaiu, M.; Naranjo, M.A.; Estrelles, E.; Belles, J.M.; Soriano, P. Responses to salt stress in the halophyte Plantago crassifolia (Plantaginaceae). J. Arid Environ. 2004, 58, 463–481. [Google Scholar] [CrossRef]
- El-Hendawy, S.E.; Hu, Y.C.; Yakout, G.M.; Awad, A.M.; Hafiz, S.E.; Schmidhalter, U. Evaluating salt tolerance of wheat genotypes using multiple parameters. Eur. J. Agron. 2005, 22, 243–253. [Google Scholar] [CrossRef]
- Kingsbury, R.W.; Epstein, E. Selection for Salt-Resistant Spring Wheat. Crop Sci. 1984, 24, 310–315. [Google Scholar] [CrossRef]
- Shannon, M.C.; Grieve, C.M. Tolerance of vegetable crops to salinity. Sci. Hortic. Amst. 1999, 78, 5–38. [Google Scholar] [CrossRef]
- Saisho, D.; Takumi, S.; Matsuoka, Y. Salt tolerance during germination and seedling growth of wild wheat Aegilops tauschii and its impact on the species range expansion. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Francois, L.E.; Maas, E.V.; Donovan, T.J.; Youngs, V.L. Effect of Salinity on Grain-Yield and Quality, Vegetative Growth, and Germination of Semidwarf and Durum-Wheat. Agron. J. 1986, 78, 1053–1058. [Google Scholar] [CrossRef]
- Chaves, M.M.; Pereira, J.S.; Maroco, J.; Rodrigues, M.L.; Ricardo, C.P.P.; Osorio, M.L.; Carvalho, I.; Faria, T.; Pinheiro, C. How plants cope with water stress in the field. Photosynthesis and growth. Ann. Bot. Lond. 2002, 89, 907–916. [Google Scholar] [CrossRef] [Green Version]
- Potters, G.; Pasternak, T.P.; Guisez, Y.; Palme, K.J.; Jansen, M.A.K. Stress-induced morphogenic responses: Growing out of trouble? Trends Plant Sci. 2007, 12, 98–105. [Google Scholar] [CrossRef]
- Lopez-Berenguer, C.; Carvajal, M.; Garcia-Viguera, C.; Alcaraz, C.F. Nitrogen, phosphorus, and sulfur nutrition in broccoli plants grown under salinity. J. Plant Nutr. 2007, 30, 1855–1870. [Google Scholar] [CrossRef]
- Sacala, E.; Demczuk, A.; Grzys, E.; Spiak, Z. Effect of salt and water stresses on growth, nitrogen and phosphorus metabolism in Cucumis sativus L. seedlings. Acta Soc. Bot. Pol. 2008, 77, 23–28. [Google Scholar] [CrossRef] [Green Version]
- Luo, Z.; Tomasi, P.; Fahlgren, N.; Abdel-Haleem, H. Genome-wide association study (GWAS) of leaf cuticular wax components in Camelina sativa identifies genetic loci related to intracellular wax transport. BMC Plant Biol. 2019, 19, 187. [Google Scholar] [CrossRef]
- Cui, Y.R.; Zhang, F.; Zhou, Y.L. The Application of Multi-Locus GWAS for the Detection of Salt-Tolerance Loci in Rice. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, Y.; Zhou, G.F.; Shabala, S.; Chen, Z.H.; Cai, S.G.; Li, C.D.; Zhou, M.X. Genome-Wide Association Study Reveals a New QTL for Salinity Tolerance in Barley (Hordeum vulgare L.). Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Zeng, A.; Chen, P.; Korth, K.; Hancock, F.; Pereira, A.; Brye, K.; Wu, C.; Shi, A. Genome-wide association study (GWAS) of salt tolerance in worldwide soybean germplasm lines. Mol. Breed. 2017, 37. [Google Scholar] [CrossRef]
- Li, D.H.; Dossa, K.; Zhang, Y.X.; Wei, X.; Wang, L.H.; Zhang, Y.J.; Liu, A.L.; Zhou, R.; Zhang, X.R. GWAS Uncovers Differential Genetic Bases for Drought and Salt Tolerances in Sesame at the Germination Stage. Genes 2018, 9, 87. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.P.; Yu, L.X. Genome-Wide Association Mapping of Loci Associated with Plant Growth and Forage Production under Salt Stress in Alfalfa (Medicago sativa L.). Front Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Luo, Z.; Brock, J.; Dyer, J.M.; Kutchan, T.; Schachtman, D.; Augustin, M.; Ge, Y.; Fahlgren, N.; Abdel-Haleem, H. Genetic Diversity and Population Structure of a Camelina sativa Spring Panel. Front. Plant Sci. 2019, 10, 184. [Google Scholar] [CrossRef] [Green Version]
- Nyquist, W.E.; Baker, R. Estimation of heritability and prediction of selection response in plant populations. Crit. Rev. Plant Sci. 1991, 10, 235–322. [Google Scholar] [CrossRef]
- Holland, J.B.; Nyquist, W.E.; Cervantes-Martínez, C.T. Estimating and interpreting heritability for plant breeding: An update. Plant Breed. Rev. 2003, 22, 9–112. [Google Scholar]
- Wei, T.; Simko, V. R Package “Corrplot”: Visualization of a Correlation Matrix (Version 0.84). 2017. Available online: https://github.com/taiyun/corrplot.
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies (vol 12, e1005767, 2016). PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [Green Version]
- Team, R.D.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- Knoch, D.; Abbadi, A.; Grandke, F.; Meyer, R.C.; Samans, B.; Werner, C.R.; Snowdon, R.J.; Altmann, T. Strong temporal dynamics of QTL action on plant growth progression revealed through high-throughput phenotyping in canola. Plant Biotechnol. J. 2020, 18, 68–82. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.Y.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w(1118); iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lageix, S.; Lanet, E.; Pouch-Pelissier, M.N.; Espagnol, M.C.; Robaglia, C.; Deragon, J.M.; Pelissier, T. Arabidopsis eIF2 alpha kinase GCN2 is essential for growth in stress conditions and is activated by wounding. BMC Plant Biol. 2008, 8. [Google Scholar] [CrossRef] [Green Version]
- Lokdarshi, A.; Morgan, P.W.; Franks, M.; Emert, Z.; Emanuel, C.; von Arnim, A.G. Light-Dependent Activation of the GCN2 Kinase Under Cold and Salt Stress Is Mediated by the Photosynthetic Status of the Chloroplast. Front. Plant Sci. 2020, 11, 431. [Google Scholar] [CrossRef]
- Hinnebusch, A.G.; Ivanov, I.P.; Sonenberg, N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 2016, 352, 1413–1416. [Google Scholar] [CrossRef]
- Kashiwagi, K.; Yokoyama, T.; Nishimoto, M.; Takahashi, M.; Sakamoto, A.; Yonemochi, M.; Shirouzu, M.; Ito, T. Structural biology Structural basis for eIF2B inhibition in integrated stress response. Science 2019, 364, 495. [Google Scholar] [CrossRef]
- Ren, W.; Liu, L.; Gu, L.; Yan, W.L.; Feng, Y.L.; Dong, D.X.; Wang, S.J.; Lyu, M.S.; Wang, C.H. Crystal Structure of GH49 Dextranase from Arthrobacter oxidans KQ11: Identification of Catalytic Base and Improvement of Thermostability Using Semirational Design Based on B-Factors. J. Agr. Food Chem. 2019, 67, 4355–4366. [Google Scholar] [CrossRef]
- Bashari, M.; Tounkara, F.; Abdelhai, M.H.; Lagnika, C.; Xu, X.M.; Jin, Z.Y. Impact of Dextranase on Sugar Manufacturing and its Kinetic on the Molecular Weights of Remaining Dextran. Sugar Tech. 2013, 15, 84–93. [Google Scholar] [CrossRef]
- Leshem, Y.; Seri, L.; Levine, A. Induction of phosphatidylinositol 3-kinase-mediated endocytosis by salt stress leads to intracellular production of reactive oxygen species and salt tolerance. Plant J. 2007, 51, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.V.; Chavan, P.D. Influence of salt stress on phosphorus metabolism in the roots and leaves of one month old Prosopis juliflora (Sw.) DC seedlings. Pharmacogn. J. 2013, 3, 48–51. [Google Scholar] [CrossRef] [Green Version]
- Zribi, O.T.; Abdelly, C.; Debez, A. Interactive effects of salinity and phosphorus availability on growth, water relations, nutritional status and photosynthetic activity of barley (Hordeum vulgare L.). Plant Biol. 2011, 13, 872–880. [Google Scholar] [CrossRef] [PubMed]
- Uhrig, R.G.; Labandera, A.M.; Moorhead, G.B. Arabidopsis PPP family of serine/threonine protein phosphatases: Many targets but few engines. Trends Plant Sci. 2013, 18, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Pais, S.M.; Gonzalez, M.A.; Tellez-Inon, M.T.; Capiati, D.A. Characterization of potato (Solanum tuberosum) and tomato (Solanum lycopersicum) protein phosphatases type 2A catalytic subunits and their involvement in stress responses. Planta 2009, 230, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Bheri, M.; Pandey, G.K. PP2A Phosphatases Take a Giant Leap in the Post-Genomics Era. Curr. Genom. 2019, 20, 154–171. [Google Scholar] [CrossRef] [PubMed]
- Almeida, D.M.; Oliveira, M.M.; Saibo, N.J.M. Regulation of Na+ and K+ homeostasis in plants: Towards improved salt stress tolerance in crop plants. Genet. Mol. Biol. 2017, 40, 326–345. [Google Scholar] [CrossRef] [Green Version]
- Hirata, D.; Harada, S.; Namba, H.; Miyakawa, T. Adaptation to High-Salt Stress in Saccharomyces-Cerevisiae Is Regulated by Ca2+/Calmodulin-Dependent Phosphoprotein Phosphatase (Calcineurin) and Camp-Dependent Protein-Kinase. Mol. Gen. Genet. 1995, 249, 257–264. [Google Scholar] [CrossRef]
- Ji, W.; Cong, R.; Li, S.; Li, R.; Qin, Z.W.; Li, Y.J.; Zhou, X.L.; Chen, S.X.; Li, J. Comparative Proteomic Analysis of Soybean Leaves and Roots by iTRAQ Provides Insights into Response Mechanisms to Short-Term Salt Stress. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.D.; Lin, K.H.; Chen, C.C.; Chiang, C.M. Oryza sativa protein phosphatase 1a (OsPP1a) involved in salt stress tolerance in transgenic rice. Mol. Breed. 2016, 36. [Google Scholar] [CrossRef]
- Maselli, G.A.; Slamovits, C.H.; Bianchi, J.I.; Vilarrasa-Blasi, J.; Cano-Delgado, A.I.; Mora-Garcia, S. Revisiting the Evolutionary History and Roles of Protein Phosphatases with Kelch-Like Domains in Plants. Plant Physiol. 2014, 164, 1527–1541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grove, M.D.; Spencer, G.F.; Rohwedder, W.K.; Mandava, N.; Worley, J.F.; Warthen, J.D.; Steffens, G.L.; Flippenanderson, J.L.; Cook, J.C. Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature 1979, 281, 216–217. [Google Scholar] [CrossRef]
- Planas-Riverola, A.; Gupta, A.; Betegon-Putze, I.; Bosch, N.; Ibanes, M.; Cano-Delgado, A.I. Brassinosteroid signaling in plant development and adaptation to stress. Development 2019, 146. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.J.; Lee, S.H.; Park, C.H.; Kim, T.W. Functional Role of BSL1 Subcellular Localization in Brassinosteroid Signaling. J. Plant Biol. 2018, 61, 40–49. [Google Scholar] [CrossRef]
- Su, Q.F.; Zheng, X.D.; Tian, Y.K.; Wang, C.H. Exogenous Brassinolide Alleviates Salt Stress in Malus hupehensis Rehd. by Regulating the Transcription of NHX-Type Na+(K+)/H+ Antiporters. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.W.; Cole, M.; Flier, A.; Werr, W. BIM1, a bHLH protein involved in brassinosteroid signalling, controls Arabidopsis embryonic patterning via interaction with DORNROSCHEN and DORNROSCHEN-LIKE. Plant Mol. Biol. 2009, 69, 57–68. [Google Scholar] [CrossRef]
- Keinanen, S.I.; Hassinen, V.H.; Karenlampi, S.O.; Tervahauta, A.I. Isolation of genes up-regulated by copper in a copper-tolerant birch (Betula pendula) clone. Tree Physiol. 2007, 27, 1243–1252. [Google Scholar] [CrossRef] [Green Version]
- Kulich, I.; Cole, R.; Drdova, E.; Cvrckova, F.; Soukup, A.; Fowler, J.; Zarsky, V. Arabidopsis exocyst subunits SEC8 and EXO70A1 and exocyst interactor ROH1 are involved in the localized deposition of seed coat pectin. New Phytol. 2010, 188, 615–625. [Google Scholar] [CrossRef]
- Sottosanto, J.B.; Saranga, Y.; Blumwald, E. Impact of AtNHX1, a vacuolar Na+/H+ antiporter, upon gene expression during short-term and long-term salt stress in Arabidopsis thaliana. BMC Plant Biol. 2007, 7. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Zhang, X.; Wan, W.T.; Zhang, H.; Liu, J.; Li, M.L.; Wang, H.Y.; Xiao, J.; Wang, X.E. Identification and Characterization of the EXO70 Gene Family in Polyploid Wheat and Related Species. Int. J. Mol. Sci. 2019, 20, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Source of Variance | Df | Mean Square |
|---|---|---|
| Replication | 2 | 0.83 ns |
| NaCl Concentration (Conc) | 7 | 495.33 *** |
| Cultivar (Gen) | 9 | 1.43 ** |
| Conc*Gen | 63 | 1.93 *** |
| Variable | Mean | Std Dev | Minimum | Maximum | Heritability (H) |
|---|---|---|---|---|---|
| Germone (%) | 78.3136 | 17.0766 | 11.76 | 100.00 | 0.8303 |
| Germtwo (%) | 82.6463 | 14.1230 | 16.00 | 100.00 | 0.7625 |
| GI | 5.3514 | 1.2363 | 0.67 | 7.50 | 0.8615 |
| Fresh weight (mg) | 18.8544 | 5.9687 | 5.16 | 53.98 | 0.7752 |
| Dry weight (mg) | 0.7835 | 0.1698 | 0.12 | 1.47 | 0.8834 |
| Dry/fresh ratio (%) | 4.4482 | 1.4694 | 0.62 | 15.43 | 0.8140 |
| Germone | Germtwo | GI | Fresh | Dry | Dry/Fresh Ratio | |
|---|---|---|---|---|---|---|
| Germone | 1 | 0.9277 | 0.8682 | 0.2899 | −0.1275 | −0.5147 |
| Germtwo | <0.0001 | 1 | 0.8455 | 0.2507 | −0.0539 | −0.4027 |
| GI | <0.0001 | <0.0001 | 1 | 0.1424 | −0.2604 | −0.4490 |
| Fresh | <0.0001 | 0.0002 | 0.0388 | 1 | 0.5525 | −0.6502 |
| Dry | 0.0644 | 0.4360 | 0.0001 | <0.0001 | 1 | 0.1644 |
| Dry/fresh ratio | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.0168 | 1 |
| SNP | Chr | Position | Effect | p-Value | FDR adj p-Value | Trait | Nearest Gene | Gene Annotation | Protein Function |
|---|---|---|---|---|---|---|---|---|---|
| S1_1910174 | 1 | 1910174 | 0.0259 | 0.00014389 | 0.00014389 | Dry weight | LOC104713675 | serine/threonine-protein phosphatase PP1 isozyme 9 | phosphatase activity toward para-nitrophenyl phosphate (pNPP) in vitro |
| S3_1462975 | 3 | 1462975 | −0.0537 | 1.67 × 10−5 | 3.97 × 10−5 | Dry weight | LOC104761613 | pre-mRNA-processing factor 39 | pre-mRNA splicing |
| S3_3511428 | 3 | 3511428 | 5.8912 | 0.00010599 | 0.00011582 | Germtwo | LOC104766624 | serine/threonine-protein phosphatase BSL2 | Phosphatase involved in elongation process, probably by acting as a regulator of brassinolide signaling |
| S7_2473684 | 7 | 2473684 | 4.9858 | 1.59 × 10−5 | 3.97 × 10−5 | Germone | LOC104699973 | eIF-2-alpha kinase GCN2-like | Metabolic-stress sensing protein kinase that phosphorylates the alpha subunit of eukaryotic translation initiation factor 2 (eIF-2-alpha/EIF2S1) on “Ser-52” in response to low amino acid availability |
| S7_2473684 | 7 | 2473684 | 3.6988 | 1.04 × 10−5 | 3.29 × 10−5 | Germtwo | LOC104699973 | eIF-2-alpha kinase GCN2-like | Metabolic-stress sensing protein kinase that phosphorylates the alpha subunit of eukaryotic translation initiation factor 2 (eIF-2-alpha/EIF2S1) on “Ser-52” in response to low amino acid availability |
| S8_24097673 | 8 | 24097673 | 0.035 | 3.53 × 10−8 | 3.35 × 10−7 | Dry weight | LOC104708282 | transcription factor BIM1-like | DNA binding; protein dimerization activity |
| S9_18205018 | 9 | 18205018 | 0.0484 | 3.20 × 10−7 | 2.03 × 10−6 | Dry weight | NA | NA | NA |
| S9_20790122 | 9 | 20790122 | 0.0225 | 0.00010973 | 0.00011582 | Dry weight | LOC104715504 | NA | NA |
| S12_13693594 | 12 | 13693594 | 47.5695 | 9.62 × 10−5 | 0.00011424 | Germone | LOC104731617 | inactive disease resistance protein RPS4 | Resistance to Pseudomonas syringae 4 |
| S12_13693600 | 12 | 13693600 | 47.5695 | 9.62 × 10−5 | 0.00011424 | Germone | LOC104731617 | inactive disease resistance protein RPS4 | Resistance to Pseudomonas syringae 4 |
| S13_23380011 | 13 | 23380011 | −5.1802 | 8.85 × 10−5 | 0.00011424 | Germone | NA | NA | NA |
| S15_20134089 | 15 | 20134089 | −0.051 | 2.10 × 10−8 | 3.35 × 10−7 | Dry weight | NA | NA | NA |
| S15_24396991 | 15 | 24396991 | 47.5695 | 9.62 × 10−5 | 0.00011424 | Germone | LOC104748747 | DNA replication licensing factor MCM4-like | DNA replication initiation and elongation |
| S15_28433235 | 15 | 28433235 | −4.4191 | 6.94 × 10−5 | 0.00011424 | Germtwo | NA | NA | NA |
| S16_2915367 | 16 | 2915367 | 0.0687 | 3.81 × 10−6 | 1.81 × 10−5 | Dry weight | LOC104749495 | DEAD-box ATP-dependent RNA helicase 21-like | mRNA splicing |
| S17_4964987 | 17 | 4964987 | −0.0395 | 3.14 × 10−5 | 6.63 × 10−5 | Dry weight | LOC104755363 | NA | NA |
| S19_4398468 | 19 | 4398468 | −0.0386 | 8.57 × 10−5 | 0.00011424 | Dry weight | LOC104764745 | exocyst complex component SEC8-like | vesicular trafficking, regulation of actin polarity and vesicle transportation |
| S20_15783417 | 20 | 15783417 | −9.7872 | 3.75 × 10−5 | 7.13 × 10−5 | Germone | LOC104772556 | dextranase-like | catalyze Endohydrolysis of (1->6)-alpha-D-glucosidic linkages in dextran |
| S20_15783417 | 20 | 15783417 | −7.7913 | 5.84 × 10−6 | 2.22 × 10−5 | Germtwo | LOC104772556 | dextranase-like | catalyze Endohydrolysis of (1->6)-alpha-D-glucosidic linkages in dextran |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, Z.; Szczepanek, A.; Abdel-Haleem, H. Genome-Wide Association Study (GWAS) Analysis of Camelina Seedling Germination under Salt Stress Condition. Agronomy 2020, 10, 1444. https://doi.org/10.3390/agronomy10091444
Luo Z, Szczepanek A, Abdel-Haleem H. Genome-Wide Association Study (GWAS) Analysis of Camelina Seedling Germination under Salt Stress Condition. Agronomy. 2020; 10(9):1444. https://doi.org/10.3390/agronomy10091444
Chicago/Turabian StyleLuo, Zinan, Aaron Szczepanek, and Hussein Abdel-Haleem. 2020. "Genome-Wide Association Study (GWAS) Analysis of Camelina Seedling Germination under Salt Stress Condition" Agronomy 10, no. 9: 1444. https://doi.org/10.3390/agronomy10091444
APA StyleLuo, Z., Szczepanek, A., & Abdel-Haleem, H. (2020). Genome-Wide Association Study (GWAS) Analysis of Camelina Seedling Germination under Salt Stress Condition. Agronomy, 10(9), 1444. https://doi.org/10.3390/agronomy10091444





