Unconventionally Secreted Manganese Superoxide Dismutase VdSOD3 Is Required for the Virulence of Verticillium dahliae
Abstract
1. Introduction
2. Materials and Methods
2.1. Growth of Microbial and Plant Material
2.2. Gene Cloning and Bioinformatics Analysis
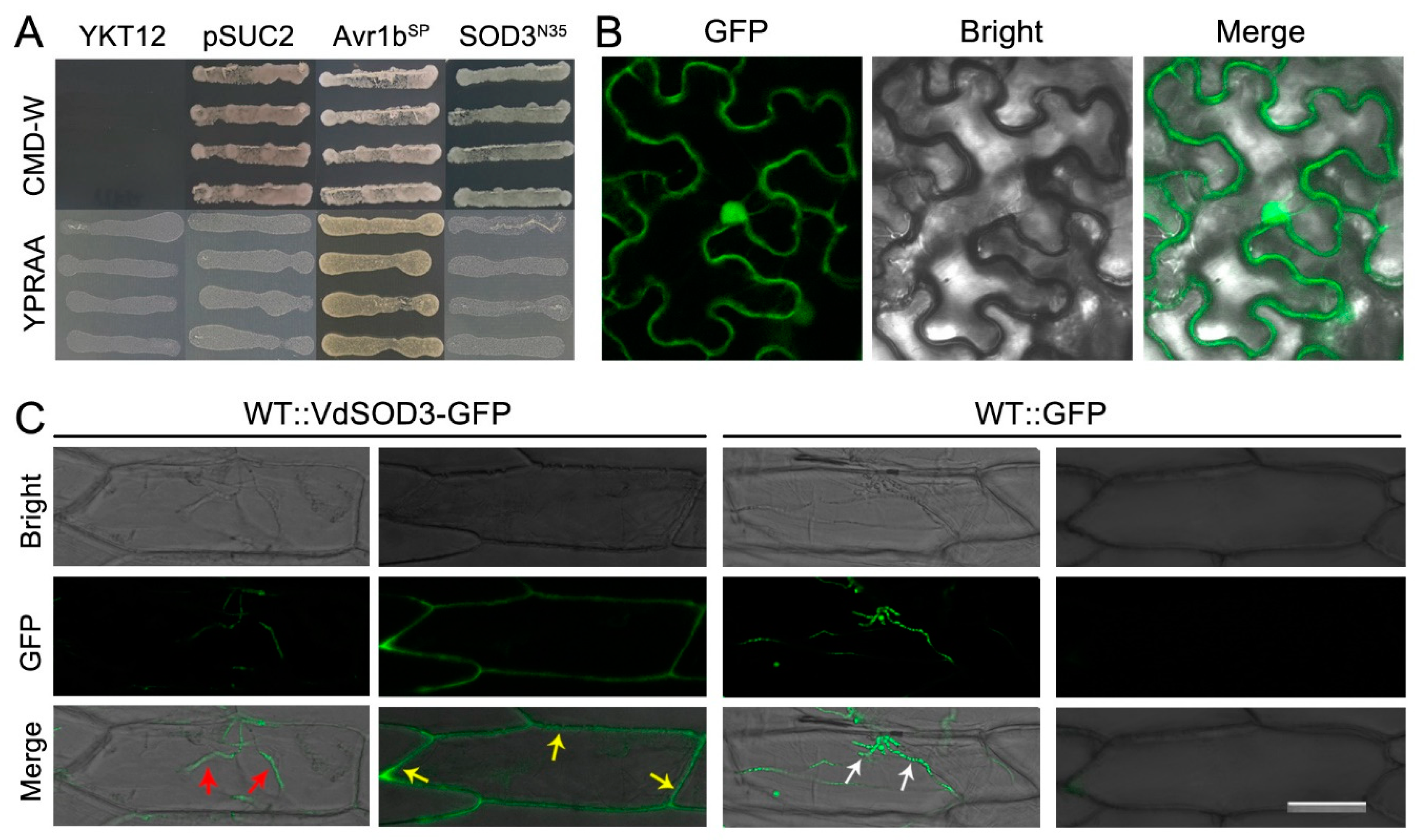
2.3. Yeast Signal Sequence Trap System
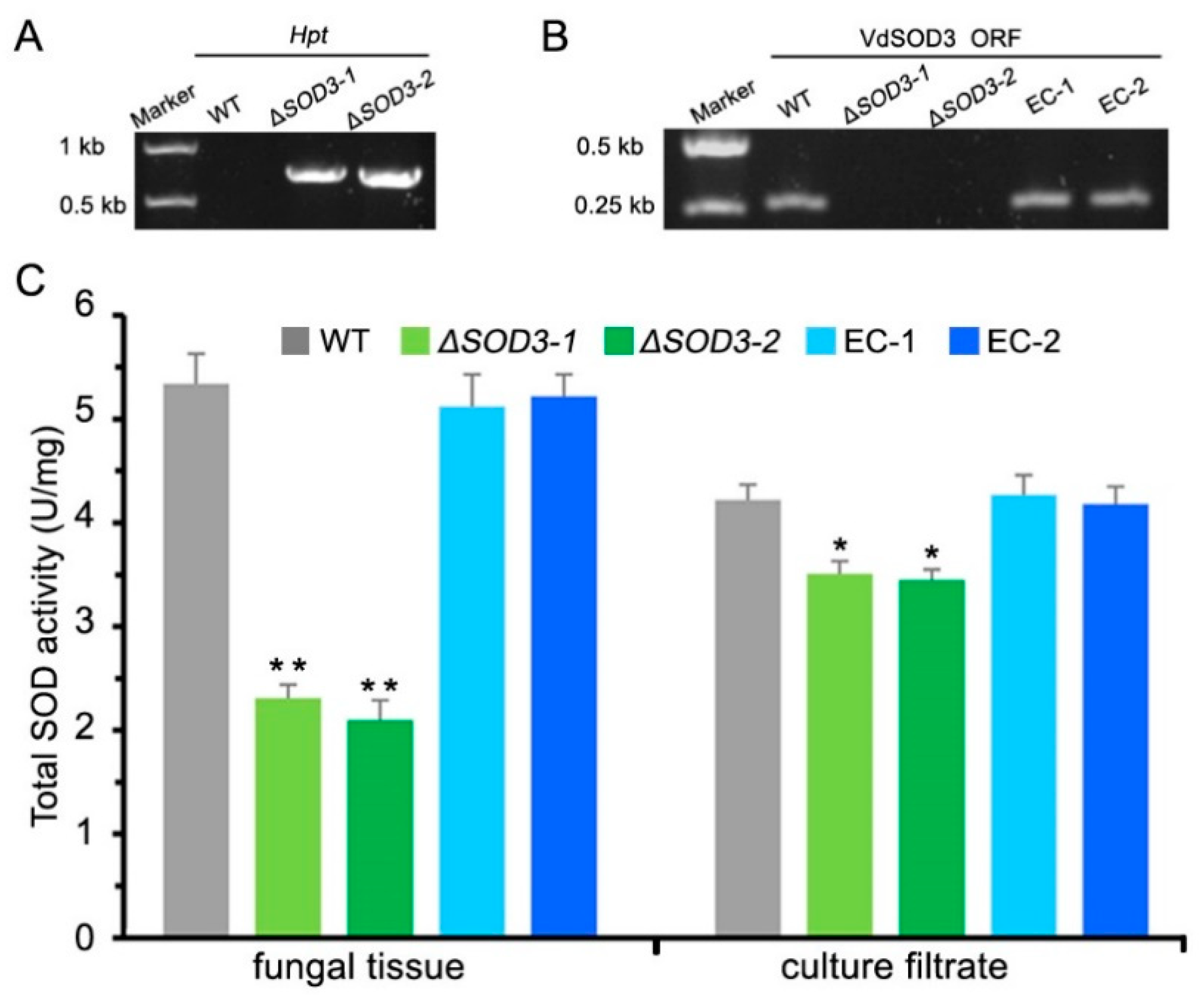
2.4. Fungal Transformation
2.5. Confocal Microscopy Analysis
2.6. VdSOD3 Expression Analysis
2.7. Measurement of SOD Enzyme Activity
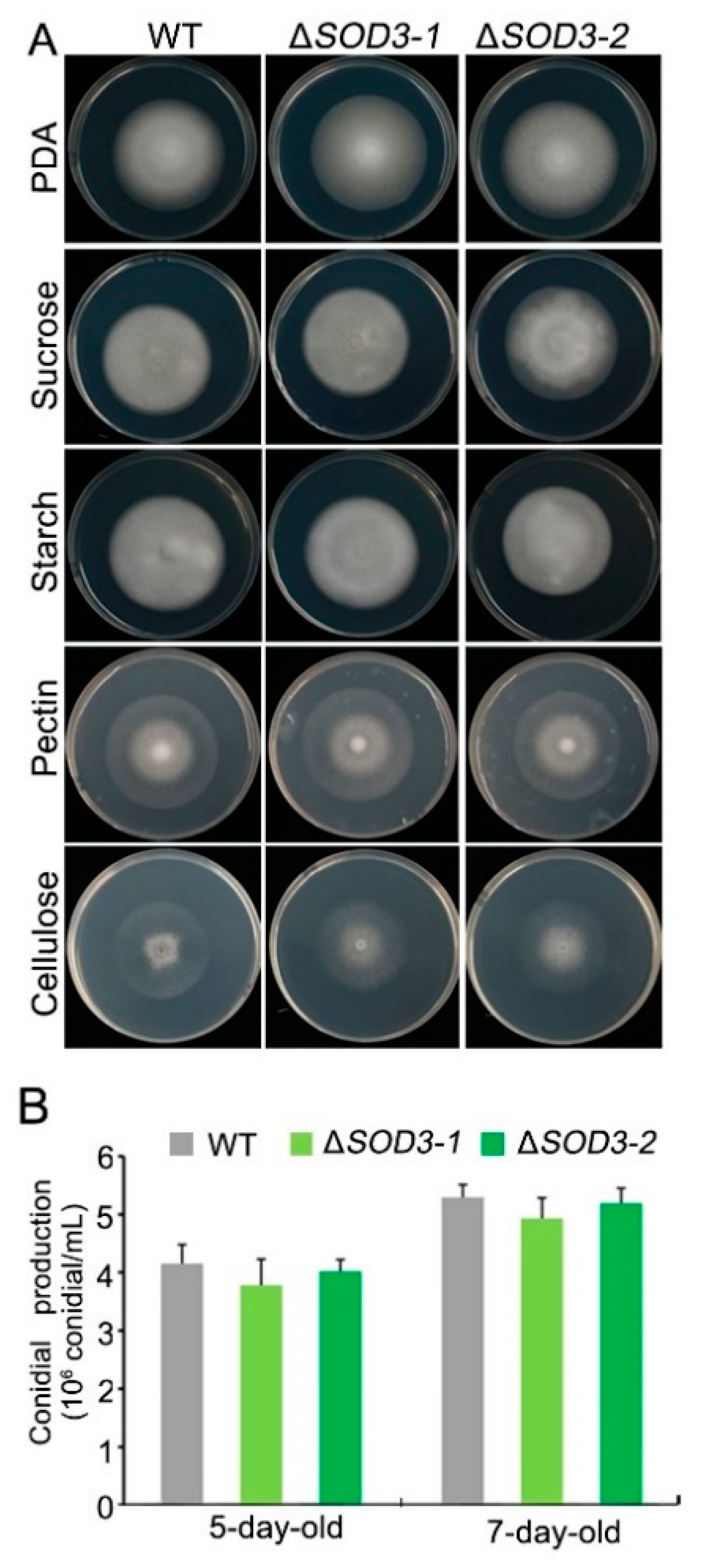
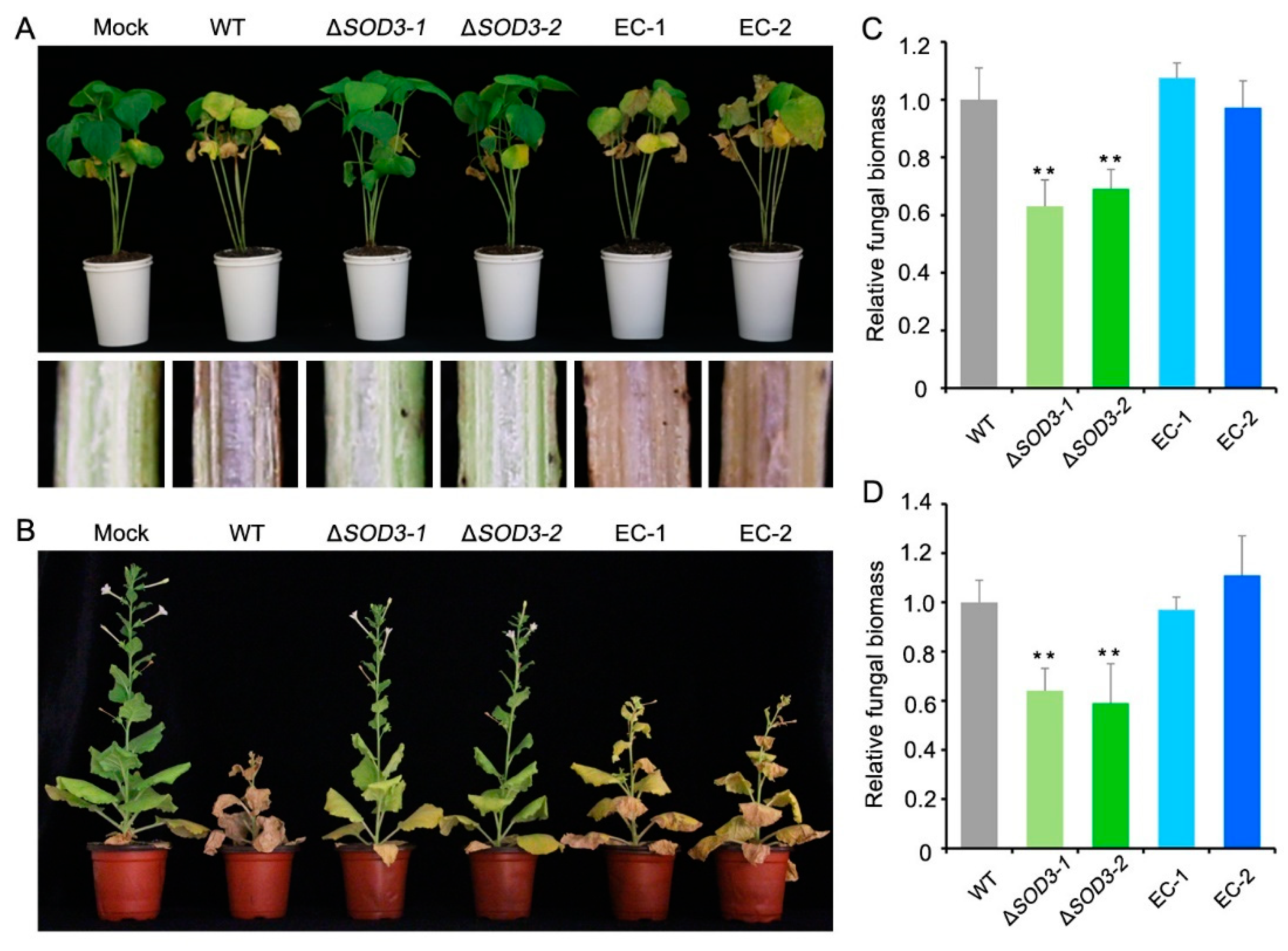
2.8. Fungal Morphology and Pathogenicity
3. Results
3.1. Bioinformatics Analysis of VdSOD3
3.2. Signal-Peptide-Lacking VdSOD3 Can Be Secreted
3.3. VdSOD3 Expression Pattern Analysis
3.4. VdSOD3 Contributes to Both Intra- and Extracellular SOD Activity of V. Dahliae
3.5. Fungal Morphology and Pathogenicity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Warris, A.; Ballou, E.R. Oxidative responses and fungal infection biology. Semin. Cell Dev. Biol. 2019, 89, 34–46. [Google Scholar] [CrossRef]
- Imlayt, J.A. Pathways of Oxidative Damage. Annu. Rev. Microbiol. 2003, 57, 395–418. [Google Scholar] [CrossRef] [PubMed]
- Broxton, C.N.; Culotta, V.C. SOD Enzymes and Microbial Pathogens: Surviving the Oxidative Storm of Infection. PLoS Pathog. 2016, 12, e1005295. [Google Scholar] [CrossRef] [PubMed]
- Fridovich, I. Superoxide Radical and Superoxide Dismutases. Annu. Rev. Biochem. 1995, 64, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Fréalle, E.; Noël, C.; Nolard, N.; Symoens, F.; Felipe, M.-S.; Dei-Cas, E.; Camus, D.; Viscogliosi, E.; Delhaes, L. Manganese superoxide dismutase based phylogeny of pathogenic fungi. Mol. Phylogenetics Evol. 2006, 41, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Staerck, C.; Gastebois, A.; Vandeputte, P.; Calenda, A.; Larcher, G.; Gillmann, L.; Papon, N.; Bouchara, J.-P.; Fleury, M.J.J. Microbial antioxidant defense enzymes. Microb. Pathog. 2017, 110, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Frealle, E.; Noel, C.; Viscogliosi, E.; Camus, D.; Dei-Cas, E.; Delhaes, L. Manganese superoxide dismutase in pathogenic fungi: An issue with pathophysiological and phylogenetic involvements. FEMS Immunol. Med Microbiol. 2005, 45, 411–422. [Google Scholar] [CrossRef]
- Leclère, V.; Bechet, M.; Blondeau, R. Functional significance of a periplasmic Mn-superoxide dismutase from Aeromonas hydrophila. J. Appl. Microbiol. 2004, 96, 828–833. [Google Scholar] [CrossRef]
- Krehenbrink, M.; Edwards, A.; Downie, J.A. The superoxide dismutase SodA is targeted to the periplasm in a SecA-dependent manner by a novel mechanism. Mol. Microbiol. 2011, 82, 164–179. [Google Scholar] [CrossRef]
- Schraw, W.; McClain, M.S.; Cover, T.L. Kinetics and Mechanisms of Extracellular Protein Release by Helicobacter pylori. Infect. Immun. 1999, 67, 5247–5252. [Google Scholar] [CrossRef]
- Chen, X.; Kodama, T.; Iida, T.; Honda, T. Demonstration and Characterization of Manganese Superoxide Dismutase ofProvidencia alcalifaciens. Microbiol. Immunol. 2007, 51, 951–961. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ebner, P.; Götz, F. Bacterial Excretion of Cytoplasmic Proteins (ECP): Occurrence, Mechanism, and Function. Trends Microbiol. 2019, 27, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Andersson, A.; Rasool, O.; Schmidt, M.; Kodzius, R.; Flückiger, S.; Zargari, A.; Crameri, R.; Scheynius, A. Cloning, expression and characterization of two new IgE-binding proteins from the yeast Malassezia sympodialis with sequence similarities to heat shock proteins and manganese superoxide dismutase. JBIC J. Biol. Inorg. Chem. 2004, 271, 1885–1894. [Google Scholar] [CrossRef] [PubMed]
- Miura, N.; Ueda, M. Evaluation of Unconventional Protein Secretion in Saccharomyces cerevisiae. Bioinform. MicroRNA Res. 2014, 1270, 51–70. [Google Scholar] [CrossRef]
- Caccia, D.; Dugo, M.; Callari, M.; Bongarzone, I. Bioinformatics tools for secretome analysis. Biochim. Biophys. Acta Proteins Proteom. 2013, 1834, 2442–2453. [Google Scholar] [CrossRef]
- Klosterman, S.J.; Subbarao, K.V.; Kang, S.; Veronese, P.; Gold, S.E.; Thomma, B.P.H.J.; Chen, Z.; Henrissat, B.; Lee, Y.-H.; Park, J.; et al. Comparative Genomics Yields Insights into Niche Adaptation of Plant Vascular Wilt Pathogens. PLoS Pathog. 2011, 7, e1002137. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Rong, W.; Yan, Z.; Li, Z.; Wu, L.; Zhang, G.; Ma, Z. Histochemical Analyses Reveal That Stronger Intrinsic Defenses in Gossypium barbadense Than in G. hirsutum Are Associated with Resistance to Verticillium dahliae. Mol. Plant Microbe Interact. 2017, 30, 984–996. [Google Scholar] [CrossRef]
- Chen, J.-Y.; Xiao, H.-L.; Gui, Y.-J.; Zhang, D.-D.; Li, L.; Bao, Y.-M.; Dai, X. Characterization of the Verticillium dahliae Exoproteome Involves in Pathogenicity from Cotton-Containing Medium. Front. Microbiol. 2016, 7, 1709. [Google Scholar] [CrossRef]
- Chen, J.-Y.; Liu, C.; Gui, Y.-J.; Si, K.-W.; Zhang, D.-D.; Wang, J.; Short, D.P.G.; Huang, J.-Q.; Li, N.-Y.; Liang, Y.; et al. Comparative genomics reveals cotton-specific virulence factors in flexible genomic regions in Verticillium dahliae and evidence of horizontal gene transfer from Fusarium. New Phytol. 2018, 217, 756–770. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Zhao, L.; Poschmann, G.; Waldera-Lupa, D.; Rafiee, N.; Kollmann, M.; Stühler, K. OutCyte: A novel tool for predicting unconventional protein secretion. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Jacobs, K.A.; Collins-Racie, L.A.; Colbert, M.; Duckett, M.; Golden-Fleet, M.; Kelleher, K.; Kriz, R.; LaVallie, E.R.; Merberg, D.; Spaulding, V.; et al. A genetic selection for isolating cDNAs encoding secreted proteins. Gene 1997, 198, 289–296. [Google Scholar] [CrossRef]
- Mullins, E.D.; Chen, X.; Romaine, P.; Raina, R.; Geiser, D.M.; Kang, S. Agrobacterium-Mediated Transformation of Fusarium oxysporum: An Efficient Tool for Insertional Mutagenesis and Gene Transfer. Phytopathology 2001, 91, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Beauchamp, C.; Fridovich, I. Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 1971, 44, 276–287. [Google Scholar] [CrossRef]
- Lamarre, C.; Lemay, J.-D.; DesLauriers, N.; Bourbonnais, Y. Candida albicansExpresses an Unusual Cytoplasmic Manganese-containing Superoxide Dismutase (SOD3Gene Product) upon the Entry and during the Stationary Phase. J. Biol. Chem. 2001, 276, 43784–43791. [Google Scholar] [CrossRef]
- Díez, B.; Schleissner, C.; Moreno, M.A.; Rodríguez, M.; Collados, A.; Barredo, J.L. The manganese superoxide dismutase from the penicillin producer Penicillium chrysogenum. Curr. Genet. 1998, 33, 387–394. [Google Scholar] [CrossRef]
- Giraldo, M.C.; Dagdas, Y.F.; Gupta, Y.K.; Mentlak, T.A.; Yi, M.; Martinez-Rocha, A.L.; Saitoh, H.; Terauchi, R.; Talbot, N.J.; Valent, B. Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae. Nat. Commun. 2013, 4, 1996. [Google Scholar] [CrossRef]
- Krombach, S.; Reissmann, S.; Kreibich, S.; Bochen, F.; Kahmann, R. Virulence function of theUstilago maydissterol carrier protein 2. New Phytol. 2018, 220, 553–566. [Google Scholar] [CrossRef]
- Liu, T.; Song, T.; Zhang, X.; Yuan, H.; Su, L.; Li, W.; Xu, J.; Liu, S.; Chen, L.; Chen, T.; et al. Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 2014, 5, 4686. [Google Scholar] [CrossRef]
- Li, C.X.; Gleason, J.E.; Zhang, S.X.; Bruno, V.M.; Cormack, B.P.; Culotta, V.C. Candida albicans adapts to host copper during infection by swapping metal cofactors for superoxide dismutase. Proc. Natl. Acad. Sci. USA 2015, 112, E5336–E5342. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, L.; Sun, W.; Li, J.; Chen, J.; Dai, X.; Qiu, N.; Zhang, D. Unconventionally Secreted Manganese Superoxide Dismutase VdSOD3 Is Required for the Virulence of Verticillium dahliae. Agronomy 2021, 11, 13. https://doi.org/10.3390/agronomy11010013
Tian L, Sun W, Li J, Chen J, Dai X, Qiu N, Zhang D. Unconventionally Secreted Manganese Superoxide Dismutase VdSOD3 Is Required for the Virulence of Verticillium dahliae. Agronomy. 2021; 11(1):13. https://doi.org/10.3390/agronomy11010013
Chicago/Turabian StyleTian, Li, Weixia Sun, Junjiao Li, Jieyin Chen, Xiaofeng Dai, Nianwei Qiu, and Dandan Zhang. 2021. "Unconventionally Secreted Manganese Superoxide Dismutase VdSOD3 Is Required for the Virulence of Verticillium dahliae" Agronomy 11, no. 1: 13. https://doi.org/10.3390/agronomy11010013
APA StyleTian, L., Sun, W., Li, J., Chen, J., Dai, X., Qiu, N., & Zhang, D. (2021). Unconventionally Secreted Manganese Superoxide Dismutase VdSOD3 Is Required for the Virulence of Verticillium dahliae. Agronomy, 11(1), 13. https://doi.org/10.3390/agronomy11010013




