Artificial microRNA-Based RNA Interference and Specific Gene Silencing for Developing Insect Resistance in Solanum lycopersicum
Abstract
:1. Introduction
2. Materials and Methods
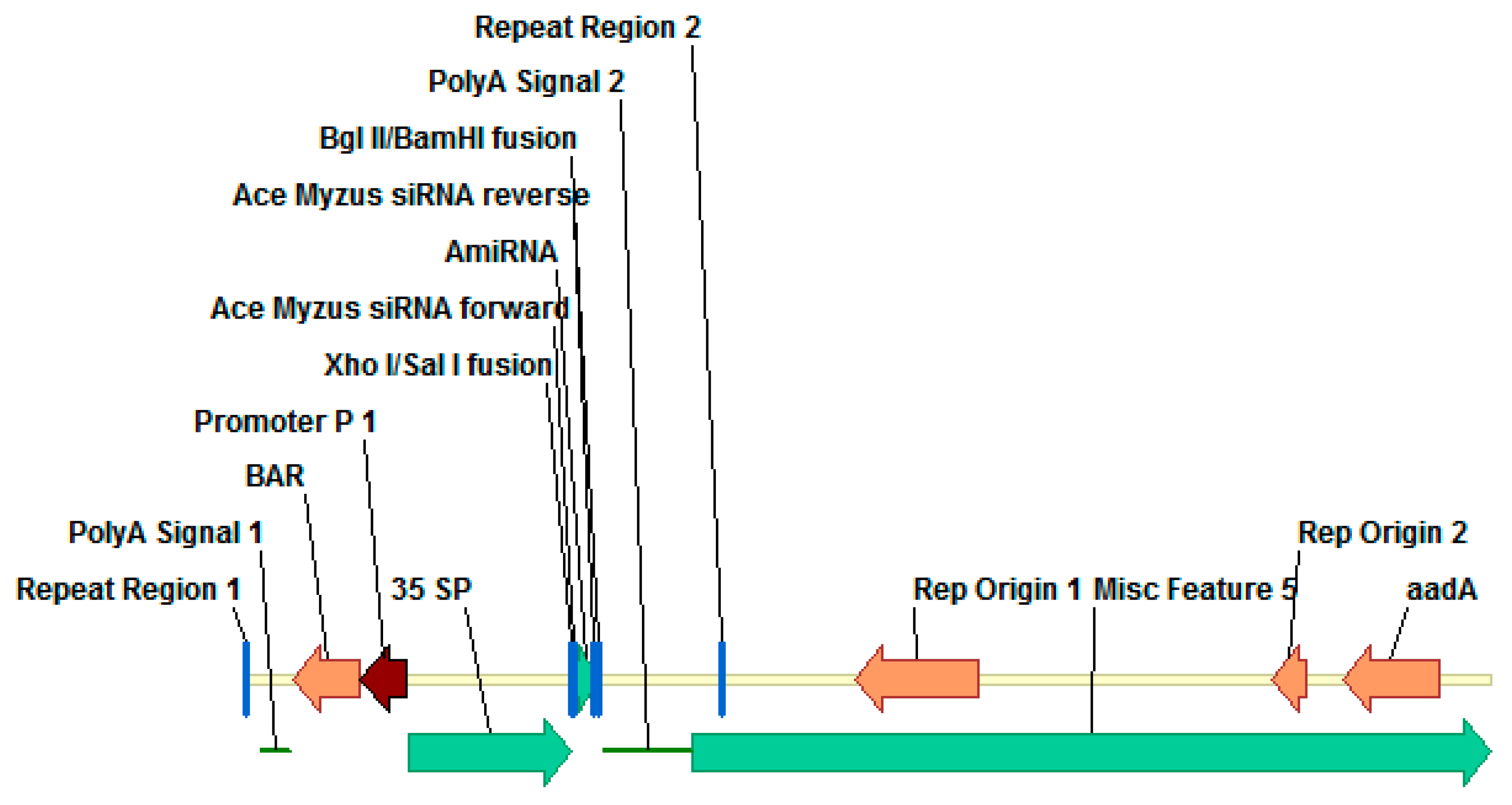
2.1. Vector Designed and Baterial Strain
2.2. Plant Materials and Genetic Transformation
2.3. Molecular Analysis of Transgenic Plants
2.4. qRT-PCR Analyses of Target Ace 1 Gene in Aphids
2.5. Aphid Performance on T1 Transgenic Plants
2.6. Data and Statistics
3. Results and Discussion
3.1. Transformation and Regeneration of Transgenic Plants
3.2. PCR Characterization of T0 Transgenic Plants
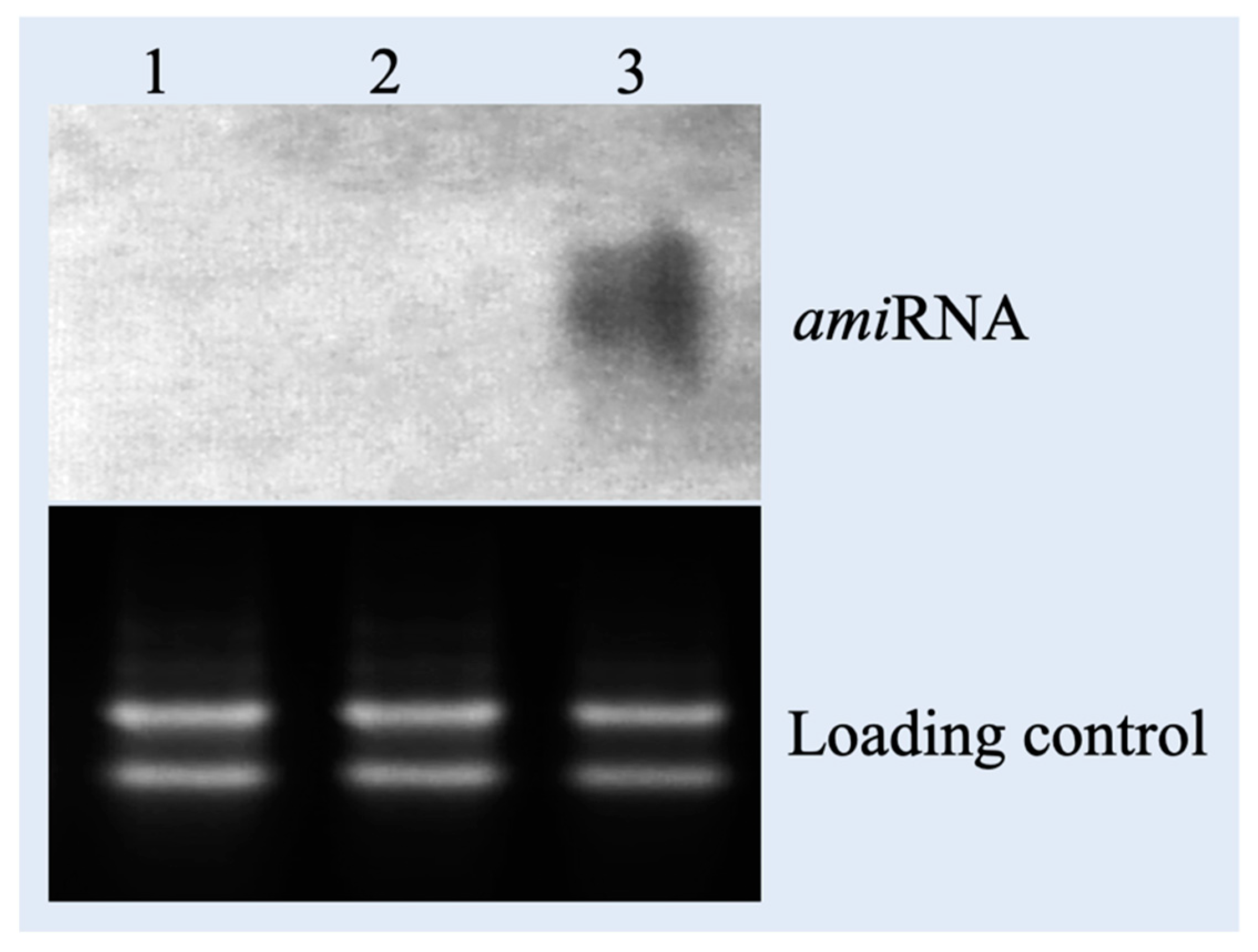
3.3. Northern Blotting of T0 Transgenic Plants
3.4. qRT-PCR Analysis of Aphids Fed on Plants Expressing amiRNA
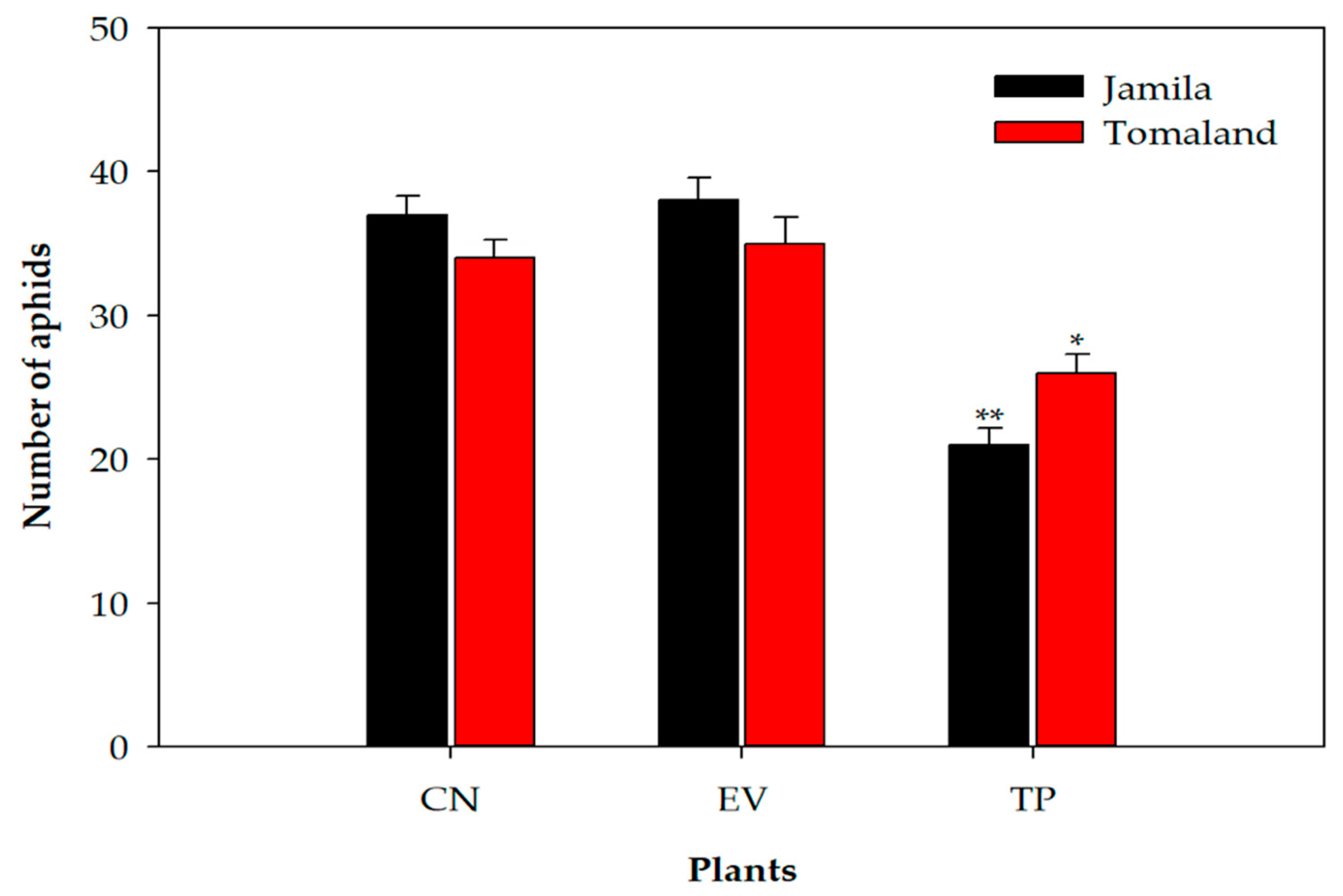
3.5. Aphid Resistance Against T1 Transgenic Tomato
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Genes Dev. 2002, 16, 1616–1626. [Google Scholar] [CrossRef] [Green Version]
- Charrier, A.; Vergne, E.; Joffrion, C.; Richer, A.; Dousset, N.; Chevreau, E. An artificial miRNA as a new tool to silence and explore gene functions in apple. Transgenic Res. 2019, 28, 611–626. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Quintero, Á.L.; López, C. Artificial microRNAs and their applications in plant molecular biology. Agron. Colomb. 2010, 28, 367–374. [Google Scholar]
- Zeng, Y.; Wagner, E.J.; Cullen, B.R. Both Natural and Designed Micro RNAs Can Inhibit the Expression of Cognate mRNAs When Expressed in Human Cells. Mol. Cell 2002, 9, 1327–1333. [Google Scholar] [CrossRef]
- Parizotto, E.A.; Dunoyer, P.; Rahm, N.; Himber, C.; Voinnet, O. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 2004, 18, 2237–2242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, Q.-W.; Lin, S.-S.; Reyes, J.L.; Chen, K.-C.; Wu, H.-W.; Yeh, S.-D.; Chua, N.-H. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 2006, 24, 1420–1428. [Google Scholar] [CrossRef]
- Eamens, A.L.; Agius, C.; Smith, N.A.; Waterhouse, P.M.; Wang, M.-B. Efficient Silencing of Endogenous MicroRNAs Using Artificial MicroRNAs in Arabidopsis thaliana. Mol. Plant 2011, 4, 157–170. [Google Scholar] [CrossRef]
- Zhang, X.; Li, H.; Zhang, J.; Zhang, C.; Gong, P.; Ziaf, K.; Xiao, F.; Ye, Z. Expression of artificial microRNAs in tomato confers efficient and stable virus resistance in a cell-autonomous manner. Transgenic Res. 2011, 20, 569–581. [Google Scholar] [CrossRef]
- Tiwari, M.; Sharma, D.; Trivedi, P.K. Artificial microRNA mediated gene silencing in plants: Progress and perspectives. Plant Mol. Biol. 2014, 86, 1–18. [Google Scholar] [CrossRef]
- Brunt, A.; Crabtree, K.; Dallwitz, M.; Gibbs, A.; Watson, L. Tomato yellow leaf curl bigeminivirus. In Virus of Plants: Description and Lists from the Vide Database; CAB International: London, UK, 1997; pp. 1319–1321. [Google Scholar]
- Hogenhout, S.A.; Ammar, E.-D.; Whitfield, A.E.; Redinbaugh, M.G. Insect Vector Interactions with Persistently Transmitted Viruses. Annu. Rev. Phytopathol. 2008, 46, 327–359. [Google Scholar] [CrossRef] [Green Version]
- Moran, P.J.; Cheng, Y.; Cassell, J.L.; Thompson, G.A. Gene expression profiling of Arabidopsis thaliana in compatible plant-aphid interactions. Arch. Insect Biochem. Physiol. 2002, 51, 182–203. [Google Scholar] [CrossRef] [PubMed]
- Emden, H.F.V.; Eastop, V.F.; Hughes, R.D.; Way, M.J. The Ecology of Myzus persicae. Annu. Rev. Entomol. 1969, 14, 197–270. [Google Scholar] [CrossRef]
- Olivera, S.; Rodriguez-Ithurralde, D.; Henley, J.M. Acetylcholinesterase promotes neurite elongation, synapse formation, and surface expression of AMPA receptors in hippocampal neurones. Mol. Cell. Neurosci. 2003, 23, 96–106. [Google Scholar] [CrossRef] [Green Version]
- Shapira, M.; Thompson, C.K.; Soreq, H.; Robinson, G.E. Changes in neuronal acetylcholinesterase gene expression and division of labor in honey bee colonies. J. Mol. Neurosci. 2001, 17, 1–12. [Google Scholar] [CrossRef]
- Kim, Y.H.; Lee, S.H. Invertebrate acetylcholinesterases: Insights into their evolution and non-classical functions. J. Asia-Pacif. Entomol. 2018, 21, 186–195. [Google Scholar] [CrossRef]
- Kim, J.I.; Jung, C.S.; Koh, Y.H.; Lee, S.H. Molecular, biochemical and histochemical characterization of two acetylcholinesterase cDNAs from the German cockroach Blattella germanica. Insect Mol. Biol. 2006, 15, 513–522. [Google Scholar] [CrossRef]
- Li, B.-L.; Chen, W.; Liu, L.; Zhang, X.-C.; Bao, Y.-Y.; Cheng, J.-A.; Zhu, Z.-R.; Zhang, C.-X. Molecular characterization of two acetylcholinesterase genes from the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Pestic. Biochem. Physiol. 2012, 102, 198–203. [Google Scholar] [CrossRef]
- Jiang, X.-C.; Jiang, X.-Y.; Liu, S. Molecular Characterization and Expression Analysis of Two Acetylcholinesterase Genes from the Small White Butterfly Pieris rapae (Lepidoptera: Pieridae). J. Insect Sci. 2018, 18, 2. [Google Scholar] [CrossRef]
- Vos, P.; Simons, G.; Jesse, T.; Wijbrandi, J.; Heinen, L.; Hogers, R.; Frijters, A.; Groenendijk, J.; Diergaarde, P.; Reijans, M.; et al. The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat. Biotechnol. 1998, 16, 1365–1369. [Google Scholar] [CrossRef]
- McCormick, S.; Niedermeyer, J.; Fry, J.; Barnason, A.; Horsch, R.; Fraley, R. Leaf disc transformation of cultivated tomato (L. esculentum) using Agrobacterium tumefaciens. Plant Cell Rep. 1986, 5, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Gal-On, A.; Wolf, D.; Wang, Y.; Faure, J.-E.; Pilowsky, M.; Zelcer, A. Transgenic resistance to cucumber mosaic virus in tomato: Blocking of long-distance movement of the virus in lines harboring a defective viral replicase gene. Phytopathology 1998, 88, 1101–1107. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Jia, L.; Goggin, F. The reliability of virus-induced gene silencing experiments using tobacco rattle virus in tomato is influenced by the size of the vector control. Mol. Plant Pathol. 2011, 12, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Picton, S.; Barton, S.L.; Bouzayen, M.; Hamilton, A.J.; Grierson, D. Altered fruit ripening and leaf senescence in tomatoes expressing an antisense ethylene-forming enzyme transgene. Plant J. 1993, 3, 469–481. [Google Scholar] [CrossRef]
- Reed, A.J.; Kretzmer, K.A.; Naylor, M.W.; Finn, R.F.; Magin, K.M.; Hammond, B.G.; Leimgruber, R.M.; Rogers, S.G.; Fuchs, R.L. Safety Assessment of 1-Aminocyclopropane-1-carboxylic Acid Deaminase Protein Expressed in Delayed Ripening Tomatoes. J. Agric. Food Chem. 1996, 44, 388–394. [Google Scholar] [CrossRef]
- Fischhoff, D.A.; Bowdish, K.S.; Perlak, F.J.; Marrone, P.G.; McCormick, S.M.; Niedermeyer, J.G.; Dean, D.A.; Kusano-Kretzmer, K.; Mayer, E.J.; Rochester, D.E.; et al. Insect Tolerant Transgenic Tomato Plants. Biotechnology 1987, 5, 807–813. [Google Scholar] [CrossRef]
- Fillatti, J.J.; Kiser, J.; Rose, R.; Comai, L. Efficient Transfer of a Glyphosate Tolerance Gene into Tomato Using a Binary Agrobacterium tumefaciens Vector. Biotechnology 1987, 5, 726–730. [Google Scholar] [CrossRef]
- Kim, J.W.; Sun, S.S.M.; German, T.L. Disease resistance in tobacco and tomato plants transformed with the tomato spotted wilt virus nucleocapsid gene. Plant Dis. 1994, 78, 615–621. [Google Scholar] [CrossRef]
- Raj, S.K.; Singh, R.; Pandey, S.K.; Singh, B.P. Agrobacterium-mediated tomato transformation and regeneration of transgenic lines expressing Tomato leaf curl virus coat protein gene for resistance against TLCV infection. Curr. Sci. 2005, 88, 1674–1679. [Google Scholar]
- Parveen, S.; Shahzad, A.; Saema, S. In vitro plant regeneration system for Cassia siamea Lam., a leguminous tree of economic importance. Agrofor. Syst. 2010, 80, 109–116. [Google Scholar] [CrossRef]
- Gisbert, C.; Rus, A.M.; Bolarín, M.C.; López-Coronado, J.M.; Arrillaga, I.; Montesinos, C.; Caro, M.; Serrano, R.; Moreno, V. The Yeast HAL1 Gene Improves Salt Tolerance of Transgenic Tomato. Plant Physiol. 2000, 123, 393. [Google Scholar] [CrossRef] [Green Version]
- Jia, G.X.; Zhu, Z.Q.; Chang, F.Q.; Li, Y.X. Transformation of tomato with the BADH gene from Atriplex improves salt tolerance. Plant Cell Rep. 2002, 21, 141–146. [Google Scholar] [CrossRef]
- Roy, R.; Purty, R.S.; Agrawal, V.; Gupta, S.C. Transformation of tomato cultivar ‘Pusa Ruby’ with bspA gene from Populus tremula for drought tolerance. Plant Cell Tissue Organ. Cult. 2005, 84, 56. [Google Scholar] [CrossRef]
- Kerschen, A.; Napoli, C.A.; Jorgensen, R.A.; Müller, A.E. Effectiveness of RNA interference in transgenic plants. FEBS Lett. 2004, 566, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Koundal, V.; Praveen, S. MicroRNA-based RNA Interference Vector for Gene Silencing in Plants. J. Plant Biochem. Biotechnol. 2010, 19, 79–82. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Faisal, M.; Abdel-Salam, E.M.; Alatar, A.A.; Saquib, Q.; Alwathnani, H.A.; Canto, T. Genetic Transformation and siRNA-Mediated Gene Silencing for Aphid Resistance in Tomato. Agronomy 2019, 9, 893. [Google Scholar] [CrossRef] [Green Version]
- Hill, K.; Schaller, G.E. Enhancing plant regeneration in tissue culture. Plant Signal. Behav. 2013, 8, e25709. [Google Scholar] [CrossRef] [Green Version]
- Ikeuchi, M.; Ogawa, Y.; Iwase, A.; Sugimoto, K. Plant regeneration: Cellular origins and molecular mechanisms. Development 2016, 143, 1442. [Google Scholar] [CrossRef] [Green Version]
- Bidabadi, S.S.; Jain, S.M. Cellular, Molecular, and Physiological Aspects of In Vitro Plant Regeneration. Plants 2020, 9, 702. [Google Scholar] [CrossRef]
- Khanna, H.K.; Paul, J.-Y.; Harding, R.M.; Dickman, M.B.; Dale, J.L. Inhibition of Agrobacterium-Induced Cell Death by Antiapoptotic Gene Expression Leads to Very High Transformation Efficiency of Banana. Mol. Plant-Microbe Interact. 2007, 20, 1048–1054. [Google Scholar] [CrossRef] [Green Version]
- Park, S.H.; Morris, J.L.; Park, J.E.; Hirschi, K.D.; Smith, R.H. Efficient and genotype-independent Agrobacterium-mediated tomato transformation. J. Plant Physiol. 2003, 160, 1253–1257. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.J.; Uchii, S.; Watanabe, S.; Ezura, H. A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol. 2006, 47, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Yasmeen, A. An improved protocol for the regeneration and transformation of tomato (cv Rio Grande). Acta Physiol. Plant. 2009, 31, 1271–1277. [Google Scholar] [CrossRef]
- Arshad, W.; Haq, I.-U.; Waheed, M.T.; Mysore, K.S.; Mirza, B. Agrobacterium-Mediated Transformation of Tomato with rolB Gene Results in Enhancement of Fruit Quality and Foliar Resistance against Fungal Pathogens. PLoS ONE 2014, 9, e96979. [Google Scholar] [CrossRef] [PubMed]
- Hansen, G.; Das, A.; Chilton, M.D. Constitutive expression of the virulence genes improves the efficiency of plant transformation by Agrobacterium. Proc. Natl. Acad. Sci. USA 1994, 91, 7603. [Google Scholar] [CrossRef] [Green Version]
- Nyaboga, E.; Tripathi, J.N.; Manoharan, R.; Tripathi, L. Agrobacterium-mediated genetic transformation of yam (Dioscorea rotundata): An important tool for functional study of genes and crop improvement. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, J.; Niu, Q.-W.; Ikeda, Y.; Chua, N.-H. Marker-free transformation: Increasing transformation frequency by the use of regeneration-promoting genes. Curr. Opin. Biotechnol. 2002, 13, 173–180. [Google Scholar] [CrossRef]
- Vu, T.V.; Roy Choudhury, N.; Mukherjee, S.K. Transgenic tomato plants expressing artificial microRNAs for silencing the pre-coat and coat proteins of a begomovirus, Tomato leaf curl New Delhi virus, show tolerance to virus infection. Virus Res. 2013, 172, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Song, X.; Wang, G.; Yang, K.; Wang, Y.; Niu, L.; Chen, X.; Fang, R. Plant-Generated Artificial Small RNAs Mediated Aphid Resistance. PLoS ONE 2014, 9, e97410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Y.; Friedrich, M. Nymphal RNAi: Systemic RNAi mediated gene knockdown in juvenile grasshopper. BMC Biotechnol. 2005, 5, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araujo, R.N.; Santos, A.; Pinto, F.S.; Gontijo, N.F.; Lehane, M.J.; Pereira, M.H. RNA interference of the salivary gland nitrophorin 2 in the triatomine bug Rhodnius prolixus (Hemiptera: Reduviidae) by dsRNA ingestion or injection. Insect Biochem. Mol. Biol. 2006, 36, 683–693. [Google Scholar] [CrossRef] [Green Version]
- Smith, M.C.; Schotzko, D.J.; Zemetra, R.S.; Souza, E.J. Categories of Resistance in Plant Introductions of Wheat Resistant to the Russian Wheat Aphid (Homoptera: Aphididae). J. Econ. Entomol. 1992, 85, 1480–1484. [Google Scholar] [CrossRef]
- Orr, D. Biological Control and Integrated Pest Management. In Integrated Pest Management: Innovation-Development Process; Peshin, R., Dhawan, A.K., Eds.; Springer: Dordrecht, The Netherlands, 2009; Volume 1, pp. 207–239. [Google Scholar] [CrossRef]
- Amer, M.; Aslam, M.; Razaq, M.; Afzal, M. Lack of plant resistance against aphids, as indicated by their seasonal abundance in canola, Brassica napus (L.) in Southern Punjab, Pakistan. Pakistan J. Bot 2009, 41, 1043–1051. [Google Scholar]
- Akbar, W.; Asif, M.U.; Muhammad, R.; Khan, M.H.; Bux, M. Abundance of Aphids on Various Canola Genotypes and Role of Abiotic Factors in its Population Fluctuation. Sci. Lett. 2019, 7, 52–58. [Google Scholar]
- Malik, H.J.; Raza, A.; Amin, I.; Scheffler, J.A.; Scheffler, B.E.; Brown, J.K.; Mansoor, S. RNAi-mediated mortality of the whitefly through transgenic expression of double-stranded RNA homologous to acetylcholinesterase and ecdysone receptor in tobacco plants. Sci. Rep. 2016, 6, 38469. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Faisal, M.; Abdel-Salam, E.M.; Alatar, A.A. Artificial microRNA-Based RNA Interference and Specific Gene Silencing for Developing Insect Resistance in Solanum lycopersicum. Agronomy 2021, 11, 136. https://doi.org/10.3390/agronomy11010136
Faisal M, Abdel-Salam EM, Alatar AA. Artificial microRNA-Based RNA Interference and Specific Gene Silencing for Developing Insect Resistance in Solanum lycopersicum. Agronomy. 2021; 11(1):136. https://doi.org/10.3390/agronomy11010136
Chicago/Turabian StyleFaisal, Mohammad, Eslam M. Abdel-Salam, and Abdulrahman A. Alatar. 2021. "Artificial microRNA-Based RNA Interference and Specific Gene Silencing for Developing Insect Resistance in Solanum lycopersicum" Agronomy 11, no. 1: 136. https://doi.org/10.3390/agronomy11010136
APA StyleFaisal, M., Abdel-Salam, E. M., & Alatar, A. A. (2021). Artificial microRNA-Based RNA Interference and Specific Gene Silencing for Developing Insect Resistance in Solanum lycopersicum. Agronomy, 11(1), 136. https://doi.org/10.3390/agronomy11010136






