Genome-Wide Identification, Characterization, and Expression Analyses of P-Type ATPase Superfamily Genes in Soybean
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Annotation of P-Type ATPase Genes in the Soybean Genome
2.2. Phylogenetic Tree Construction and Chromosome Distribution
2.3. Gene Duplication Analysis and Calculation of Ka/Ks Values
2.4. In Silico Expression Analysis of P-Type ATPase Genes in Soybean
2.5. Analysis of Cis-Acting Regulatory Elements
2.6. Plant Growth and qRT-PCR Analysis
3. Results
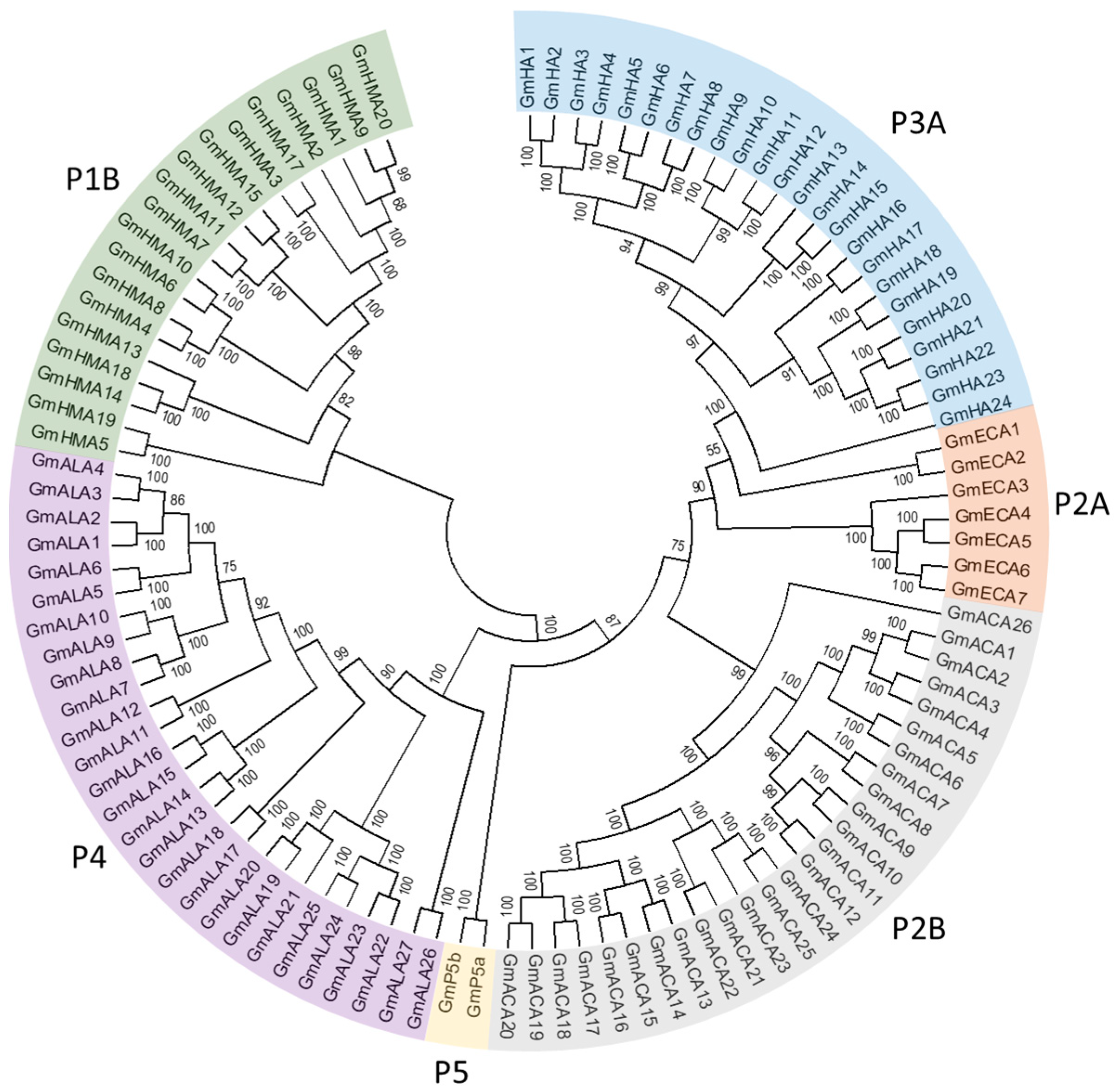
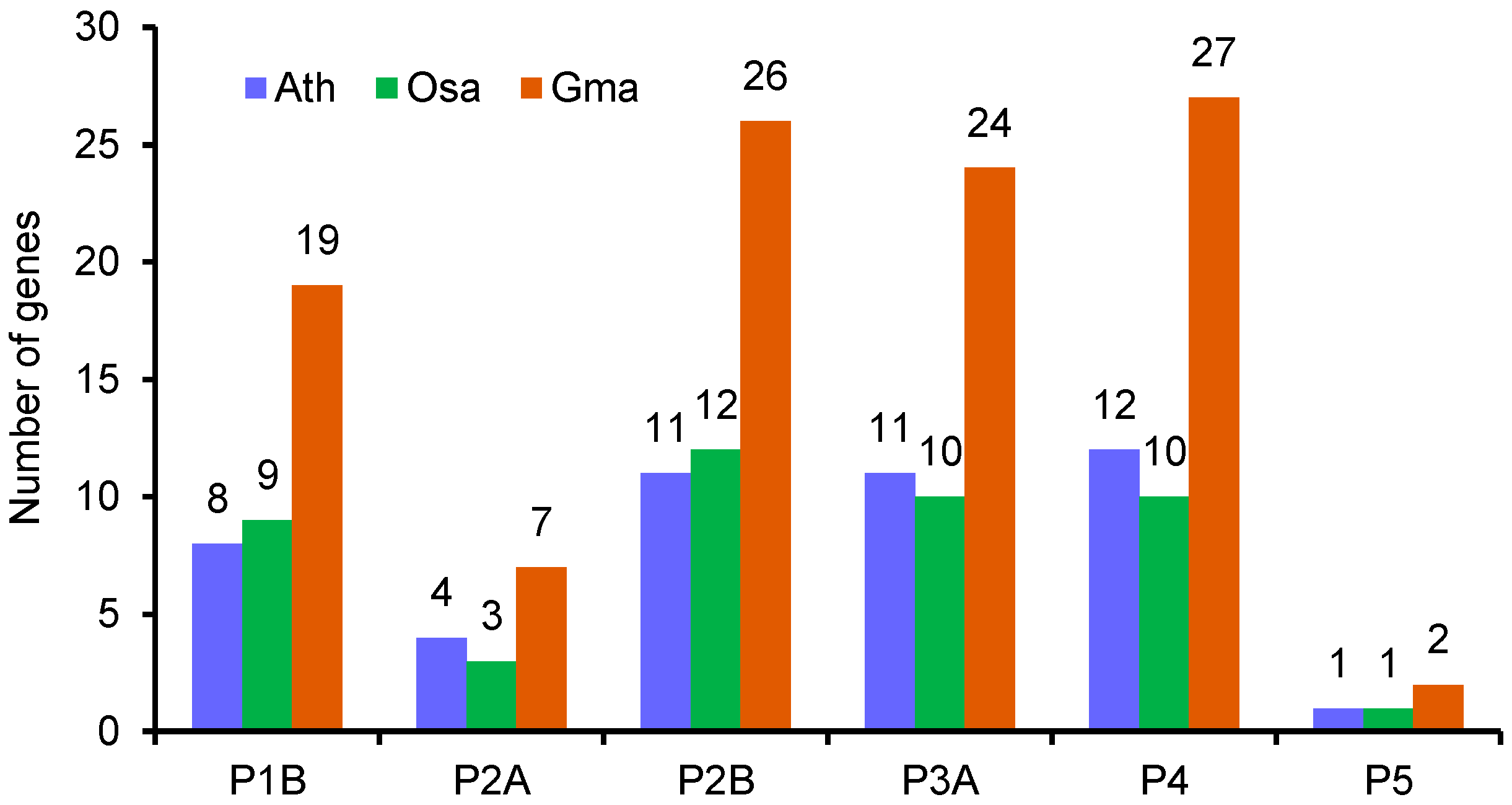
3.1. Identification of Putative Genes Encoding P-Type ATPase Proteins in the Soybean Genome
3.2. P1B Subfamily Genes in Soybean
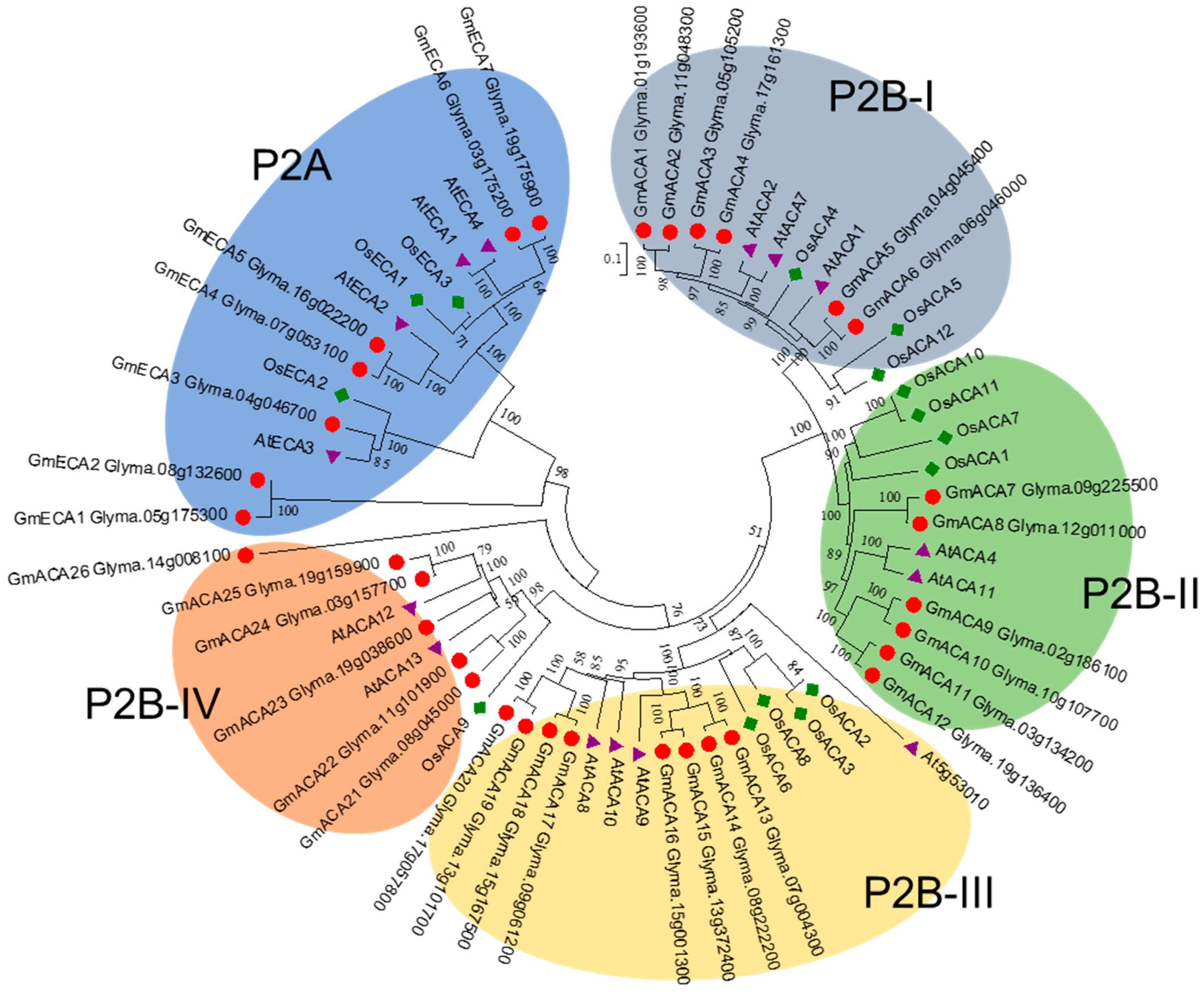
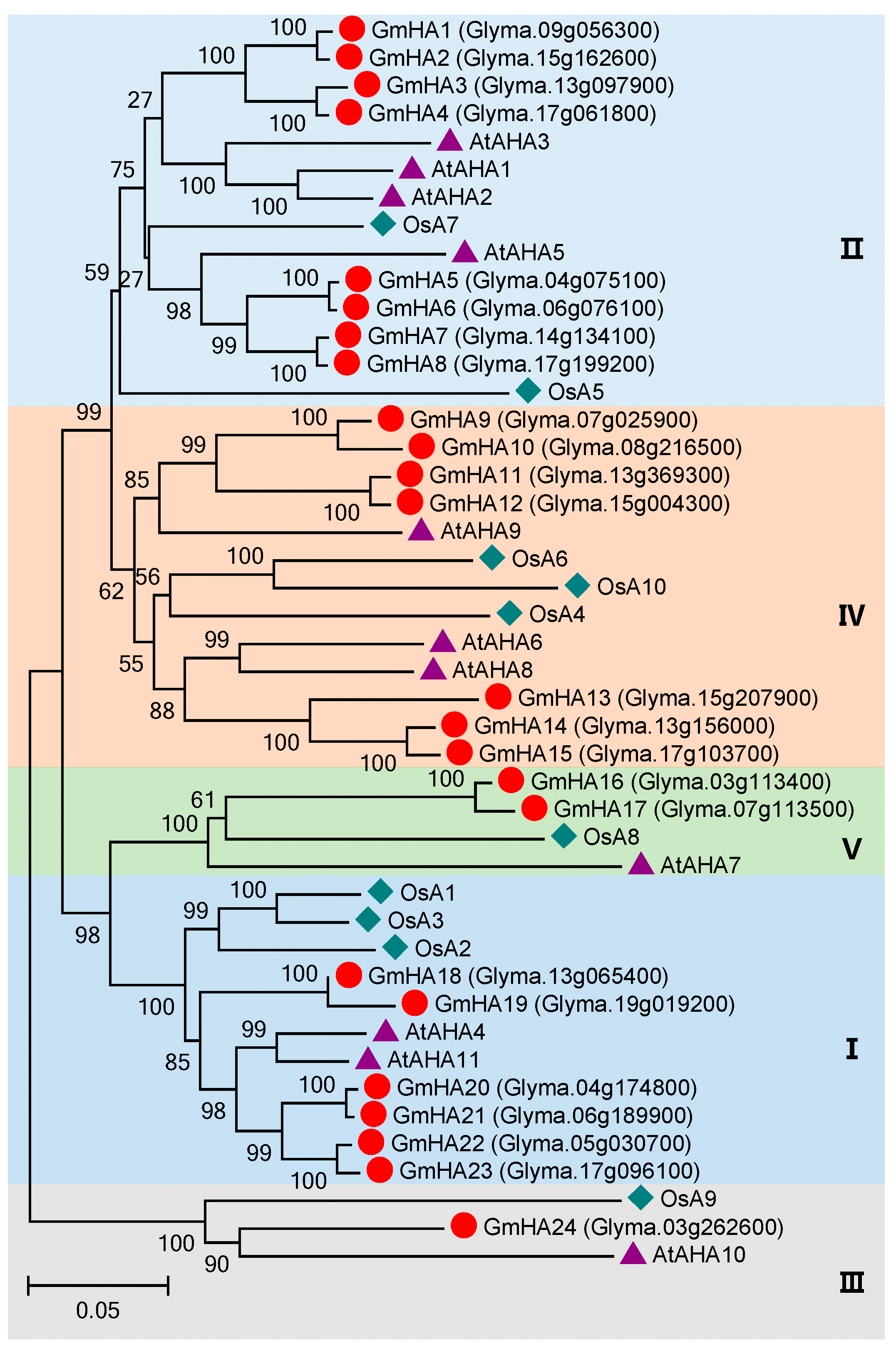
3.3. P2 Subfamily Genes in Soybean
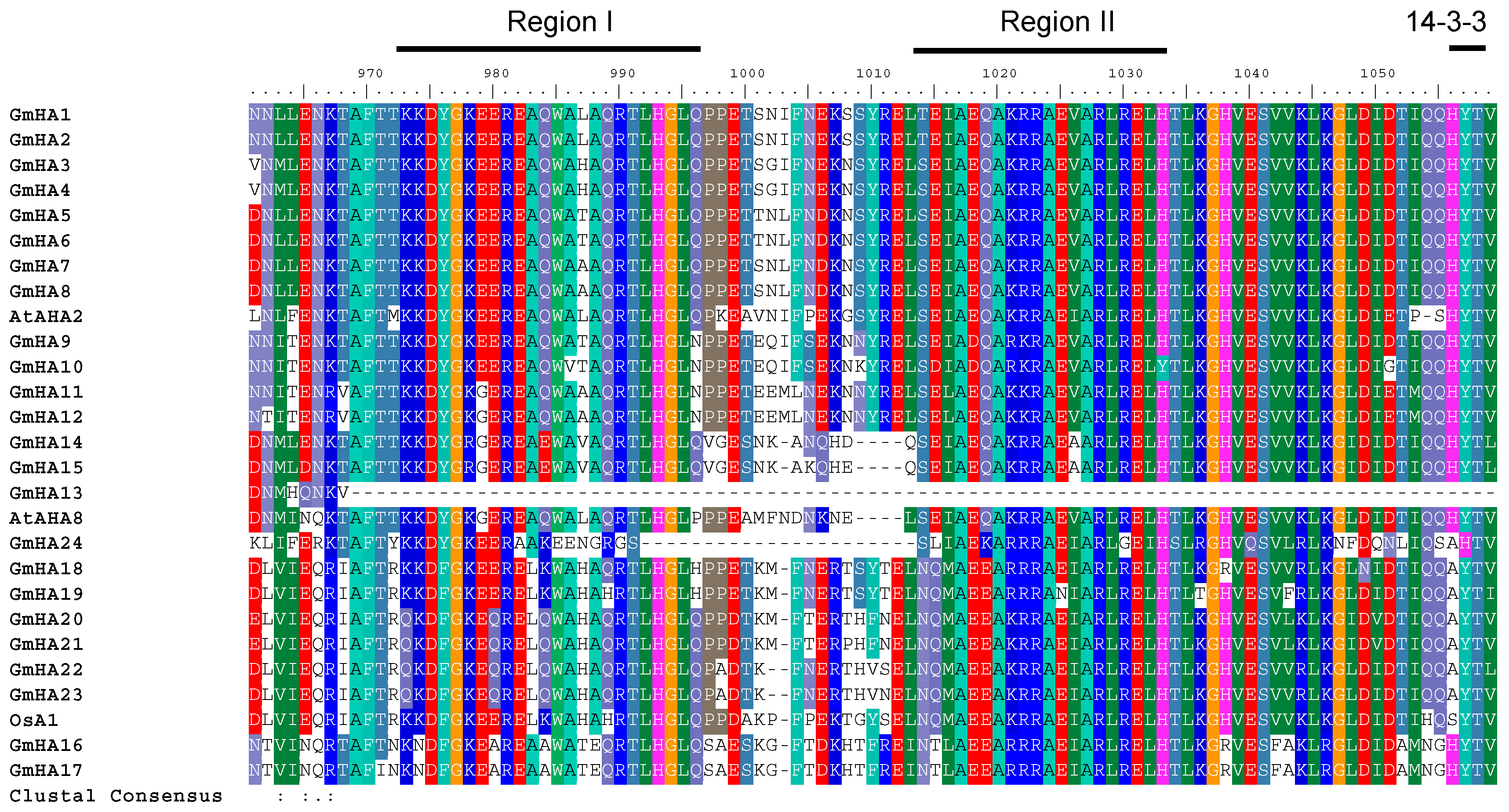
3.4. P3 Subfamily Genes in Soybean
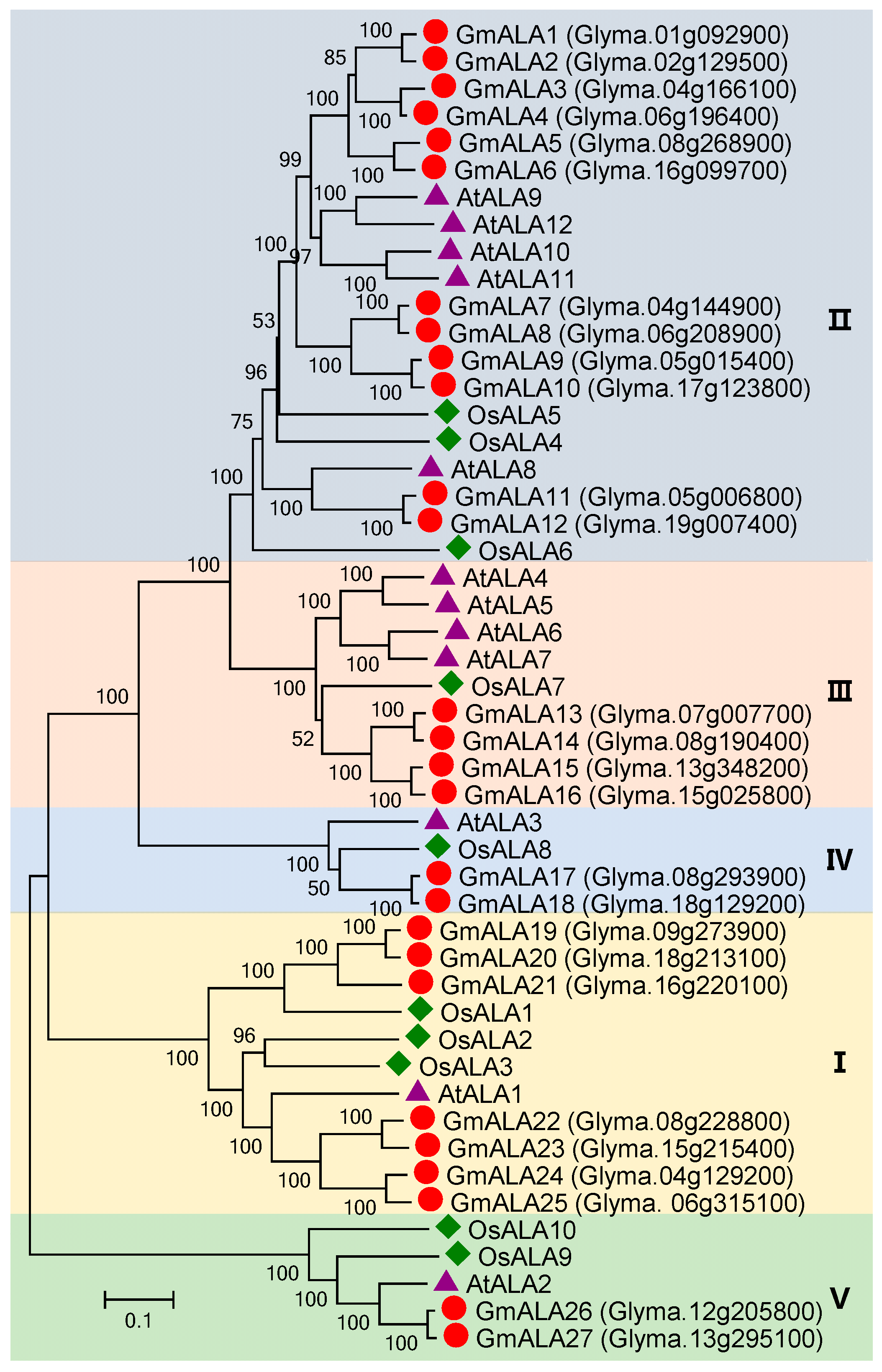
3.5. P4 Subfamily Genes in Soybean
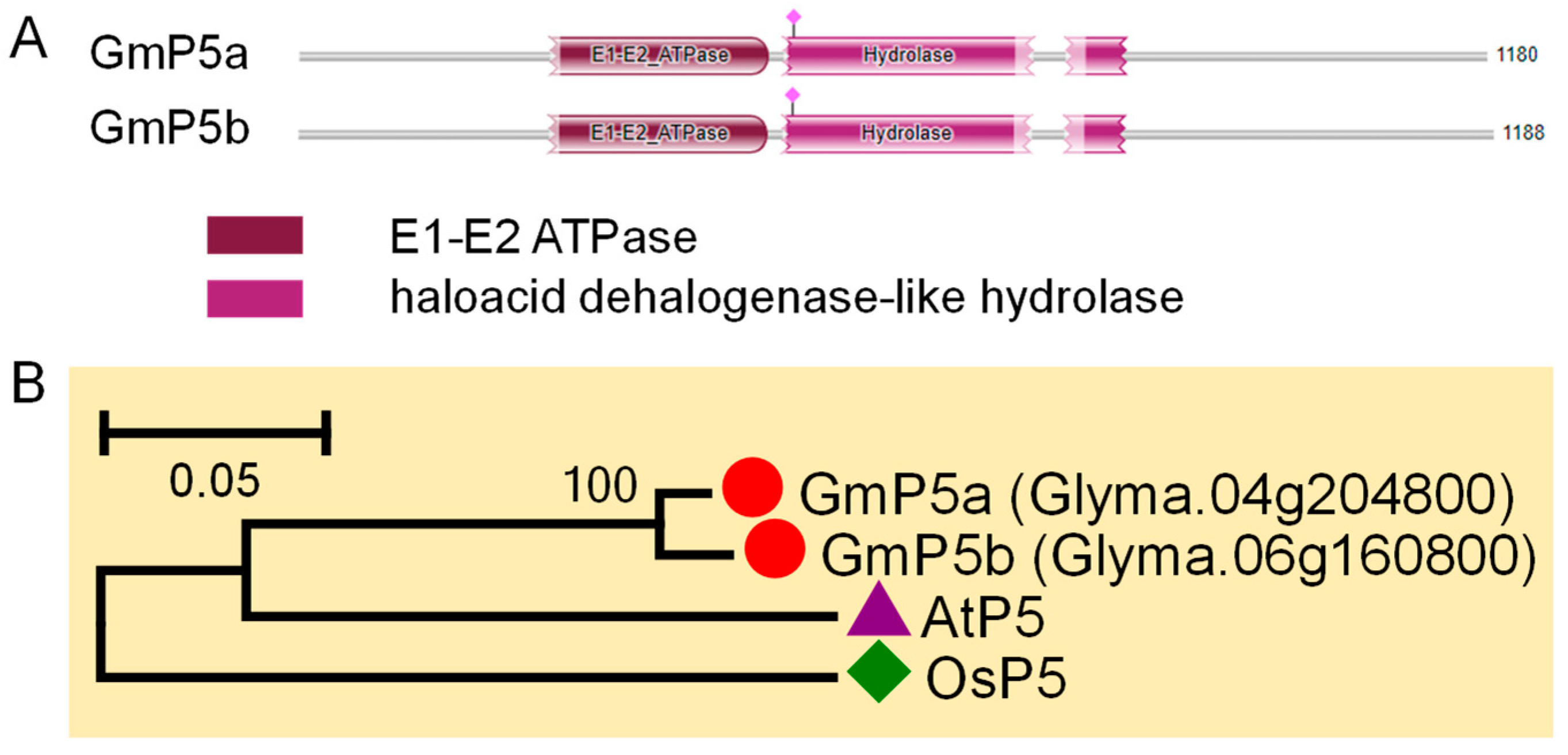
3.6. P5 Subfamily Genes in Soybean
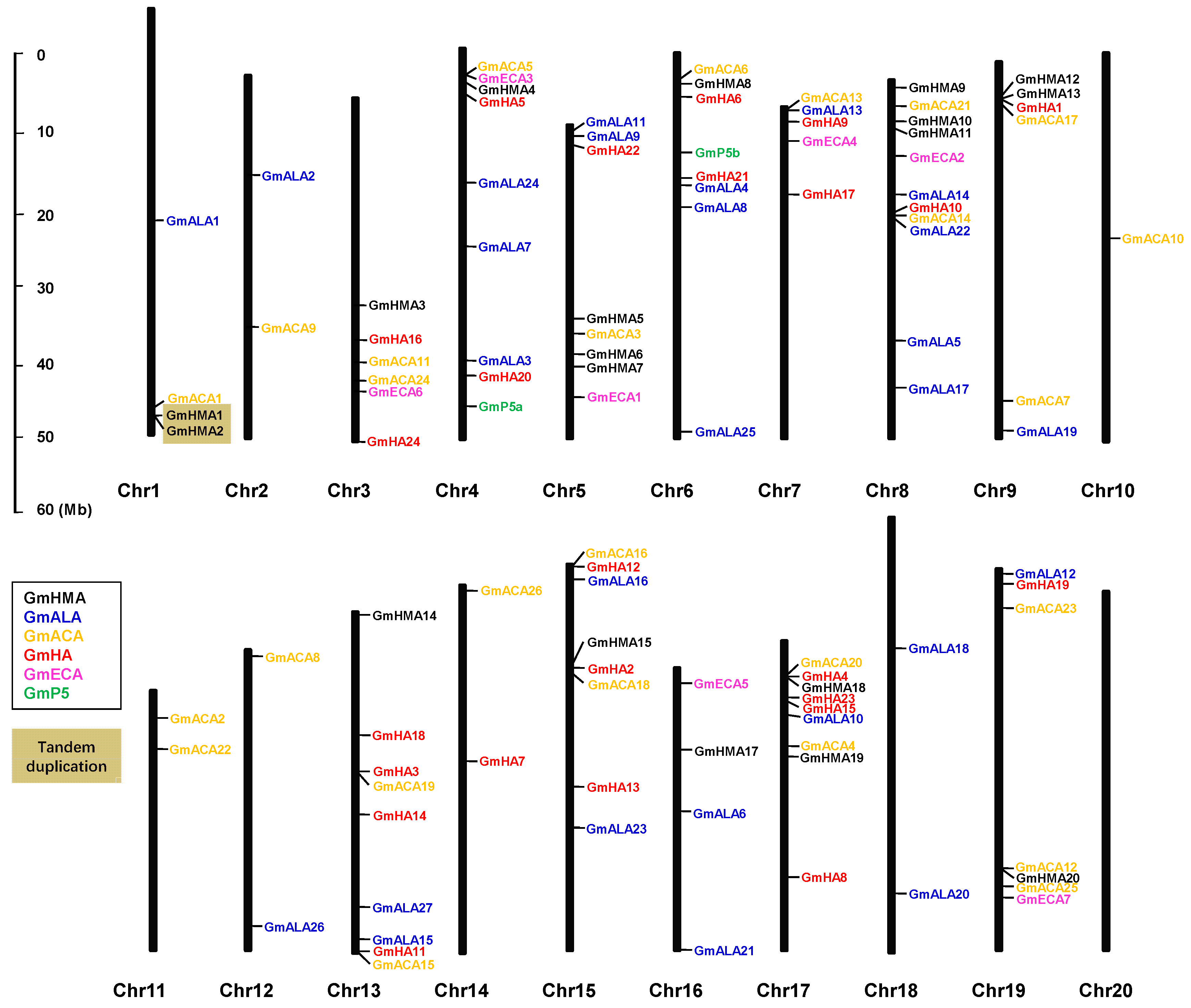
3.7. Chromosomal Distribution and Gene Duplication of Soybean P-Type ATPase Genes
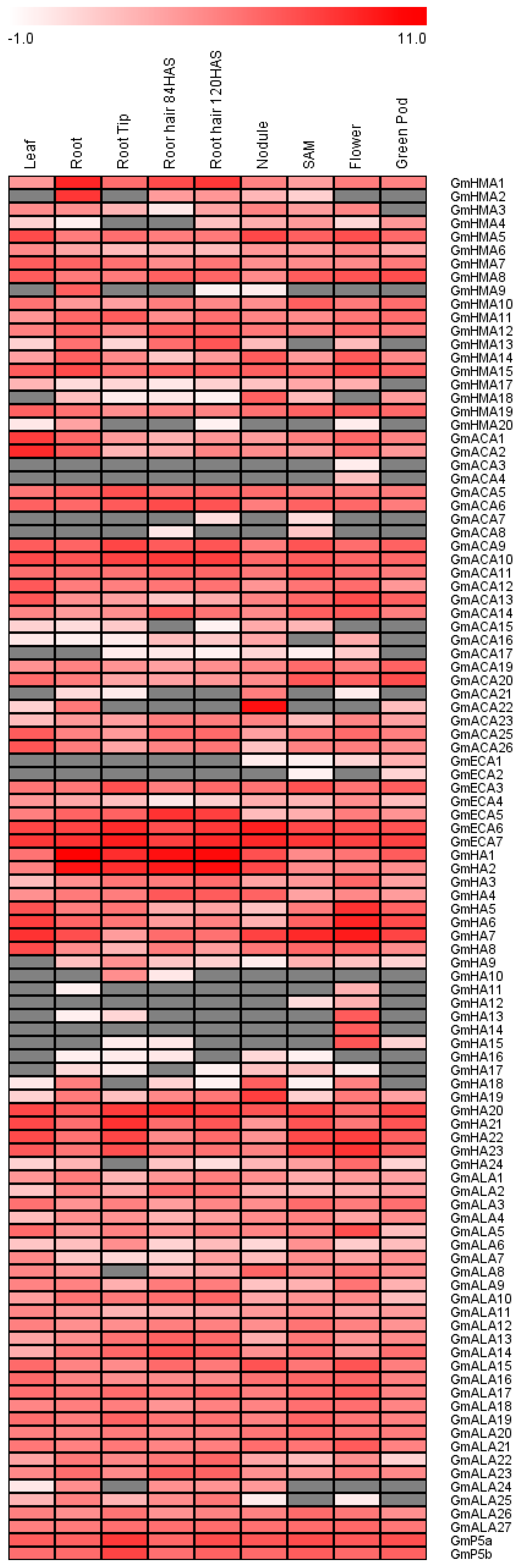
3.8. Tissue Expression Patterns of Soybean P-Type ATPase Genes
3.9. Analysis of Cis-Acting Element in the Promoter Regions of Soybean P-Type ATPase Genes
3.10. Expression Analysis of Soybean P-Type ATPase Genes in Response to Stresses and Rhizobia Inoculation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Palmgren, M.G.; Nissen, P. P-Type ATPases. Annu. Rev. Biophys. 2011, 40, 243–266. [Google Scholar] [CrossRef] [PubMed]
- Axelsen, B.K.; Palmgren, G.M. Evolution of Substrate Specificities in the P-Type ATPase Superfamily. J. Mol. Evol. 1998, 46, 84–101. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, C.N.; Axelsen, K.B.; Harper, J.F.; Palmgren, M.G. Evolution of plant P-type ATPases. Front. Plant Sci. 2012, 3, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmgren, M.; Østerberg, J.T.; Nintemann, S.J.; Poulsen, L.R.; López-Marqués, R.L. Evolution and a revised nomenclature of P4 ATPases, a eukaryotic family of lipid flippases. Biochim. Et Biophys. Acta (Bba) Biomembr. 2019, 1861, 1135–1151. [Google Scholar] [CrossRef]
- Sørensen, D.M.; Holen, H.W.; Holemans, T.; Vangheluwe, P.; Palmgren, M.G. Towards defining the substrate of orphan P5A-ATPases. Biochim. Et Biophys. Acta (Bba) Gen. Subj. 2015, 1850, 524–535. [Google Scholar]
- Inoue, S.I.; Kinoshita, T. Blue Light Regulation of Stomatal Opening and the Plasma Membrane H(+)-ATPase. Plant Physiol. 2017, 174, 531–538. [Google Scholar] [CrossRef] [Green Version]
- Falhof, J.; Pedersen, J.T.; Fuglsang, A.T.; Palmgren, M. Plasma Membrane H(+)-ATPase Regulation in the Center of Plant Physiology. Mol. Plant 2016, 9, 323–337. [Google Scholar] [CrossRef] [Green Version]
- Huda, K.M.K.; Banu, M.S.A.; Tuteja, R.; Tuteja, N. Global calcium transducer P-type Ca2+-ATPases open new avenues for agriculture by regulating stress signalling. J. Exp. Bot. 2013, 64, 3099–3109. [Google Scholar] [CrossRef]
- Williams, L.E.; Mills, R.F. P1B-ATPases—An ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci. 2005, 10, 491–502. [Google Scholar] [CrossRef]
- Poulsen, L.R.; Lopez-Marques, R.L.; McDowell, S.C.; Okkeri, J.; Licht, D.; Schulz, A.; Pomorski, T.; Harper, J.F.; Palmgren, M.G. The Arabidopsis P4-ATPase ALA3 localizes to the golgi and requires a beta-subunit to function in lipid translocation and secretory vesicle formation. Plant Cell 2008, 20, 658–676. [Google Scholar] [CrossRef] [Green Version]
- Baxter, I.; Tchieu, J.; Sussman, M.R.; Boutry, M.; Palmgren, M.G.; Gribskov, M.; Harper, J.F.; Axelsen, K.B. Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol. 2003, 132, 618–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Axelsen, K.B.; Palmgren, M.G. Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiol. 2001, 126, 696–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huda, K.M.K.; Yadav, S.; Akhter Banu, M.S.; Trivedi, D.K.; Tuteja, N. Genome-wide analysis of plant-type II Ca(2+)ATPases gene family from rice and Arabidopsis: Potential role in abiotic stresses. Plant Physiol. Biochem. Ppb Soc. Fr. Physiol. Veg. 2013, 65, 32–47. [Google Scholar] [CrossRef] [PubMed]
- Rensing, C.; Ghosh, M.; Rosen, B.P. Families of Soft-Metal-Ion-Transporting ATPases. J. Bacteriol. 1999, 181, 5891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argüello, J.M.; Eren, E.; González-Guerrero, M. The structure and function of heavy metal transport P1B-ATPases. Biometals 2007, 20, 233. [Google Scholar] [CrossRef]
- Huang, X.-Y.; Deng, F.; Yamaji, N.; Pinson, S.R.; Fujii-Kashino, M.; Danku, J.; Douglas, A.; Guerinot, M.L.; Salt, D.E.; Ma, J.F. A heavy metal P-type ATPase OsHMA4 prevents copper accumulation in rice grain. Nat. Commun. 2016, 7, 12138. [Google Scholar] [CrossRef] [Green Version]
- Morel, M.; Crouzet, J.; Gravot, A.; Auroy, P.; Leonhardt, N.; Vavasseur, A.; Richaud, P. AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 2009, 149, 894–904. [Google Scholar] [CrossRef] [Green Version]
- Eren, E.; Argüello, J.M. Arabidopsis HMA2, a divalent heavy metal-transporting P(IB)-type ATPase, is involved in cytoplasmic Zn2+ homeostasis. Plant Physiol. 2004, 136, 3712–3723. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, R.; Bashir, K.; Ishimaru, Y.; Nishizawa, N.K.; Nakanishi, H. The role of heavy-metal ATPases, HMAs, in zinc and cadmium transport in rice. Plant Signal. Behav. 2012, 7, 1605–1607. [Google Scholar] [CrossRef]
- Cai, H.; Huang, S.; Che, J.; Yamaji, N.; Ma, J.F. The tonoplast-localized transporter OsHMA3 plays an important role in maintaining Zn homeostasis in rice. J. Exp. Bot. 2019, 70, 2717–2725. [Google Scholar] [CrossRef] [Green Version]
- Bonza, M.C.; De Michelis, M.I. The plant Ca2+-ATPase repertoire: Biochemical features and physiological functions. Plant Biol. 2011, 13, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Bonza, M.C.; Morandini, P.; Luoni, L.; Geisler, M.; Palmgren, M.G.; De Michelis, M.I. At-ACA8 encodes a plasma membrane-localized calcium-ATPase of Arabidopsis with a calmodulin-binding domain at the N terminus. Plant Physiol. 2000, 123, 1495–1506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, W.S.; Lee, S.H.; Kim, J.C.; Heo, W.D.; Kim, M.C.; Park, C.Y.; Park, H.C.; Lim, C.O.; Kim, W.B.; Harper, J.F.; et al. Identification of a calmodulin-regulated soybean Ca(2+)-ATPase (SCA1) that is located in the plasma membrane. Plant Cell 2000, 12, 1393–1407. [Google Scholar] [CrossRef] [PubMed]
- Huda, K.M.; Banu, M.; Akhter, S.; Garg, B.; Tula, S.; Tuteja, R.; Tuteja, N. OsACA6, a P-type IIB Ca2+ ATPase promotes salinity and drought stress tolerance in tobacco by ROS scavenging and enhancing stress-responsive genes. Plant J. 2013, 76, 997–1015. [Google Scholar] [CrossRef] [PubMed]
- Schiøtt, M.; Romanowsky, S.M.; Baekgaard, L.; Jakobsen, M.K.; Palmgren, M.G.; Harper, J.F. A plant plasma membrane Ca2+ pump is required for normal pollen tube growth and fertilization. Proc. Natl. Acad. Sci. USA 2004, 101, 9502–9507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, M.; Jia, B.; Cui, N.; Wen, Y.; Duanmu, H.; Yu, Q.; Xiao, J.; Sun, X.; Zhu, Y. Functional characterization of a Glycine soja Ca2+ATPase in salt–alkaline stress responses. Plant Mol. Biol. 2016, 90, 419–434. [Google Scholar] [CrossRef]
- Hilleary, R.; Paez-Valencia, J.; Vens, C.; Toyota, M.; Palmgren, M.; Gilroy, S. Tonoplast-localized Ca(2+) pumps regulate Ca(2+) signals during pattern-triggered immunity in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2020, 117, 18849–18857. [Google Scholar] [CrossRef]
- Palmgren, M.G. Plant Plasma Membrane H+-ATPases: Powerhouses for Nutrient Uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 817–845. [Google Scholar] [CrossRef] [Green Version]
- Arango, M.; Gevaudant, F.; Oufattole, M.; Boutry, M. The plasma membrane proton pump ATPase: The significance of gene subfamilies. Planta 2003, 216, 355–365. [Google Scholar] [CrossRef]
- Duby, G.; Boutry, M. The plant plasma membrane proton pump ATPase: A highly regulated P-type ATPase with multiple physiological roles. Pflug Arch. Eur. J. Physiol. 2009, 457, 645–655. [Google Scholar] [CrossRef]
- Kinoshita, T.; Shimazaki, K. Blue light activates the plasma membrane H(+)-ATPase by phosphorylation of the C-terminus in stomatal guard cells. Embo J. 1999, 18, 5548–5558. [Google Scholar] [CrossRef] [Green Version]
- Olsson, A.; Svennelid, F.; Ek, B.; Sommarin, M.; Larsson, C. A phosphothreonine residue at the C-terminal end of the plasma membrane H+-ATPase is protected by fusicoccin-induced 14-3-3 binding. Plant Physiol. 1998, 118, 551–555. [Google Scholar] [CrossRef] [Green Version]
- Zeng, H.Q.; Di, T.J.; Zhu, Y.Y.; Subbarao, G.V. Transcriptional response of plasma membrane H+-ATPase genes to ammonium nutrition and its functional link to the release of biological nitrification inhibitors from sorghum roots. Plant Soil 2016, 398, 301–312. [Google Scholar] [CrossRef]
- Yan, F.; Zhu, Y.; Muller, C.; Zorb, C.; Schubert, S. Adaptation of H+-pumping and plasma membrane H+ ATPase activity in proteoid roots of white lupin under phosphate deficiency. Plant Physiol. 2002, 129, 50–63. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, K.; Hayashi, K.-I.; Kinoshita, T. Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol. 2012, 159, 632–641. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.X.; Ding, M.; Xu, F.Y.; Afzal, M.R.; Chen, X.; Zeng, H.Q.; Yan, F.; Zhu, Y.Y. Involvement of plasma membrane H+-ATPase in the ammonium-nutrition response of barley roots. J. Plant Nutr. Soil Sci. 2018, 181, 878–885. [Google Scholar] [CrossRef]
- Lopez-Marques, R.L.; Theorin, L.; Palmgren, M.G.; Pomorski, T.G. P4-ATPases: Lipid flippases in cell membranes. Pflug. Arch. Eur. J. Physiol. 2014, 466, 1227–1240. [Google Scholar] [CrossRef] [Green Version]
- Baldridge, R.D.; Graham, T.R. Identification of residues defining phospholipid flippase substrate specificity of type IV P-type ATPases. Proc. Natl. Acad. Sci. USA 2012, 109, E290–E298. [Google Scholar] [CrossRef] [Green Version]
- Gomès, E.; Jakobsen, M.K.; Axelsen, K.B.; Geisler, M.; Palmgren, M.G. Chilling Tolerance in Arabidopsis Involves ALA1, a Member of a New Family of Putative Aminophospholipid Translocases. Plant Cell 2000, 12, 2441. [Google Scholar] [CrossRef]
- Poulsen, L.R.; López-Marqués, R.L.; Pedas, P.R.; McDowell, S.C.; Brown, E.; Kunze, R.; Harper, J.F.; Pomorski, T.G.; Palmgren, M. A phospholipid uptake system in the model plant Arabidopsis thaliana. Nat. Commun. 2015, 6, 7649. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Li, Q.; Xu, L.; Qiao, X.; Liu, C.; Zhang, S. Comparative analysis of the P-type ATPase gene family in seven Rosaceae species and an expression analysis in pear (Pyrus bretschneideri Rehd.). Genomics 2020, 112, 2550–2563. [Google Scholar] [CrossRef]
- Chen, W.; Si, G.-Y.; Zhao, G.; Abdullah, M.; Guo, N.; Li, D.-H.; Sun, X.; Cai, Y.-P.; Lin, Y.; Gao, J.-S. Genomic Comparison of the P-ATPase Gene Family in Four Cotton Species and Their Expression Patterns in Gossypium hirsutum. Molecules 2018, 23, 5. [Google Scholar] [CrossRef] [Green Version]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [Green Version]
- Chan, C.; Qi, X.; Li, M.-W.; Wong, F.-L.; Lam, H.-M. Recent Developments of Genomic Research in Soybean. J. Genet. Genom. 2012, 39, 317–324. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, M.; Feng, F.; Tian, Z. Toward a “Green Revolution” for Soybean. Mol. Plant 2020, 13, 688–697. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [Green Version]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L.; et al. InterPro: The integrative protein signature database. Nucleic Acids Res. 2009, 37 (Suppl. 1), D211–D215. [Google Scholar] [CrossRef] [Green Version]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes11Edited by F. Cohen. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.H.; Tang, H.; Wang, X.; Paterson, A.H. PGDD: A database of gene and genome duplication in plants. Nucleic Acids Res. 2013, 41, D1152–D1158. [Google Scholar] [CrossRef]
- Zeng, H.; Zhang, Y.; Zhang, X.; Pi, E.; Zhu, Y. Analysis of EF-Hand proteins in soybean genome suggests their potential roles in environmental and nutritional stress signaling. Front. Plant Sci. 2017, 8, 877. [Google Scholar] [CrossRef] [Green Version]
- Patil, G.; Valliyodan, B.; Deshmukh, R.; Prince, S.; Nicander, B.; Zhao, M.; Sonah, H.; Song, L.; Lin, L.; Chaudhary, J.; et al. Soybean (Glycine max) SWEET gene family: Insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genom. 2015, 16, 520. [Google Scholar] [CrossRef] [Green Version]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Severin, A.J.; Woody, J.L.; Bolon, Y.-T.; Joseph, B.; Diers, B.W.; Farmer, A.D.; Muehlbauer, G.J.; Nelson, R.T.; Grant, D.; Specht, J.E. RNA-Seq Atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant Biol. 2010, 10, 160. [Google Scholar] [CrossRef] [Green Version]
- Libault, M.; Farmer, A.; Joshi, T.; Takahashi, K.; Langley, R.J.; Franklin, L.D.; He, J.; Xu, D.; May, G.; Stacey, G. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 2010, 63, 86–99. [Google Scholar] [CrossRef]
- Belamkar, V.; Weeks, N.T.; Bharti, A.K.; Farmer, A.D.; Graham, M.A.; Cannon, S.B. Comprehensive characterization and RNA-Seq profiling of the HD-Zip transcription factor family in soybean (Glycine max) during dehydration and salt stress. BMC Genom. 2014, 15, 950. [Google Scholar] [CrossRef] [Green Version]
- Maruyama, K.; Todaka, D.; Mizoi, J.; Yoshida, T.; Kidokoro, S.; Matsukura, S.; Takasaki, H.; Sakurai, T.; Yamamoto, Y.Y.; Yoshiwara, K.; et al. Identification of cis-acting promoter elements in cold- and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2012, 19, 37–49. [Google Scholar] [CrossRef]
- Chen, W.; Yao, Q.; Patil, G.B.; Agarwal, G.; Deshmukh, R.K.; Lin, L.; Wang, B.; Wang, Y.; Prince, S.J.; Song, L.; et al. Identification and Comparative Analysis of Differential Gene Expression in Soybean Leaf Tissue under Drought and Flooding Stress Revealed by RNA-Seq. Front. Plant Sci. 2016, 7, 1044. [Google Scholar] [CrossRef] [Green Version]
- Zeng, H.Q.; Wang, G.P.; Zhang, Y.Q.; Hu, X.Y.; Pi, E.X.; Zhu, Y.Y.; Wang, H.Z.; Du, L.Q. Genome-wide identification of phosphate-deficiency-responsive genes in soybean roots by high-throughput sequencing. Plant Soil 2016, 398, 207–227. [Google Scholar] [CrossRef]
- Libault, M.; Farmer, A.; Brechenmacher, L.; Drnevich, J.; Langley, R.J.; Bilgin, D.D.; Radwan, O.; Neece, D.J.; Clough, S.J.; May, G.D.; et al. Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol. 2010, 152, 541–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medina-Rivera, A.; Defrance, M.; Sand, O.; Herrmann, C.; Castro-Mondragon, J.A.; Delerce, J.; Jaeger, S.; Blanchet, C.; Vincens, P.; Caron, C.; et al. RSAT 2015: Regulatory Sequence Analysis Tools. Nucleic Acids Res. 2015, 43, W50–W56. [Google Scholar] [CrossRef]
- Zeng, H.; Zhang, X.; Zhang, X.; Pi, E.; Xiao, L.; Zhu, Y. Early Transcriptomic Response to Phosphate Deprivation in Soybean Leaves as Revealed by RNA-Sequencing. Int. J. Mol. Sci. 2018, 19, 2145. [Google Scholar] [CrossRef] [Green Version]
- Zeng, H.; Zhang, X.; Ding, M.; Zhang, X.; Zhu, Y. Transcriptome profiles of soybean leaves and roots in response to zinc deficiency. Physiol. Plant 2019, 167, 330–351. [Google Scholar] [CrossRef] [Green Version]
- Fang, X.; Wang, L.; Deng, X.; Wang, P.; Ma, Q.; Nian, H.; Wang, Y.; Yang, C. Genome-wide characterization of soybean P1B -ATPases gene family provides functional implications in cadmium responses. BMC Genom. 2016, 17, 376. [Google Scholar] [CrossRef] [Green Version]
- Zeng, H.; Zhao, B.; Wu, H.; Zhu, Y.; Chen, H. Comprehensive in Silico Characterization and Expression Profiling of Nine Gene Families Associated with Calcium Transport in Soybean. Agronomy 2020, 10, 1539. [Google Scholar] [CrossRef]
- Palmgren, M.; Sommarin, M.; Serrano, R.; Larsson, C. Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H(+)-ATPase. J. Biol. Chem. 1991, 266, 20470–20475. [Google Scholar]
- Kinoshita, T.; Shimazaki, K.-I. Biochemical evidence for the requirement of 14-3-3 protein binding in activation of the guard-cell plasma membrane H+-ATPase by blue light. Plant Cell Physiol. 2002, 43, 1359–1365. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Poovaiah, B.W. A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J. Biol. Chem. 2002, 277, 45049–45058. [Google Scholar] [CrossRef] [Green Version]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-Binding Specificity of the ERF/AP2 Domain of Arabidopsis DREBs, Transcription Factors Involved in Dehydration- and Cold-Inducible Gene Expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Osakabe, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S. ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 2014, 202, 35–49. [Google Scholar] [CrossRef]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. ARF1, a Transcription Factor That Binds to Auxin Response Elements. Science 1997, 276, 1865. [Google Scholar] [CrossRef]
- Williams, M.E.; Foster, R.; Chua, N.H. Sequences flanking the hexameric G-box core CACGTG affect the specificity of protein binding. Plant Cell 1992, 4, 485–496. [Google Scholar]
- Rubio, V.; Linhares, F.; Solano, R.; Martin, A.C.; Iglesias, J.; Leyva, A.; Paz-Ares, J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001, 15, 2122–2133. [Google Scholar] [CrossRef] [Green Version]
- Maruyama-Nakashita, A.; Nakamura, Y.; Watanabe-Takahashi, A.; Inoue, E.; Yamaya, T.; Takahashi, H. Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J. 2005, 42, 305–314. [Google Scholar] [CrossRef]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Et Biophys. Acta 2012, 1819, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.J.; Van Loon, L.C. NPR1: The spider in the web of induced resistance signaling pathways. Curr. Opin. Plant Biol. 2004, 7, 456–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manzara, T.; Carrasco, P.; Gruissem, W. Developmental and organ-specific changes in promoter DNA-protein interactions in the tomato rbcS gene family. Plant Cell 1991, 3, 1305. [Google Scholar] [PubMed] [Green Version]
- Li, N.; Xiao, H.; Sun, J.; Wang, S.; Wang, J.; Chang, P.; Zhou, X.; Lei, B.; Lu, K.; Luo, F.; et al. Genome-wide analysis and expression profiling of the HMA gene family in Brassica napus under cd stress. Plant Soil. 2018, 426, 365–381. [Google Scholar] [CrossRef]
- Liu, J.; Liu, J.; Chen, A.; Ji, M.; Chen, J.; Yang, X.; Gu, M.; Qu, H.; Xu, G. Analysis of tomato plasma membrane H+-ATPase gene family suggests a mycorrhiza-mediated regulatory mechanism conserved in diverse plant species. Mycorrhiza 2016, 26, 645–656. [Google Scholar] [CrossRef]
- Taneja, M.; Upadhyay, S.K. Molecular characterization and differential expression suggested diverse functions of P-type II Ca2+ATPases in Triticum aestivum L. BMC Genom. 2018, 19, 389. [Google Scholar] [CrossRef] [Green Version]
- Yin, G.; Xu, H.; Xiao, S.; Qin, Y.; Li, Y.; Yan, Y.; Hu, Y. The large soybean (Glycine max) WRKY TF family expanded by segmental duplication events and subsequent divergent selection among subgroups. BMC Plant Biol. 2013, 13, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eren, E.; González-Guerrero, M.; Kaufman, B.M.; Argüello, J.M. Novel Zn2+ Coordination by the Regulatory N-Terminus Metal Binding Domain of Arabidopsis thaliana Zn2+-ATPase HMA2. Biochemistry 2007, 46, 7754–7764. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, R.D.; Portes, M.T.; Olsen, L.I.; Damineli, D.S.C.; Hayashi, M.; Nunes, C.O.; Pedersen, J.T.; Lima, P.T.; Campos, C.; Feijó, J.A.; et al. Plasma membrane H+-ATPases sustain pollen tube growth and fertilization. Nat. Commun. 2020, 11, 2395. [Google Scholar] [CrossRef] [PubMed]
- Toda, Y.; Kawai, Y.; Kinoshita, T.; Wang, Y.; Takahashi, A.; Tada, Y.; Feng Ma, J.; Yamaji, N.; Ashikari, M. Oryza sativa H+ -ATPase (OSA) is Involved in the Regulation of Dumbbell-Shaped Guard Cells of Rice. Plant Cell Physiol. 2016, 57, 1220–1230. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Yan, J.; Du, X.; Hua, J. Overlapping and differential roles of plasma membrane calcium ATPases in Arabidopsis growth and environmental responses. J. Exp. Bot. 2018, 69, 2693–2703. [Google Scholar] [CrossRef] [Green Version]
- Verweij, W.; Spelt, C.; Di Sansebastiano, G.P.; Vermeer, J.; Reale, L.; Ferranti, F.; Koes, R.; Quattrocchio, F. An H+ P-ATPase on the tonoplast determines vacuolar pH and flower colour. Nat. Cell Biol. 2008, 10, 1456–1462. [Google Scholar] [CrossRef]
- Lang, V.; Pertl-Obermeyer, H.; Safiarian, M.J.; Obermeyer, G. Pump up the volume—A central role for the plasma membrane H+ pump in pollen germination and tube growth. Protoplasma 2014, 251, 477–488. [Google Scholar] [CrossRef]
- Wang, E.; Yu, N.; Bano, S.A.; Liu, C.; Miller, A.J.; Cousins, D.; Zhang, X.; Ratet, P.; Tadege, M.; Mysore, K.S.; et al. A H+-ATPase That Energizes Nutrient Uptake during Mycorrhizal Symbioses in Rice and Medicago truncatula. Plant Cell 2014, 26, 1818–1830. [Google Scholar] [CrossRef] [Green Version]
- Krajinski, F.; Courty, P.E.; Sieh, D.; Franken, P.; Zhang, H.; Bucher, M.; Gerlach, N.; Kryvoruchko, I.; Zoeller, D.; Udvardi, M.; et al. The H+-ATPase HA1 of Medicago truncatula Is Essential for Phosphate Transport and Plant Growth during Arbuscular Mycorrhizal Symbiosis. Plant Cell 2014, 26, 1808–1817. [Google Scholar] [CrossRef] [Green Version]
- Capoen, W.; Sun, J.; Wysham, D.; Otegui, M.S.; Venkateshwaran, M.; Hirsch, S.; Miwa, H.; Downie, J.A.; Morris, R.J.; Ane, J.M.; et al. Nuclear membranes control symbiotic calcium signaling of legumes. Proc. Natl. Acad. Sci. USA 2011, 108, 14348–14353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Chen, J.; Xie, K.; Tian, Y.; Yan, A.; Liu, J.; Huang, Y.; Wang, S.; Zhu, Y.; Chen, A.; et al. A mycorrhiza-specific H+-ATPase is essential for arbuscule development and symbiotic phosphate and nitrogen uptake. Plant Cell Environ. 2020, 43, 1069–1083. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Zhang, D.; Song, T.; Xu, F.; Lin, S.; Xu, W.; Li, Q.; Zhu, Y.; Liang, J.; Zhang, J. Arabidopsis plasma membrane H+-ATPase genes AHA2 and AHA7 have distinct and overlapping roles in the modulation of root tip H+ efflux in response to low-phosphorus stress. J. Exp. Bot. 2017, 68, 1731–1741. [Google Scholar] [CrossRef] [PubMed]
- Kumari, A.; Chetelat, A.; Nguyen, C.T.; Farmer, E.E. Arabidopsis H(+)-ATPase AHA1 controls slow wave potential duration and wound-response jasmonate pathway activation. Proc. Natl. Acad. Sci. USA 2019, 116, 20226–20231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.-Y.; Choi, H.; Segami, S.; Cho, H.-T.; Martinoia, E.; Maeshima, M.; Lee, Y. AtHMA1 contributes to the detoxification of excess Zn(II) in Arabidopsis. Plant J. 2009, 58, 737–753. [Google Scholar] [CrossRef] [Green Version]
- Seigneurin-Berny, D.; Gravot, A.; Auroy, P.; Mazard, C.; Kraut, A.; Finazzi, G.; Grunwald, D.; Rappaport, F.; Vavasseur, A.; Joyard, J.; et al. HMA1, A New Cu-ATPase of the Chloro plast Envelope, Is Essential for Growth under Adverse Light Conditions. J. Biol. Chem. 2006, 281, 2882–2892. [Google Scholar] [CrossRef] [Green Version]
- Gévaudant, F.; Duby, G.; von Stedingk, E.; Zhao, R.; Morsomme, P.; Boutry, M. Expression of a constitutively activated plasma membrane H+-ATPase alters plant development and increases salt tolerance. Plant Physiol. 2007, 144, 1763–1776. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Wu, Y.; Ma, L.; Yang, Z.; Dong, Q.; Li, Q.; Ni, X.; Kudla, J.; Song, C.; Guo, Y. The Ca(2+) Sensor SCaBP3/CBL7 Modulates Plasma Membrane H(+)-ATPase Activity and Promotes Alkali Tolerance in Arabidopsis. Plant Cell 2019, 31, 1367–1384. [Google Scholar] [CrossRef]
- Muchhal, U.S.; Liu, C.; Raghothama, K.G. Ca2+-ATPase is expressed differentially in phosphate-starved roots of tomato. Physiol. Plant. 1997, 101, 540–544. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, B.; Wu, H.; Xu, W.; Zhang, W.; Chen, X.; Zhu, Y.; Chen, H.; Zeng, H. Genome-Wide Identification, Characterization, and Expression Analyses of P-Type ATPase Superfamily Genes in Soybean. Agronomy 2021, 11, 71. https://doi.org/10.3390/agronomy11010071
Zhao B, Wu H, Xu W, Zhang W, Chen X, Zhu Y, Chen H, Zeng H. Genome-Wide Identification, Characterization, and Expression Analyses of P-Type ATPase Superfamily Genes in Soybean. Agronomy. 2021; 11(1):71. https://doi.org/10.3390/agronomy11010071
Chicago/Turabian StyleZhao, Bingqian, Haicheng Wu, Wenjing Xu, Wei Zhang, Xi Chen, Yiyong Zhu, Huatao Chen, and Houqing Zeng. 2021. "Genome-Wide Identification, Characterization, and Expression Analyses of P-Type ATPase Superfamily Genes in Soybean" Agronomy 11, no. 1: 71. https://doi.org/10.3390/agronomy11010071
APA StyleZhao, B., Wu, H., Xu, W., Zhang, W., Chen, X., Zhu, Y., Chen, H., & Zeng, H. (2021). Genome-Wide Identification, Characterization, and Expression Analyses of P-Type ATPase Superfamily Genes in Soybean. Agronomy, 11(1), 71. https://doi.org/10.3390/agronomy11010071




