Growth Promotion of Rapeseed (Brassica napus L.) and Blackleg Disease (Leptosphaeria maculans) Suppression Mediated by Endophytic Bacteria
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Samples
2.2. Isolation of Endophytic Bacteria
2.3. Primary Screening of Endophytic Bacteria
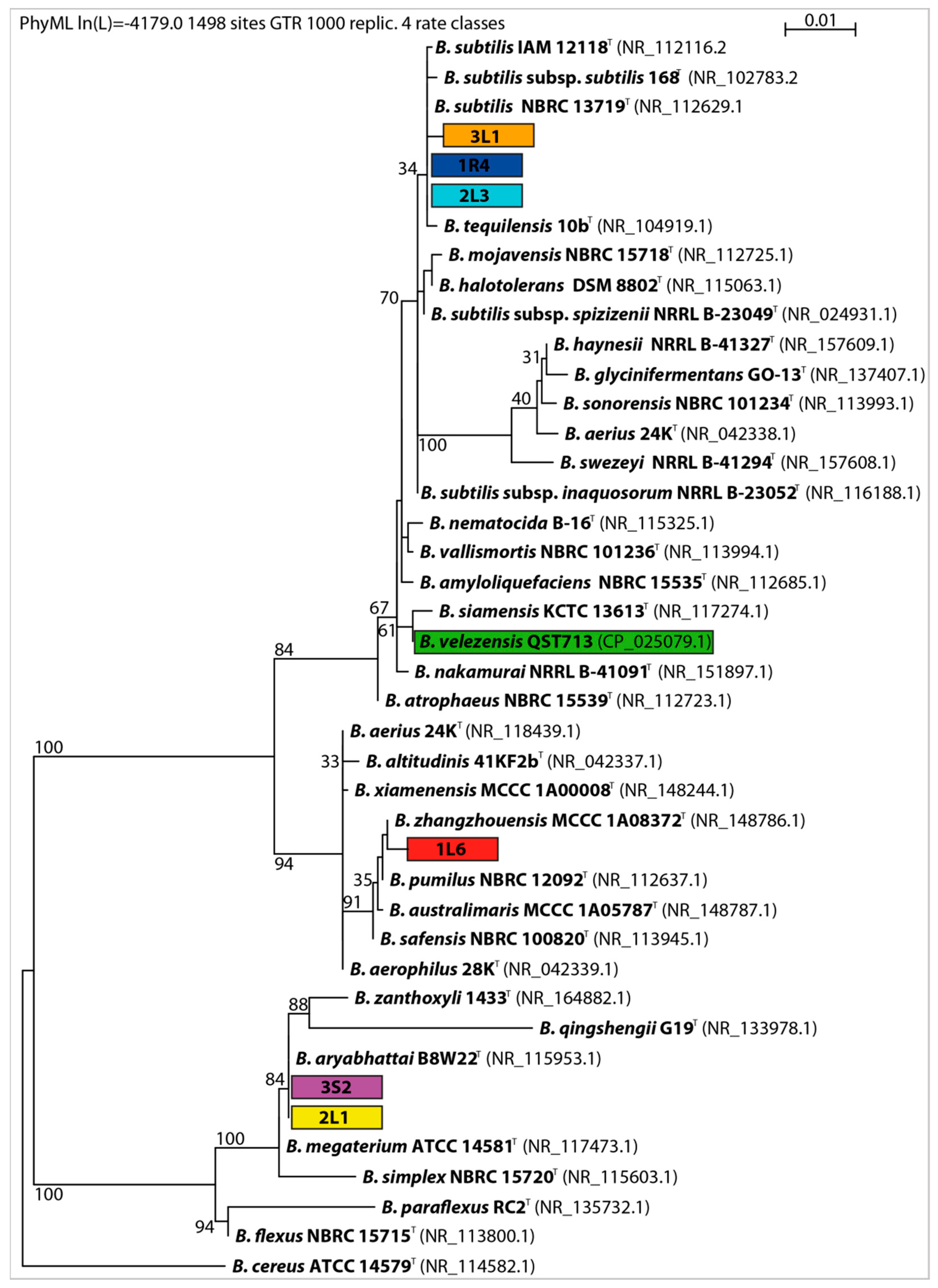
2.4. Molecular Characterization
2.5. Enzymatic and Physiological Characterization
2.6. Quantitative Analysis of PGP Traits
2.7. In Planta Assay
2.8. Growth Promotion Experiment
2.9. Leptosphaeria Suppression Experiment
2.10. Statistical Analysis
3. Results
3.1. Primary Screening
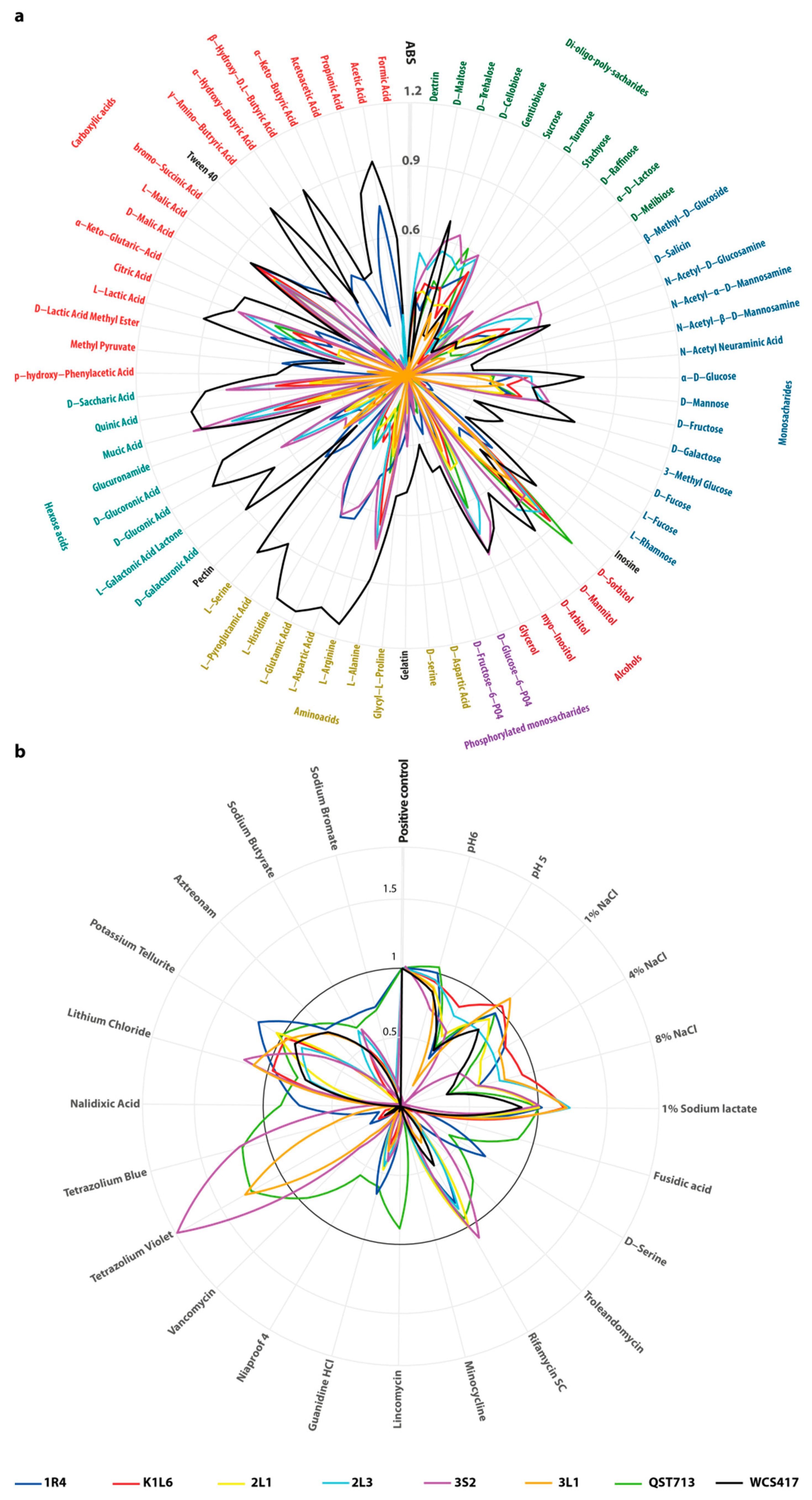
3.2. PGP Traits and Physiological Characterization of Selected Isolates
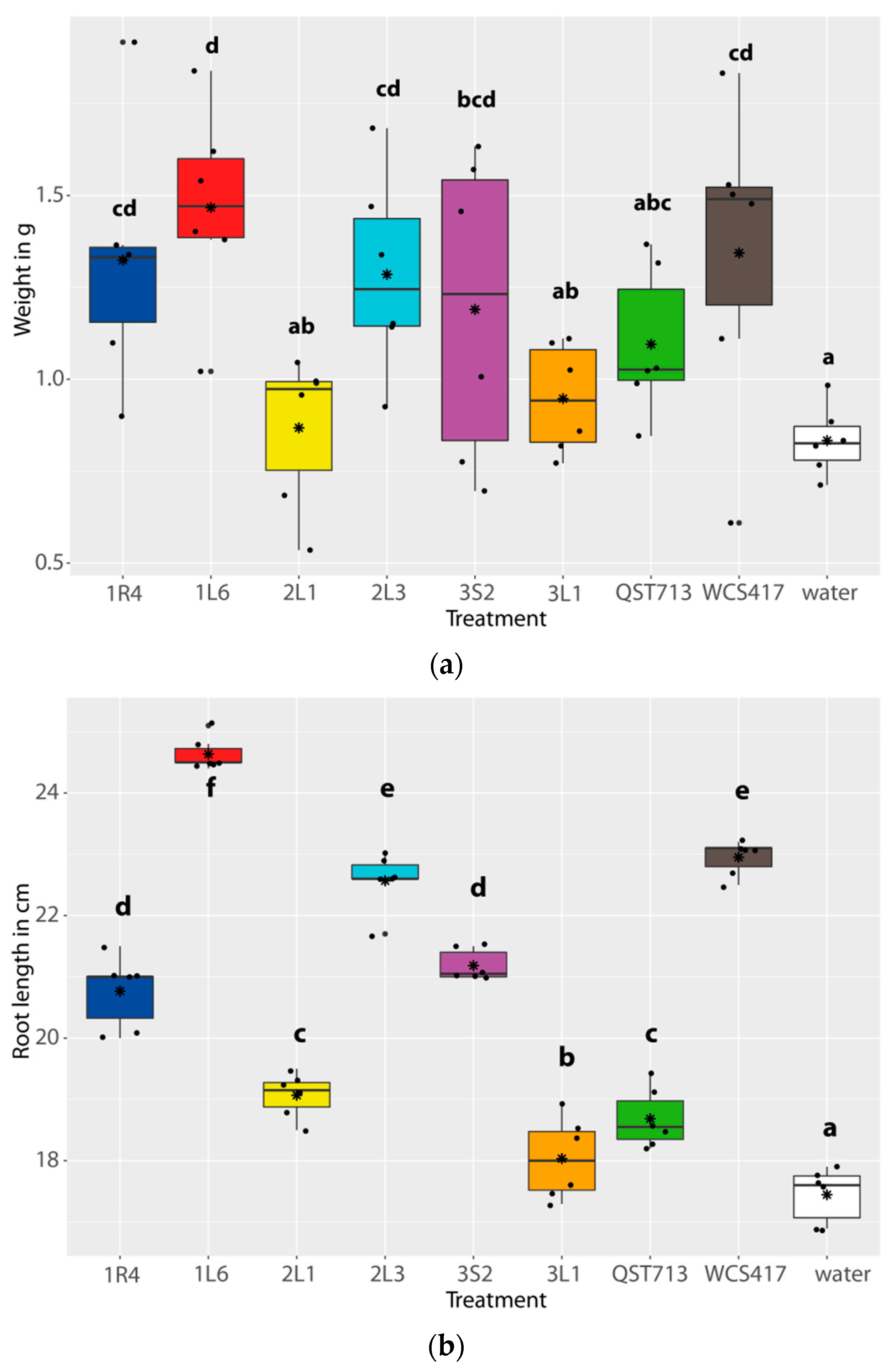
3.3. Plant Growth Promoting Properties of Isolates
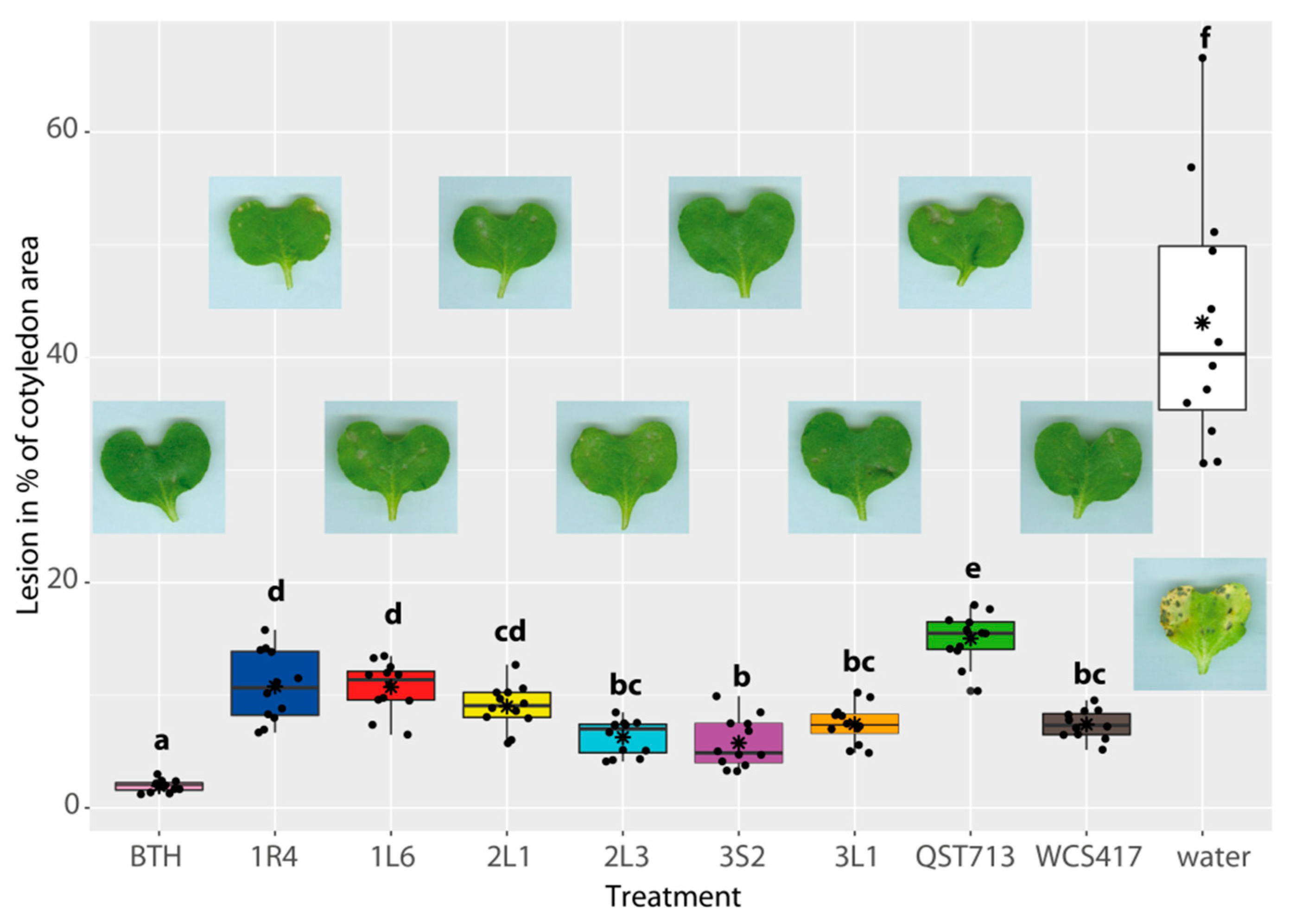
3.4. Suppression of L. maculans
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carré, P.; Pouzet, A. Rapeseed market, worldwide and in Europe. OCL 2014, 21, D102. [Google Scholar] [CrossRef]
- Konur, O. Rapeseed oil-based biodiesel fuels: A review of the research. In Biodiesel Fuels Based Edible Nonedible Feedstocks Wastes Algae; Konur, O., Ed.; CRC Press: Boca Raton, FL, USA, 2021; pp. 497–514. [Google Scholar]
- Bunzel, K.; Kattwinkel, M.; Schauf, M.; Thrän, D. Energy crops and pesticide contamination: Lessons learnt from the development of energy crop cultivation in Germany. Biomass Bioenergy 2014, 70, 416–428. [Google Scholar] [CrossRef]
- Souza, R.d.; Ambrosini, A.; Passaglia, L.M. Plant growth-promoting bacteria as inoculants in agricultural soils. Genet. Mol. Biol. 2015, 38, 401–419. [Google Scholar] [CrossRef] [PubMed]
- Card, S.D.; Hume, D.E.; Roodi, D.; McGill, C.R.; Millner, J.P.; Johnson, R.D. Beneficial endophytic microorganisms of Brassica–A review. Biol. Control 2015, 90, 102–112. [Google Scholar] [CrossRef]
- West, J.S.; Kharbanda, P.; Barbetti, M.; Fitt, B.D. Epidemiology and management of Leptosphaeria maculans (phoma stem canker) on oilseed rape in Australia, Canada and Europe. Plant Pathol. 2001, 50, 10–27. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.-J.; Qi, A.; King, G.J.; Fitt, B.D. Assessing quantitative resistance against Leptosphaeria maculans (phoma stem canker) in Brassica napus (oilseed rape) in young plants. PLoS ONE 2014, 9, e84924. [Google Scholar] [CrossRef] [Green Version]
- Toscano-Underwood, C.; West, J.S.; Fitt, B.D.; Todd, A.; Jedryczka, M. Development of phoma lesions on oilseed rape leaves inoculated with ascospores of A-group or B-group Leptosphaeria maculans (stem canker) at different temperatures and wetness durations. Plant Pathol. 2001, 50, 28–41. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Fitt, B.D.; Hall, A. Survival of A-group and B-group Leptosphaeria maculans (phoma stem canker) ascospores in air and mycelium on oilseed rape stem debris. Ann. Appl. Biol. 2003, 143, 359–369. [Google Scholar] [CrossRef]
- Deb, D.; Khan, A.; Dey, N. Phoma diseases: Epidemiology and control. Plant Pathol. 2020, 69, 1203–1217. [Google Scholar] [CrossRef]
- Syed Ab Rahman, S.F.; Singh, E.; Pieterse, C.M.J.; Schenk, P.M. Emerging microbial biocontrol strategies for plant pathogens. Plant Sci. 2018, 267, 102–111. [Google Scholar] [CrossRef] [Green Version]
- Berg, G. Plant–microbe interactions promoting plant growth and health: Perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 2009, 84, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Hardoim, P.R.; van Overbeek, L.S.; van Elsas, J.D. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008, 16, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Clément, C.; Sessitsch, A. Plant growth-promoting bacteria in the rhizo-and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 2010, 42, 669–678. [Google Scholar] [CrossRef] [Green Version]
- Riaz, U.; Murtaza, G.; Anum, W.; Samreen, T.; Sarfraz, M.; Nazir, M.Z. Plant Growth-Promoting Rhizobacteria (PGPR) as biofertilizers and biopesticides. In Microbiota and Biofertilizers; Springer: Berlin/Heidelberg, Germany, 2021; pp. 181–196. [Google Scholar]
- Ahmad, F.; Ahmad, I.; Khan, M. Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol. Res. 2008, 163, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Boubekri, K.; Soumare, A.; Mardad, I.; Lyamlouli, K.; Hafidi, M.; Ouhdouch, Y.; Kouisni, L. The Screening of potassium-and phosphate-solubilizing actinobacteria and the assessment of their ability to promote wheat growth parameters. Microorganisms 2021, 9, 470. [Google Scholar] [CrossRef] [PubMed]
- Raymaekers, K.; Ponet, L.; Holtappels, D.; Berckmans, B.; Cammue, B.P. Screening for novel biocontrol agents applicable in plant disease management—A review. Biol. Control 2020, 144, 104240. [Google Scholar] [CrossRef]
- Duca, D.; Lorv, J.; Patten, C.L.; Rose, D.; Glick, B.R. Indole-3-acetic acid in plant–microbe interactions. Antonie Van Leeuwenhoek 2014, 106, 85–125. [Google Scholar] [CrossRef]
- Etesami, H.; Alikhani, H.A.; Hosseini, H.M. Indole-3-acetic acid (IAA) production trait, a useful screening to select endophytic and rhizosphere competent bacteria for rice growth promoting agents. MethodsX 2015, 2, 72–78. [Google Scholar] [CrossRef]
- Raghothama, K.G. Phosphorus and plant nutrition: An overview. Phosphorus Agric. Environ. 2005, 46, 353–378. [Google Scholar]
- Frossard, E.; Condron, L.M.; Oberson, A.; Sinaj, S.; Fardeau, J. Processes governing phosphorus availability in temperate soils. J. Environ. Qual. 2000, 29, 15–23. [Google Scholar] [CrossRef] [Green Version]
- Pizzol, M.; Smart, J.C.; Thomsen, M. External costs of cadmium emissions to soil: A drawback of phosphorus fertilizers. J. Clean. Prod. 2014, 84, 475–483. [Google Scholar] [CrossRef]
- Huang, J.; Xu, C.-C.; Ridoutt, B.G.; Wang, X.-C.; Ren, P.-A. Nitrogen and phosphorus losses and eutrophication potential associated with fertilizer application to cropland in China. J. Clean. Prod. 2017, 159, 171–179. [Google Scholar] [CrossRef]
- Colombo, C.; Palumbo, G.; He, J.-Z.; Pinton, R.; Cesco, S. Review on iron availability in soil: Interaction of Fe minerals, plants, and microbes. J. Soils Sedim. 2014, 14, 538–548. [Google Scholar] [CrossRef]
- Lemanceau, P.; Bauer, P.; Kraemer, S.; Briat, J.-F. Iron dynamics in the rhizosphere as a case study for analyzing interactions between soils, plants and microbes. Plant Soil 2009, 321, 513–535. [Google Scholar] [CrossRef]
- Loaces, I.; Ferrando, L.; Scavino, A.F. Dynamics, diversity and function of endophytic siderophore-producing bacteria in rice. Microb. Ecol. 2011, 61, 606–618. [Google Scholar] [CrossRef] [PubMed]
- Höfte, M.; Bakker, P.A. Competition for iron and induced systemic resistance by siderophores of plant growth promoting rhizobacteria. In Microbial Siderophores; Springer: Berlin/Heidelberg, Germany, 2007; pp. 121–133. [Google Scholar]
- Ramette, A.; Frapolli, M.; Défago, G.; Moënne-Loccoz, Y. Phylogeny of HCN synthase-encoding hcnBC genes in biocontrol fluorescent pseudomonads and its relationship with host plant species and HCN synthesis ability. Mol. Plant Microbe Interact. 2003, 16, 525–535. [Google Scholar] [CrossRef] [Green Version]
- Blom, D.; Fabbri, C.; Connor, E.; Schiestl, F.; Klauser, D.; Boller, T.; Eberl, L.; Weisskopf, L. Production of plant growth modulating volatiles is widespread among rhizosphere bacteria and strongly depends on culture conditions. Environ. Microbiol. 2011, 13, 3047–3058. [Google Scholar] [CrossRef]
- Bakker, A.W.; Schippers, B. Microbial cyanide production in the rhizosphere in relation to potato yield reduction and Pseudomonas spp-mediated plant growth-stimulation. Soil Biol. Biochem. 1987, 19, 451–457. [Google Scholar] [CrossRef]
- Rijavec, T.; Lapanje, A. Hydrogen cyanide in the rhizosphere: Not suppressing plant pathogens, but rather regulating availability of phosphate. Front. Microbiol. 2016, 7, 1785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- von Rohr, M.R.; Furrer, G.; Brandl, H. Effect of iron and phosphate on bacterial cyanide formation determined by methemoglobin in two-dimensional gradient microcultivations. J. Microbiol. Methods 2009, 79, 71–75. [Google Scholar] [CrossRef]
- Santos, M.S.; Nogueira, M.A.; Hungria, M. Microbial inoculants: Reviewing the past, discussing the present and previewing an outstanding future for the use of beneficial bacteria in agriculture. AMB Express 2019, 9, 1–22. [Google Scholar] [CrossRef]
- Afzal, I.; Shinwari, Z.K.; Sikandar, S.; Shahzad, S. Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 2019, 221, 36–49. [Google Scholar] [CrossRef]
- Sun, L.; Qiu, F.; Zhang, X.; Dai, X.; Dong, X.; Song, W. Endophytic bacterial diversity in rice (Oryza sativa L.) roots estimated by 16S rDNA sequence analysis. Microb. Ecol. 2008, 55, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Schwyn, B.; Neilands, J. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Gupta, R.; Singal, R.; Shankar, A.; Kuhad, R.C.; Saxena, R.K. A modified plate assay for screening phosphate solubilizing microorganisms. J. Gen. Appl. Microbiol. 1994, 40, 255–260. [Google Scholar] [CrossRef]
- Fernando, W.; Pierson III, L. The effect of increased phenazine antibiotic production on the inhibition of economically important soil-borne plant pathogens by pseudomonas a ureofaciens 30–84. Arch. Phytopathol. Plant Prot. 1999, 32, 491–502. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef] [Green Version]
- Collins, C.H.; Grange, J.; Falkinham, J. Collins and Lyne’s Microbiological Methods, 8th ed.; Hodder Arnold: London, UK, 2003; p. 480. [Google Scholar]
- Kumar, V.; Kumar, V.; Umrao, V.; Singh, M. Effect of GA3 and IAA on growth and flowering of carnation. Hortflora Res. Spectr. 2012, 89, 69. [Google Scholar]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Arora, N.K.; Verma, M. Modified microplate method for rapid and efficient estimation of siderophore produced by bacteria. 3 Biotech 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Šašek, V.; Nováková, M.; Dobrev, P.I.; Valentová, O.; Burketová, L. β-aminobutyric acid protects Brassica napus plants from infection by Leptosphaeria maculans. Resistance induction or a direct antifungal effect? Eur. J. Plant Pathol. 2012, 133, 279–289. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Maggini, V.; Mengoni, A.; Gallo, E.R.; Biffi, S.; Fani, R.; Firenzuoli, F.; Bogani, P. Tissue specificity and differential effects on in vitro plant growth of single bacterial endophytes isolated from the roots, leaves and rhizospheric soil of Echinacea purpurea. BMC Plant Biol. 2019, 19, 284. [Google Scholar] [CrossRef] [Green Version]
- Costa, L.E.D.O.; Queiroz, M.V.D.; Borges, A.C.; Moraes, C.A.D.; Araújo, E.F.D. Isolation and characterization of endophytic bacteria isolated from the leaves of the common bean (Phaseolus vulgaris). Braz. J. Microbiol. 2012, 43, 1562–1575. [Google Scholar] [CrossRef]
- Forchetti, G.; Masciarelli, O.; Alemano, S.; Alvarez, D.; Abdala, G. Endophytic bacteria in sunflower (Helianthus annuus L.): Isolation, characterization, and production of jasmonates and abscisic acid in culture medium. Appl. Microbiol. Biotechnol. 2007, 76, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- ALKahtani, M.D.F.; Fouda, A.; Attia, K.A.; Al-Otaibi, F.; Eid, A.M.; Ewais, E.E.-D.; Hijri, M.; St-Arnaud, M.; Hassan, S.E.-D.; Khan, N.; et al. Isolation and Characterization of Plant Growth Promoting Endophytic Bacteria from Desert Plants and Their Application as Bioinoculants for Sustainable Agriculture. Agronomy 2020, 10, 1325. [Google Scholar] [CrossRef]
- Fouda, A.; Eid, A.M.; Elsaied, A.; El-Belely, E.F.; Barghoth, M.G.; Azab, E.; Gobouri, A.A.; Hassan, S.E.-D. Plant Growth-Promoting Endophytic Bacterial Community Inhabiting the Leaves of Pulicaria incisa (Lam.) DC Inherent to Arid Regions. Plants 2021, 10, 76. [Google Scholar] [CrossRef] [PubMed]
- Tarroum, M.; Ben Romdhane, W.; Ali, A.A.M.; Al-Qurainy, F.; Al-Doss, A.; Fki, L.; Hassairi, A. Harnessing the Rhizosphere of the Halophyte Grass Aeluropus littoralis for Halophilic Plant-Growth-Promoting Fungi and Evaluation of Their Biostimulant Activities. Plants 2021, 10, 784. [Google Scholar] [CrossRef] [PubMed]
- Kloepper, J.; Hume, D.; Scher, F.; Singleton, C.; Tipping, B.; Laliberte, M.; Frauley, K.; Kutchaw, T.; Simonson, C.; Lifshitz, R. Plant growth-promoting rhizobacteria on canola (rapeseed). Plant Dis. 1988, 72, 42–46. [Google Scholar] [CrossRef]
- De Freitas, J.; Banerjee, M.; Germida, J. Phosphate-solubilizing rhizobacteria enhance the growth and yield but not phosphorus uptake of canola (Brassica napus L.). Biol. Fertil. Soils 1997, 24, 358–364. [Google Scholar] [CrossRef]
- Bertrand, H.; Nalin, R.; Bally, R.; Cleyet-Marel, J.-C. Isolation and identification of the most efficient plant growth-promoting bacteria associated with canola (Brassica napus). Biol. Fertil. Soils 2001, 33, 152–156. [Google Scholar] [CrossRef]
- Schmidt, C.; Mrnka, L.; Lovecká, P.; Frantík, T.; Fenclová, M.; Demnerová, K.; Vosátka, M. Bacterial and fungal endophyte communities in healthy and diseased oilseed rape and their potential for biocontrol of Sclerotinia and Phoma disease. Sci. Rep. 2021, 11, 3810. [Google Scholar] [CrossRef]
- Sridevi, M.; Mallaiah, K.V. Bioproduction of indole acetic acid by Rhizobium strains isolated from root nodules of green manure crop, Sesbania sesban (L.) Merr. Brief Rep. 2007, 5, 178–182. [Google Scholar]
- Khan, A.L.; Halo, B.A.; Elyassi, A.; Ali, S.; Al-Hosni, K.; Hussain, J.; Al-Harrasi, A.; Lee, I.-J. Indole acetic acid and ACC deaminase from endophytic bacteria improves the growth of Solanum lycopersicum. Electron. J. Biotechnol. 2016, 21, 58–64. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.; Singh, P.; Jorquera, M.A.; Sangwan, P.; Kumar, P.; Verma, A.; Agrawal, S. Isolation of phytase-producing bacteria from Himalayan soils and their effect on growth and phosphorus uptake of Indian mustard (Brassica juncea). World J. Microbiol. Biotechnol. 2013, 29, 1361–1369. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef] [Green Version]
- Kunkel, B.N.; Chen, Z. Virulence strategies of plant pathogenic bacteria. Prokaryotes 2006, 2, 421–440. [Google Scholar]
- Kumar, P.; Thakur, S.; Dhingra, G.; Singh, A.; Pal, M.K.; Harshvardhan, K.; Dubey, R.; Maheshwari, D. Inoculation of siderophore producing rhizobacteria and their consortium for growth enhancement of wheat plant. Biocatal. Agric. Biotechnol. 2018, 15, 264–269. [Google Scholar] [CrossRef]
- Husen, E. Screening of soil bacteria for plant growth promotion activities in vitro. Indones. J. Agric. Sci. 2003, 4, 27–31. [Google Scholar] [CrossRef]
- Cinkocki, R.; Lipková, N.; Javoreková, S.; Petrová, J.; Maková, J.; Medo, J.; Ducsay, L. The Impact of Growth-Promoting Streptomycetes Isolated from Rhizosphere and Bulk Soil on Oilseed Rape (Brassica napus L.) Growth Parameters. Sustainability 2021, 13, 5704. [Google Scholar] [CrossRef]
- Krewulak, K.D.; Vogel, H.J. Structural biology of bacterial iron uptake. Biochim. Biophys. Acta BBA Biomembr. 2008, 1778, 1781–1804. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, J.; Satyan, K. Isolation and selection of fluorescent pseudomonads based on multiple plant growth promotion traits and siderotyping. Chil. J. Agric. Res. 2014, 74, 319–325. [Google Scholar] [CrossRef] [Green Version]
- Kotasthane, A.S.; Agrawal, T.; Zaidi, N.W.; Singh, U. Identification of siderophore producing and cynogenic fluorescent Pseudomonas and a simple confrontation assay to identify potential bio-control agent for collar rot of chickpea. 3 Biotech 2017, 7, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Xu, Y.; Lai, X.-H.; Shan, C.; Deng, Z.; Ji, Y. Screening and characterization of endophytic Bacillus and Paenibacillus strains from medicinal plant Lonicera japonica for use as potential plant growth promoters. Braz. J. Microbiol. 2015, 46, 977–989. [Google Scholar] [CrossRef]
- Santoyo, G.; Moreno-Hagelsieb, G.; del Carmen Orozco-Mosqueda, M.; Glick, B.R. Plant growth-promoting bacterial endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Perez-Rosales, E.; Alcaraz-Meléndez, L.; Puente, M.E.; Vázquez-Juárez, R.; Quiroz-Guzmán, E.; Zenteno-Savín, T.; Morales-Bojórquez, E. Isolation and characterization of endophytic bacteria associated with roots of jojoba (Simmondsia chinensis (Link) Schneid). Curr. Sci. 2017, 396–401. [Google Scholar]
- Wahyudi, A.T.; Astuti, R.P.; Widyawati, A.; Mery, A.; Nawangsih, A.A. Characterization of Bacillus sp. strains isolated from rhizosphere of soybean plants for their use as potential plant growth for promoting rhizobacteria. J. Microbiol. Antimicrob. 2011, 3, 34–40. [Google Scholar]
- Arya, N.; Rana, A.; Rajwar, A.; Sahgal, M.; Sharma, A. Biocontrol efficacy of siderophore producing indigenous Pseudomonas strains against Fusarium Wilt in Tomato. Natl. Acad. Sci. Lett. 2018, 41, 133–136. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Y.C.; Zhang, S.; Fu, Y.; Fan, X.; Patel, J.S.; Zhang, M. Characterization of phosphate-solubilizing bacteria isolated from calcareous soils. Appl. Soil Ecol. 2015, 96, 217–224. [Google Scholar] [CrossRef]
- Stephen, J.; Jisha, M. Gluconic acid production as the principal mechanism of mineral phosphate solubilization by Burkholderia sp. (MTCC 8369). J. Trop. Agric. 2011, 49, 99–103. [Google Scholar]
- Surapat, W.; Pukahuta, C.; Rattanachaikunsopon, P.; Aimi, T.; Boonlue, S. Characteristics of phosphate solubilization by phosphate-solubilizing bacteria isolated from agricultural chili soil and their efficiency on the growth of chili (Capsicum frutescens L. cv. Hua Rua). Chiang Mai J. Sci. 2013, 40, 11–25. [Google Scholar]
- Chaiharn, M.; Lumyong, S. Screening and optimization of indole-3-acetic acid production and phosphate solubilization from rhizobacteria aimed at improving plant growth. Curr. Microbiol. 2011, 62, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.d.A.; Silveira, J.A.G.d.; Bonifacio, A.; Rodrigues, A.C.; Figueiredo, M.d.V.B. Antioxidant response of cowpea co-inoculated with plant growth-promoting bacteria under salt stress. Braz. J. Microbiol. 2018, 49, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Nosheen, A.; Bano, A. Potential of plant growth promoting rhizobacteria and chemical fertilizers on soil enzymes and plant growth. Pak. J. Bot. 2014, 46, 1521–1530. [Google Scholar]
- Woźniak, M.; Gałązka, A.; Tyśkiewicz, R.; Jaroszuk-Ściseł, J. Endophytic Bacteria Potentially Promote Plant Growth by Synthesizing Different Metabolites and their Phenotypic/Physiological Profiles in the Biolog GEN III MicroPlateTM Test. Int. J. Mol. Sci. 2019, 20, 5283. [Google Scholar] [CrossRef] [Green Version]
- Altuntaş, Ö. A comparative study on the effects of different conventional, organic and bio-fertilizers on broccoli yield and quality. Appl. Ecol. Environ. Res. 2018, 16, 1595–1608. [Google Scholar] [CrossRef]
- McGehee, C.S.; Raudales, R.E.; Elmer, W.H.; McAvoy, R.J. Efficacy of biofungicides against root rot and damping-off of microgreens caused by Pythium spp. Crop Prot. 2019, 121, 96–102. [Google Scholar] [CrossRef]
- Lahlali, R.; Peng, G.; McGregor, L.; Gossen, B.D.; Hwang, S.F.; McDonald, M. Mechanisms of the biofungicide Serenade (Bacillus subtilis QST713) in suppressing clubroot. Biocontrol Sci. Technol. 2011, 21, 1351–1362. [Google Scholar] [CrossRef]
- Bach, E.; dos Santos Seger, G.D.; de Carvalho Fernandes, G.; Lisboa, B.B.; Passaglia, L.M.P. Evaluation of biological control and rhizosphere competence of plant growth promoting bacteria. Appl. Soil Ecol. 2016, 99, 141–149. [Google Scholar] [CrossRef]
- Seema, K.; Mehta, K.; Singh, N. Studies on the effect of plant growth promoting rhizobacteria (PGPR) on growth, physiological parameters, yield and fruit quality of strawberry cv. chandler. Chandler. J. Pharmacogn. Phytochem. 2018, 7, 383–387. [Google Scholar]
- Pieterse, C.M.J.; Berendsen, R.L.; de Jonge, R.; Stringlis, I.A.; Van Dijken, A.J.H.; Van Pelt, J.A.; Van Wees, S.C.M.; Yu, K.; Zamioudis, C.; Bakker, P.A.H.M. Pseudomonas simiae WCS417: Star track of a model beneficial rhizobacterium. Plant Soil 2021, 461, 245–263. [Google Scholar] [CrossRef]
- van der Wolf, J.M.; Michta, A.; van der Zouwen, P.S.; de Boer, W.J.; Davelaar, E.; Stevens, L.H. Seed and leaf treatments with natural compounds to induce resistance against Peronospora parasitica in Brassica oleracea. Crop Prot. 2012, 35, 78–84. [Google Scholar] [CrossRef]
- Haney, C.H.; Wiesmann, C.L.; Shapiro, L.R.; Melnyk, R.A.; O’Sullivan, L.R.; Khorasani, S.; Xiao, L.; Han, J.; Bush, J.; Carrillo, J.; et al. Rhizosphere-associated Pseudomonas induce systemic resistance to herbivores at the cost of susceptibility to bacterial pathogens. Mol. Ecol. 2018, 27, 1833–1847. [Google Scholar] [CrossRef]
| Strain | Origin | BBCH Stage | MALDI Best Match | MALDI Score * | SP | PS | AFA |
|---|---|---|---|---|---|---|---|
| 1R1 | root | 15 | Pseudomonas mendocina DSM 50017T | 1.997 | + | 0 | 9.26 |
| 1R2 | root | 15 | Erwinia amylovora CFBP 1232T | 1.912 | + | 0 | 19.63 |
| 1R3 | root | 15 | Acinetobacter baumannii B389 | 1.632 | + | 0 | 0 |
| 1R4 | root | 15 | Bacillus pumilus DSM 1794 | 1.998 | + | 1.56 | 60 |
| 1R5 | root | 15 | Microbacterium liquefaciens HKI 11374 | 1.808 | + | 0 | 0 |
| 1R6 | root | 15 | Chryseobacterium joostei LMG 18212T | 1.769 | + | 1.97 | 60 |
| 1R7 | root | 15 | Staphylococcus xylosus DSM 20267 | 1.984 | + | 0 | 68.89 |
| 1S1 | stem | 15 | Xanthomonas codiaei DSM 18812TB | 1.444 | + | 0 | 0 |
| 1S2 | stem | 15 | Citrobacter freundii 22054_1 | 1.945 | + | 1.28 | 14.81 |
| 1S3 | stem | 15 | Flavobacterium hibernum DSM 12611T | 2.033 | + | 0 | 51.85 |
| 1S4 | stem | 15 | Enterobacter ludwigii DSM 16688T | 2.085 | + | 1.39 | 18.52 |
| 1S5 | stem | 15 | Pseudomonas graminis DSM 11363T | 1.398 | + | 0 | 0 |
| 1L1 | leaf | 15 | Pantoea agglomerans DSM 8570 | 1.795 | + | 0 | 9.26 |
| 1L2 | leaf | 15 | Microbacterium liquefaciens DSM 20638T | 1.686 | + | 0 | 19.63 |
| 1L3 | leaf | 15 | Bacillus licheniformis DSM 13T | 1.718 | + | 0 | 5.56 |
| 1L4 | leaf | 15 | Pseudomonas aeruginosa 8147_2 | 2.162 | + | 0 | 0 |
| 1L5 | leaf | 15 | Acinetobacter junii DSM 14968 | 2.023 | + | 0 | 62.22 |
| 1L6 | leaf | 15 | Bacillus pumilus DSM 354 | 1.896 | + | 1.09 | 38.89 |
| 1L7 | leaf | 15 | Serratia marcescens DSM 12485 | 1.926 | + | 0 | 0 |
| 1L8 | leaf | 15 | Xanthomonas codiaei DSM 18812TB | 1.995 | + | 0 | 10.37 |
| 2R1 | root | 55 | Microbacterium liquefaciens DSM 20638T | 1.828 | + | 1.48 | 57.04 |
| 2R2 | root | 55 | Staphylococcus lutrae DSM 10246 | 1.437 | + | 1.37 | 46.3 |
| 2R3 | root | 55 | Serratia marcescens DSM 30122 | 2.192 | + | 0 | 0 |
| 2S1 | stem | 55 | Pantoea eucrina DSM 24231T | 1.814 | + | 0 | 60.74 |
| 2S2 | stem | 55 | Serratia fonticola CCUG 38570 | 2.114 | − | 0 | 40.74 |
| 2L1 | leaf | 55 | Bacillus subtilis DSM 5611 | 1.998 | + | 1.18 | 44.81 |
| 2L2 | leaf | 55 | Chryseobacterium joostei LMG 18212T | 1.818 | + | 1.31 | 0 |
| 2L3 | leaf | 55 | Bacillus subtilis 107_W_7_QSA | 1.962 | + | 1.59 | 62.22 |
| 3R1 | root | 85 | Pantoea agglomerans CCM 2406 | 1.697 | + | 1.18 | 0 |
| 3R2 | root | 85 | Pseudomonas oleovorans DSM 50188 | 1.598 | + | 1.5 | 10.37 |
| 3S1 | stem | 85 | Acinetobacter calcoaceticus DSM 30006T | 1.653 | + | 0 | 0 |
| 3S2 | stem | 85 | Bacillus megaterium DSM 32T DSM | 2.118 | + | 1.09 | 81.48 |
| 3S3 | stem | 85 | Klebsiella oxytoca ATCC 700324 THL | 1.415 | − | 1.1 | 13.33 |
| 3S4 | stem | 85 | Pantoea agglomerans DSM 8570 DSM | 2.088 | − | 1.28 | 64.07 |
| 3S5 | stem | 85 | Bacillus megaterium DSM 32T DSM | 1.477 | + | 1.7 | 27.04 |
| 3S6 | stem | 85 | Enterobacter kobei S58 ADRIA | 1.773 | + | 0 | 0 |
| 3L1 | leaf | 85 | Bacillus subtilis DSM 5552 DSM | 1.982 | + | 1.13 | 61.85 |
| 3L2 | leaf | 85 | Sphingobium chlorophenolicum DSM 7098T HAM | 1.407 | + | 0 | 64.81 |
| Isolate | 1R4 | 1L6 | 2L1 | 2L3 | 3S2 | 3L1 | QST713 | WCS417 |
|---|---|---|---|---|---|---|---|---|
| Identification/Best BLAST hit | B. subtilis NR_112116.2 100% | B. pumilus NR_148786.1 99.73% | B. arybhattai NR_115953.1 100% | B. subtilis NR_112116.2 100% | B. arybhattai NR_115953.1 100% | B. subtilis NR_112116.2 99.88% | Bacillus velezensis | Pseudomonas simiae |
| 16S rRNA Accession | MZ947229 | MZ947231 | MZ947227 | MZ947228 | MZ947230 | MZ947232 | CP025079.1 | CP007637.1 |
| Catalase | + | + | + | + | + | + | + | + |
| Urease | + | − | − | + | − | − | − | + |
| Oxidase | + | - | − | + | − | + | + | + |
| HCN (mg·L−1) | 124.5 ± 0.21 a 1 | 221.8 ± 1.55 g | 158.2 ± 0.77 c | 170.1 ± 0.57 d | 170.6 ± 0.98 d | 173.6 ± 0.74 e | 198.0 ± 0.57 f | 145.3 ± 0.74 b |
| IAA (mg·L−1) | 31.8 ± 0.83 b | 58.0 ± 0.77 g | 26.6 ± 0.10 a | 40.4 ± 0.63 e | 31.6 ± 0.36 b | 38.7 ± 0.66 d | 36.5 ± 0.46 c | 49.9 ± 0.63 f |
| Solubilized P (mg·L−1) | 350.2 ± 2.34 e | 457.5 ± 6.37 g | 305.4 ± 2.66 b | 337.7 ± 0.58 d | 363.9 ± 2.52 f | 284.8 ± 0.98 a | 322.6 ± 0.58 c | 366.9 ± 0.98 f |
| Siderophores (%) | 87.5 ± 0.82 f | 67.1 ± 1.46 b | 70.2 ± 1.48 c | 78.4 ± 0.68 d | 87.1 ± 0.43 f | 84.5 ± 0.33 ef | 83.0 ± 1.57 e | 59.8 ± 1.22 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lipková, N.; Medo, J.; Artimová, R.; Maková, J.; Petrová, J.; Javoreková, S.; Michalko, J. Growth Promotion of Rapeseed (Brassica napus L.) and Blackleg Disease (Leptosphaeria maculans) Suppression Mediated by Endophytic Bacteria. Agronomy 2021, 11, 1966. https://doi.org/10.3390/agronomy11101966
Lipková N, Medo J, Artimová R, Maková J, Petrová J, Javoreková S, Michalko J. Growth Promotion of Rapeseed (Brassica napus L.) and Blackleg Disease (Leptosphaeria maculans) Suppression Mediated by Endophytic Bacteria. Agronomy. 2021; 11(10):1966. https://doi.org/10.3390/agronomy11101966
Chicago/Turabian StyleLipková, Nikola, Juraj Medo, Renata Artimová, Jana Maková, Jana Petrová, Soňa Javoreková, and Jaroslav Michalko. 2021. "Growth Promotion of Rapeseed (Brassica napus L.) and Blackleg Disease (Leptosphaeria maculans) Suppression Mediated by Endophytic Bacteria" Agronomy 11, no. 10: 1966. https://doi.org/10.3390/agronomy11101966
APA StyleLipková, N., Medo, J., Artimová, R., Maková, J., Petrová, J., Javoreková, S., & Michalko, J. (2021). Growth Promotion of Rapeseed (Brassica napus L.) and Blackleg Disease (Leptosphaeria maculans) Suppression Mediated by Endophytic Bacteria. Agronomy, 11(10), 1966. https://doi.org/10.3390/agronomy11101966






