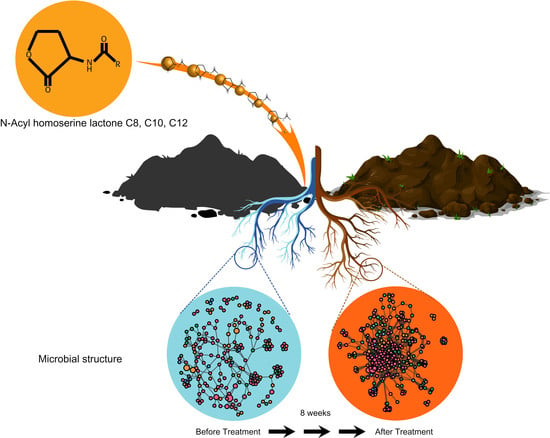
Use of Acyl-Homoserine Lactones Leads to Improved Growth of Ginseng Seedlings and Shifts in Soil Microbiome Structure
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ginseng Soil Sampling
2.2. In Vitro Pot Experiment and Treatment of Quorum-Sensing Signaling Molecules in Ginseng
2.3. DNA Extraction, PCR Amplification, and Sequencing
2.4. Bioinformatic Analysis
2.5. Statistical Analysis
3. Results
3.1. Phenotypic Effect of the AHL Soil Treatments on Ginseng Seedlings
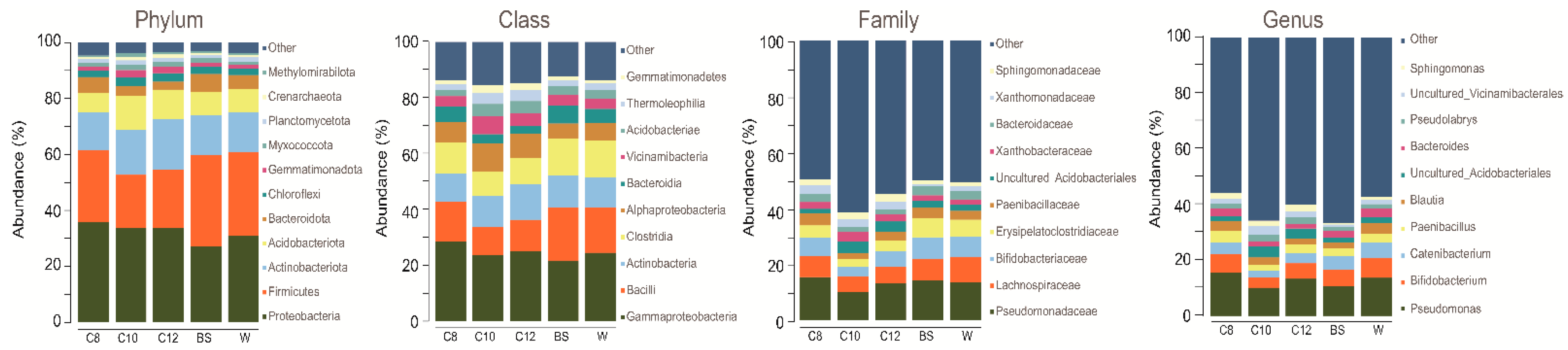
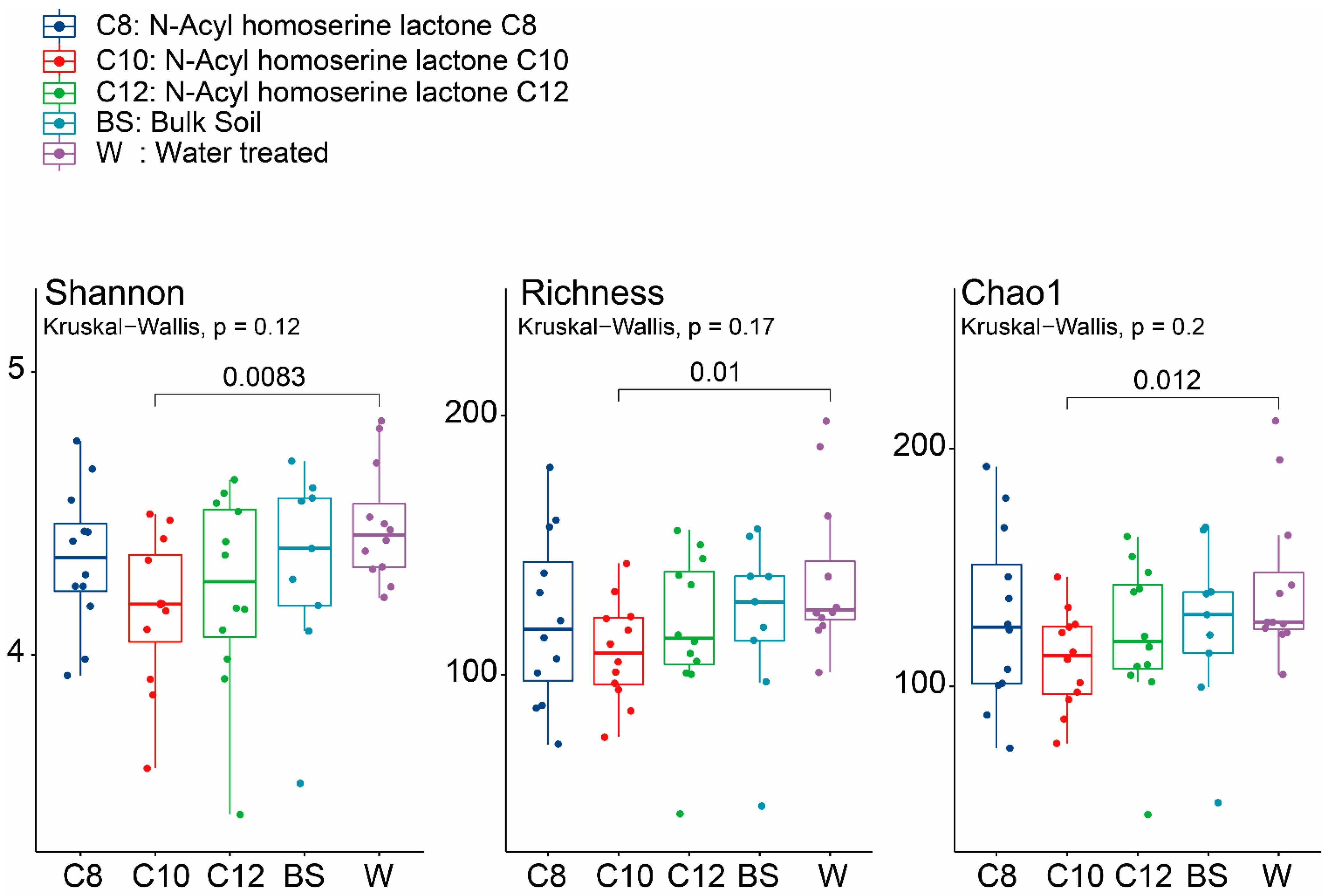
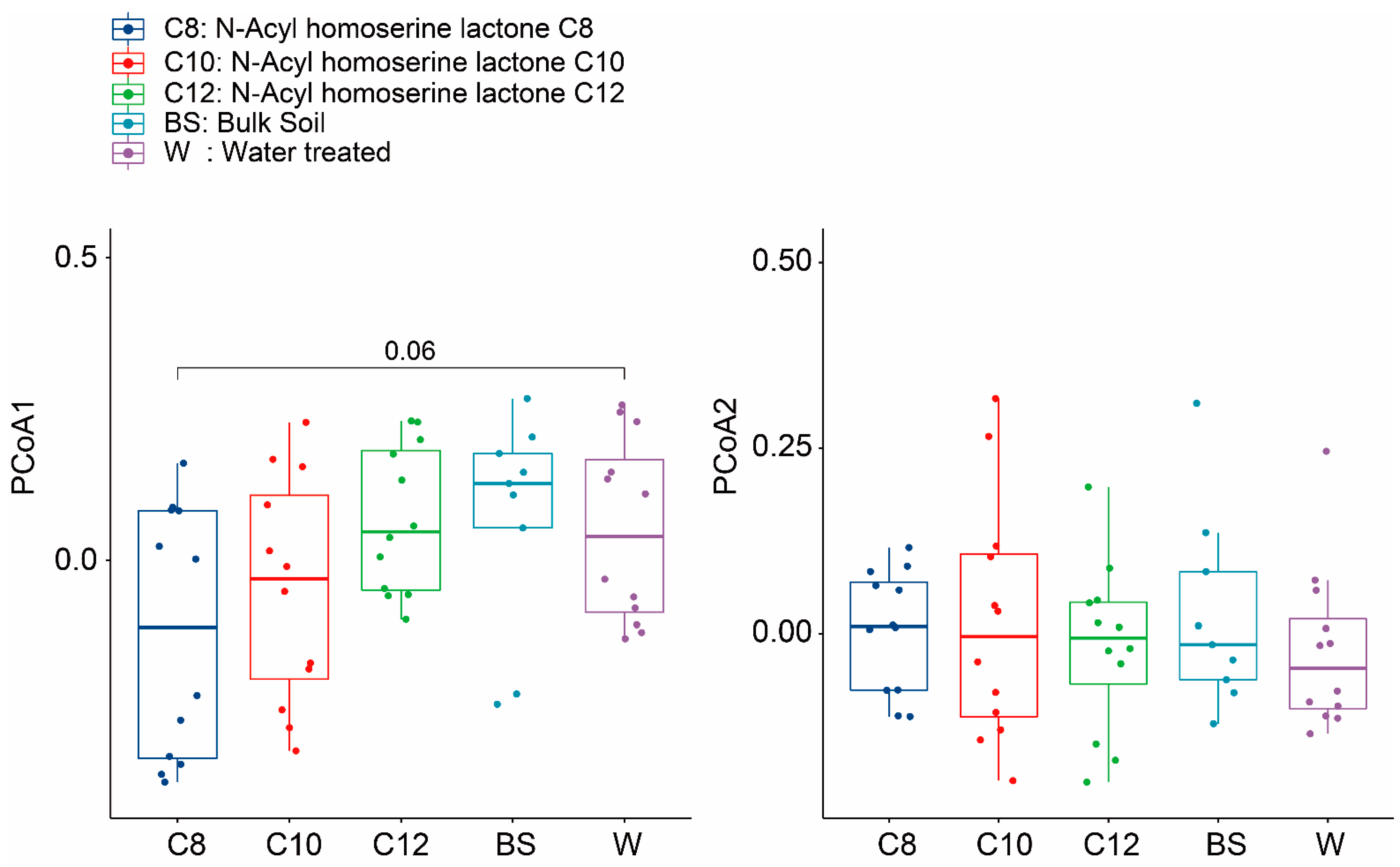
3.2. Microbial Community Shifts Involving AHL Treatments on Ginseng Soil
3.3. Functional Gene Analysis Using PICRUSt2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wen, J.; Zimmer, E.A. Phylogeny and biogeography of Panax L. (the ginseng genus, Araliaceae): Inferences from ITS sequences of nuclear ribosomal DNA. Mol. Phylogenet. Evol. 1996, 6, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Yun, T.K. Brief introduction of Panax ginseng CA Meyer. J. Korean Med. Sci. 2001, 16, S3–S5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H. Cardiovascular diseases and Panax ginseng: A review on molecular mechanisms and medical applications. J. Ginseng Res. 2012, 36, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, H.; Liu, D.; Sun, H. Deciphering microbiome related to rusty roots of Panax ginseng and evaluation of antagonists against pathogenic Ilyonectria. Front. Microbiol. 2019, 10, 1350. [Google Scholar]
- Wei, X.; Wang, X.; Cao, P.; Gao, Z.; Chen, A.J.; Han, J. Microbial community changes in the rhizosphere soil of healthy and rusty Panax ginseng and discovery of pivotal fungal genera associated with rusty roots. Biomed Res. Int. 2020. [Google Scholar] [CrossRef] [Green Version]
- Atkinson, D.; Watson, C.A. The beneficial rhizosphere: A dynamic entity. Appl. Soil Ecol. 2000, 15, 99–104. [Google Scholar] [CrossRef]
- Whipps, J.M. Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 2001, 52 (Suppl. 1), 487–511. [Google Scholar] [CrossRef]
- Song, C.; Zhu, F.; Carrión, V.J.; Cordovez, V. Beyond plant microbiome Composition: Exploiting microbial functions and plant traits via integrated approaches. Front. Bioeng. Biotechnol. 2020, 8. [Google Scholar] [CrossRef]
- Dong, L.L.; Niu, W.H.; Wang, R.; Xu, J.; Zhang, L.J.; Zhang, J.; Chen, S.L. Changes of diversity and composition of fungal communities in rhizosphere of Panax ginseng. Zhongguo Zhong Yao Za Zhi 2017, 42, 443–449. [Google Scholar] [CrossRef]
- Korenblum, E.; Dong, Y.; Szymanski, J.; Panda, S.; Jozwiak, A.; Massalha, H.; Meir, S.; Rogachev, I.; Aharoni, A. Rhizosphere microbiome mediates systemic root metabolite exudation by root-to-root signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 3874–3883. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; Paulitz, T.C.; Steinberg, C.; Alabouvette, C.; Moënne-Loccoz, Y. The rhizosphere: A playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil. 2009, 321, 341–361. [Google Scholar] [CrossRef] [Green Version]
- Lareen, A.; Burton, F.; Schäfer, P. Plant root-microbe communication in shaping root microbiomes. Plant Mol. Biol. 2016, 90, 575–587. [Google Scholar] [CrossRef] [Green Version]
- Antoun, H.; Kloepper, J.W. Encyclopedia of Genetics; Academic: New York, NY, USA, 2001. [Google Scholar]
- Mohanram, S.; Kumar, P. Rhizosphere microbiome: Revisiting the synergy of plant-microbe interactions. Ann. Clin. Microbiol. 2019, 69, 307–320. [Google Scholar] [CrossRef]
- Shrestha, A.; Schikora, A. AHL-priming for enhanced resistance as a tool in sustainable agriculture. FEMS Microbiol. Ecol. 2020, 96, fiaa226. [Google Scholar] [CrossRef]
- Ng, W.L.; Bassler, B.L. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009, 43, 197–222. [Google Scholar] [CrossRef] [Green Version]
- Lazazzera, B.A.; Grossman, A.D. The ins and outs of peptide signaling. Trends Microbiol. 1998, 6, 288–294. [Google Scholar] [CrossRef]
- Von Bodman, S.B.; Bauer, W.D.; Coplin, D.L. Quorum sensing in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 2003, 41, 455–482. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal–response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576. [Google Scholar] [CrossRef] [PubMed]
- Zarkani, A.A.; Stein, E.; Röhrich, C.R.; Schikora, M.; Evguenieva-Hackenberg, E.; Degenkolb, T.; Vilcinskas, A.; Klug, G.; Kogel, K.-H.; Schikora, A. Homoserine lactones influence the reaction of plants to rhizobia. Int. J. Mol. Sci. 2013, 14, 17122–17146. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, A.; Elhady, A.; Adss, S.; Wehner, G.; Böttcher, C.; Heuer, H.; Ordon, F.; Schikora, A. Genetic differences in barley govern the responsiveness to N-Acyl homoserine lactone. Phytobiomes J. 2019, 3, 191–202. [Google Scholar] [CrossRef] [Green Version]
- Shrestha, A.; Grimm, M.; Ojiro, I.; Krumwiede, J.; Schikora, A. Impact of quorum sensing molecules on plant growth and immune system. Front. Microbiol. 2020, 11, 1545. [Google Scholar] [CrossRef]
- Liang, X.; Wagner, R.E.; Li, B.; Zhang, N.; Radosevich, M. Quorum sensing signals alter in vitro soil virus abundance and bacterial community composition. Front. Microbiol. 2020, 11, 1287. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Bai, Y. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Amir, A.; McDonald, D.; Navas-Molina, J.A.; Kopylova, E.; Morton, J.T.; Zech Xu, Z.; Kightley, E.P.; Thompson, L.R.; Hyde, E.R.; Gonzalez, A.; et al. Deblur rapidly resolves single-nucleotide community sequence patterns. MSystems 2017, 2, e00191-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef] [Green Version]
- Dixon, P. VEGAN, a package of R functions for community ecology. Appl. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2. Comput. Stat. Data Anal. 2011, 3, 180–185. [Google Scholar]
- Lei, H.; Liu, A.; Hou, Q.; Zhao, Q.; Guo, J.; Wang, Z. Diversity patterns of soil microbial communities in the Sophora flavescens rhizosphere in response to continuous monocropping. BMC Microbiol. 2020, 20, 272. [Google Scholar] [CrossRef]
- Hartmann, A. Quorum sensing N-acyl-homoserine lactone signal molecules of plant beneficial Gram-negative rhizobacteria support plant growth and resistance to pathogens. Rhizosphere 2020, 16, 100258. [Google Scholar] [CrossRef]
- Schenk, S.T.; Schikora, A. AHL-priming functions via oxylipin and salicylic acid. Front. Plant Sci. 2015, 5, 784. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, A.; Rothballer, M.; Hense, B.A.; Schröder, P. Bacterial quorum sensing compounds are important modulators of microbe-plant interactions. Front. Plant Sci. 2014, 5, 131. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Bian, Z.; Jia, Z.; Zhao, Q.; Song, S. The GCR1 and GPA1 participate in promotion of Arabidopsis primary root elongation induced by N-acyl-homoserine lactones, the bacterial quorum-sensing signals. Mol. Plant-Microbe Interact. 2012, 25, 677–683. [Google Scholar] [CrossRef] [Green Version]
- Götz-Rösch, C.; Sieper, T.; Fekete, A.; Schmitt-Kopplin, P.; Hartmann, A.; Schröder, P. Influence of bacterial N-acyl-homoserine lactones on growth parameters, pigments, antioxidative capacities and the xenobiotic phase II detoxification enzymes in barley and yam bean. Front. Plant Sci. 2015, 6, 205. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Martínez, I.; Walter, J.; Keshavarzian, A.; Rose, D.J. In vitro characterization of the impact of selected dietary fibers on fecal microbiota composition and short chain fatty acid production. Anaerobe 2013, 23, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Maquia, I.S.; Fareleira, P.; Videira E Castro, I.; Brito, D.; Soares, R.; Chaúque, A.; Ferreira-Pinto, M.M.; Lumini, E.; Berruti, A.; Ribeiro, N.S.; et al. Mining the Microbiome of Key Species from African Savanna Woodlands: Potential for Soil Health Improvement and Plant Growth Promotion. Microorganisms 2020, 8, 1291. [Google Scholar] [CrossRef] [PubMed]
- Kämpfer, P.; Young, C.C.; Arun, A.B.; Shen, F.T.; Jäckel, U.; Rossello-Mora, R.; Rekha, P.D. Pseudolabrys taiwanensis gen. nov., sp. nov., an alphaproteobacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 2006, 56, 2469–2472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Q.; Song, X.; Deng, X.; Luo, J.; Wang, J.; Min, K.; Song, R. Effects of Plant Growth Promoting Rhizobacteria Microbial Inoculants on the Growth, Rhizosphere Soil Properties, and Bacterial Community of Pinus sylvestris var. mongolica Annual Seedlings. Res. Sq. 2020. [Google Scholar] [CrossRef]
- Cipriano, M.A.; Lupatini, M.; Lopes-Santos, L.; da Silva, M.J.; Roesch, L.F.; Destéfano, S.A.; Freitas, S.S.; Kuramae, E.E. Lettuce and rhizosphere microbiome responses to growth promoting Pseudomonas species under field conditions. FEMS Microbiol. Ecol. 2016, 92, fiw197. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Hao, Z.; Li, K.; Sha, Y.; Wang, E.; Sui, X.; Mi, G.; Tian, C.; Chen, W. Effectsof growth-promoting rhizobacteria on maize growth and rhizosphere microbial community under conservation tillage in Northeast China. Microbial Biotechnol. 2021, 14, 535–550. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.; Rychahou, P.; Sviripa, V.M.; Weiss, H.L.; Liu, C.; Watt, D.S.; Evers, B.M. Characterizing changes in soil microbiome abundance and diversity due to different cover crop techniques. PLoS ONE 2020, 15, e0232453. [Google Scholar]
- Wagg, C.; Schlaeppi, K.; Banerjee, S.; Kuramae, E.E.; van der Heijden, M.G. Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 2019, 10, 4841. [Google Scholar] [CrossRef] [PubMed]
- Lau, J.A.; Lennon, J.T. Evolutionary ecology of plant–microbe interactions: Soil microbial structure alters selection on plant traits. New Phytol. 2011, 192, 215–224. [Google Scholar] [CrossRef] [PubMed]
- van der Heijden, M.G.; Wagg, C. Soil microbial diversity and agro-ecosystem functioning. Plant Soil. 2013, 363, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Jousset, A.; Schmid, B.; Scheu, S.; Eisenhauer, N. Genotypic richness and dissimilarity opposingly affect ecosystem functioning. Ecol. Lett. 2011, 14, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Kruijt, M.; De Bruijn, I.; Dekkers, E.; van der Voort, M.; Schneider, J.H.; Raaijmakers, J.M. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef]
- Fierer, N.; Ladau, J.; Clemente, J.C.; Leff, J.W.; Owens, S.M.; Pollard, K.S.; McCulley, R.L. Reconstructing the microbial diversity and function of pre-agricultural tallgrass prairie soils in the United States. Science 2013, 342, 621–624. [Google Scholar] [CrossRef] [Green Version]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Zhalnina, K.; Louie, K.B.; Hao, Z.; Mansoori, N.; da Rocha, U.N.; Shi, S.; Brodie, E.L. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 2018, 3, 470–480. [Google Scholar] [CrossRef] [Green Version]
- Abisado, R.G.; Benomar, S.; Klaus, J.R.; Dandekar, A.A.; Chandler, J.R. Bacterial quorum sensing and microbial community interactions. MBio 2018, 9, e02331-17. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.J.; Lim, J.; Sukweenadhi, J.; Seok, J.W.; Lee, S.W.; Park, J.C.; Taizhanova, A.; Kim, D.; Yang, D.C. Genomic characterization of a newly isolated rhizobacteria Sphingomonas panacis reveals plant growth promoting effect to rice. Biotechnol. Bioprocess Eng. 2019, 24, 119–125. [Google Scholar] [CrossRef]
- Khan, A.L.; Waqas, M.; Kang, S.M.; Al-Harrasi, A.; Hussain, J.; Al-Rawahi, A.; Al-Khiziri, S.; Ullah, I.; Ali, L.; Jung, H.Y.; et al. Bacterial endophyte Sphingomonas sp. LK11 produces gibberellins and IAA and promotes tomato plant growth. J. Microbiol. 2014, 52, 689–695. [Google Scholar] [CrossRef]
- Von Rad, U.; Klein, I.; Dobrev, P.I.; Kottova, J.; Zazimalova, E.; Fekete, A.; Hartmann, A.; Schmitt-Kopplin, P.; Durner, J. Response of Arabidopsis thaliana to N-hexanoyl-DL-homoserine-lactone, a bacterial quorum sensing molecule produced in the rhizosphere. Planta 2008, 229, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Jung, B.K.; Ibal, J.C.; Pham, H.Q.; Kim, M.C.; Park, G.S.; Hong, S.J.; Jo, H.W.; Park, C.E.; Choi, S.D.; Jung, Y.; et al. Quorum sensing system affects the plant growth promotion traits of serratia fonticola GS2. Front. Microbiol. 2020, 11, 536865. [Google Scholar] [CrossRef] [PubMed]


| Treatments | Shoot Length (cm) | Root Length (cm) | Fresh Shoot Biomass (g/plant) | Fresh Root Biomass (g/plant) | Dry Shoot Biomass (g/plant) | Dry Root Biomass (g/plant) |
|---|---|---|---|---|---|---|
| W | 4.87 ± 0.14 a | 14.67 ± 0.95 a | 0.45 ± 0.09 ab | 0.51 ± 0.03 a | 0.08 ± 0.01 a | 0.18 ± 0.01 a |
| C8 | 5.94 ± 0.83 a | 13.02 ± 1.04 a | 0.38 ± 0.06 a | 0.53 ± 0.06 a | 0.11 ± 0.02 ad | 0.19 ± 0.01 a |
| C10 | 6.05 ± 0.52 a | 13.54 ± 0.67 a | 0.61 ± 0.07 b | 0.83 ± 0.09 c | 0.15 ± 0.02 d | 0.32 ± 0.04 c |
| C12 | 4.62 ± 0.46 a | 13.27 ± 0.59 a | 0.39 ± 0.05 a | 0.53 ± 0.06 a | 0.09 ± 0.01 a | 0.20 ± 0.02 a |
| DF | Sums of Sqs | Means Sqs | F. Model | R2 | Pr (>F) | ||
|---|---|---|---|---|---|---|---|
| Overall | Treatment | 4 | 1.8139 | 0.45346 | 1.1343 | 0.08025 | 0.021 * |
| Residuals | 52 | 20.7888 | 0.39978 | 0.91975 | |||
| Total | 56 | 22.6026 | 1.00000 | ||||
| 2 weeks | Treatment | 4 | 1.9865 | 0.49661 | 1.2304 | 0.32983 | 0.00 ** |
| Residuals | 10 | 4.0362 | 0.40362 | 0.67017 | |||
| Total | 14 | 6.0226 | 1.00000 | ||||
| 4 weeks | Treatment | 4 | 1.2441 | 0.31103 | 0.98797 | 0.30512 | 0.539 |
| Residuals | 9 | 2.8334 | 0.31482 | 0.69488 | |||
| Total | 13 | 4.0775 | 1.00000 | ||||
| 8 weeks | Treatment | 4 | 1.5079 | 0.37697 | 1.0388 | 0.34185 | 0.27 |
| Residuals | 8 | 2.9030 | 0.36288 | 0.65815 | |||
| Total | 12 | 4.4109 | 1.00000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibal, J.-C.; Park, M.-K.; Park, G.-S.; Jung, B.-K.; Park, T.-H.; Kim, M.-S.; Kang, G.-U.; Park, Y.-J.; Shin, J.-H. Use of Acyl-Homoserine Lactones Leads to Improved Growth of Ginseng Seedlings and Shifts in Soil Microbiome Structure. Agronomy 2021, 11, 2177. https://doi.org/10.3390/agronomy11112177
Ibal J-C, Park M-K, Park G-S, Jung B-K, Park T-H, Kim M-S, Kang G-U, Park Y-J, Shin J-H. Use of Acyl-Homoserine Lactones Leads to Improved Growth of Ginseng Seedlings and Shifts in Soil Microbiome Structure. Agronomy. 2021; 11(11):2177. https://doi.org/10.3390/agronomy11112177
Chicago/Turabian StyleIbal, Jerald-Conrad, Min-Kyu Park, Gun-Seok Park, Byung-Kwon Jung, Tae-Hyung Park, Min-Sueng Kim, Gi-Ung Kang, Yeong-Jun Park, and Jae-Ho Shin. 2021. "Use of Acyl-Homoserine Lactones Leads to Improved Growth of Ginseng Seedlings and Shifts in Soil Microbiome Structure" Agronomy 11, no. 11: 2177. https://doi.org/10.3390/agronomy11112177
APA StyleIbal, J.-C., Park, M.-K., Park, G.-S., Jung, B.-K., Park, T.-H., Kim, M.-S., Kang, G.-U., Park, Y.-J., & Shin, J.-H. (2021). Use of Acyl-Homoserine Lactones Leads to Improved Growth of Ginseng Seedlings and Shifts in Soil Microbiome Structure. Agronomy, 11(11), 2177. https://doi.org/10.3390/agronomy11112177