Novel Loci for Kernel Hardness Appeared as a Response to Heat and Combined Heat-Drought Conditions in Wheat Harboring Aegilops tauschii Diversity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Field Experiment
2.3. Hardness and Protein Content Measurements and SEM Observation
2.4. Statistical Analysis and Hardness Stress Indexes
2.5. Association Analysis
2.6. Bioinformatics Analysis
3. Results
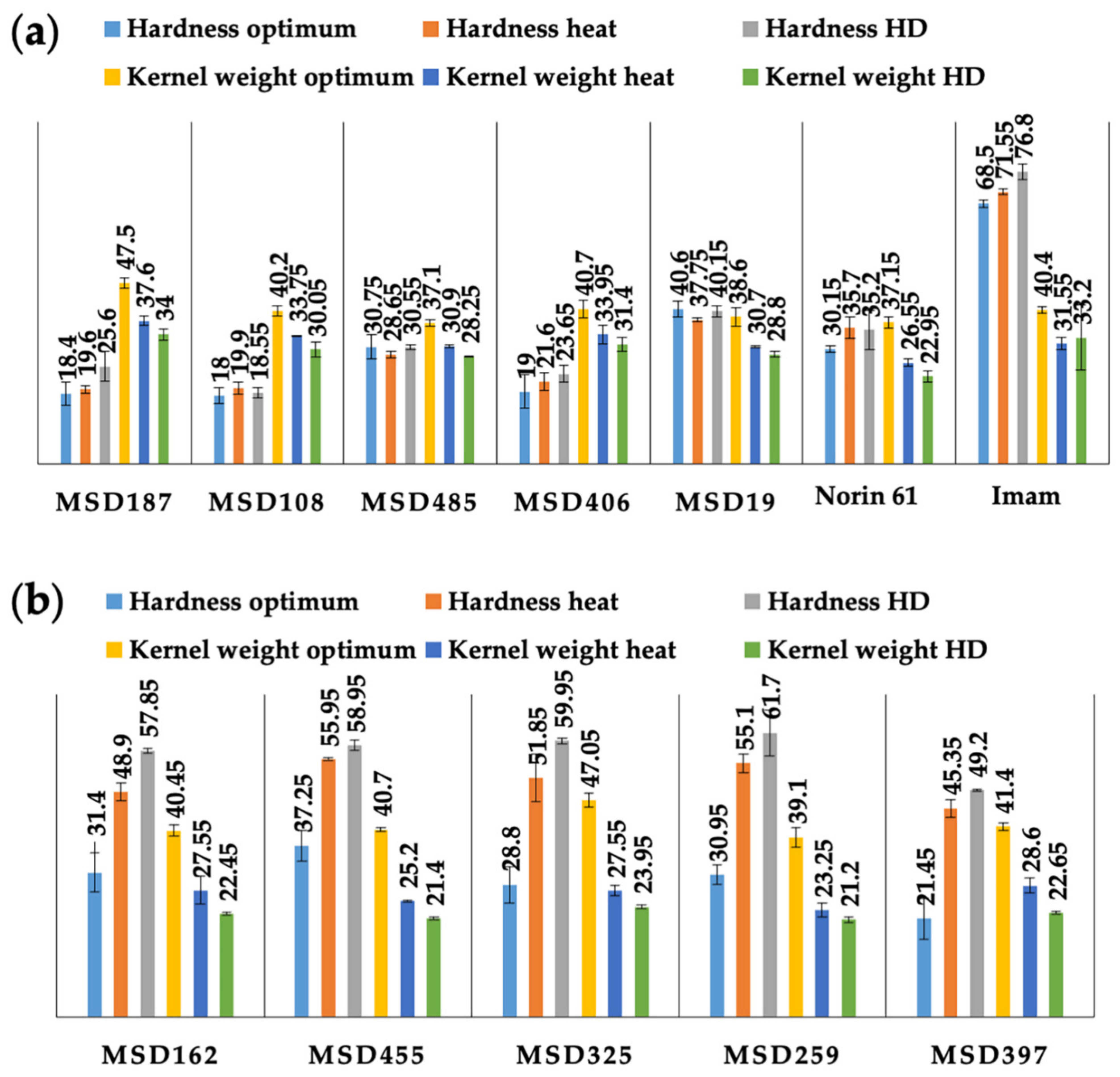
3.1. Phenotypic Variation and Diversity in Hardness among MSD Lines under Optimum and Stress Conditions
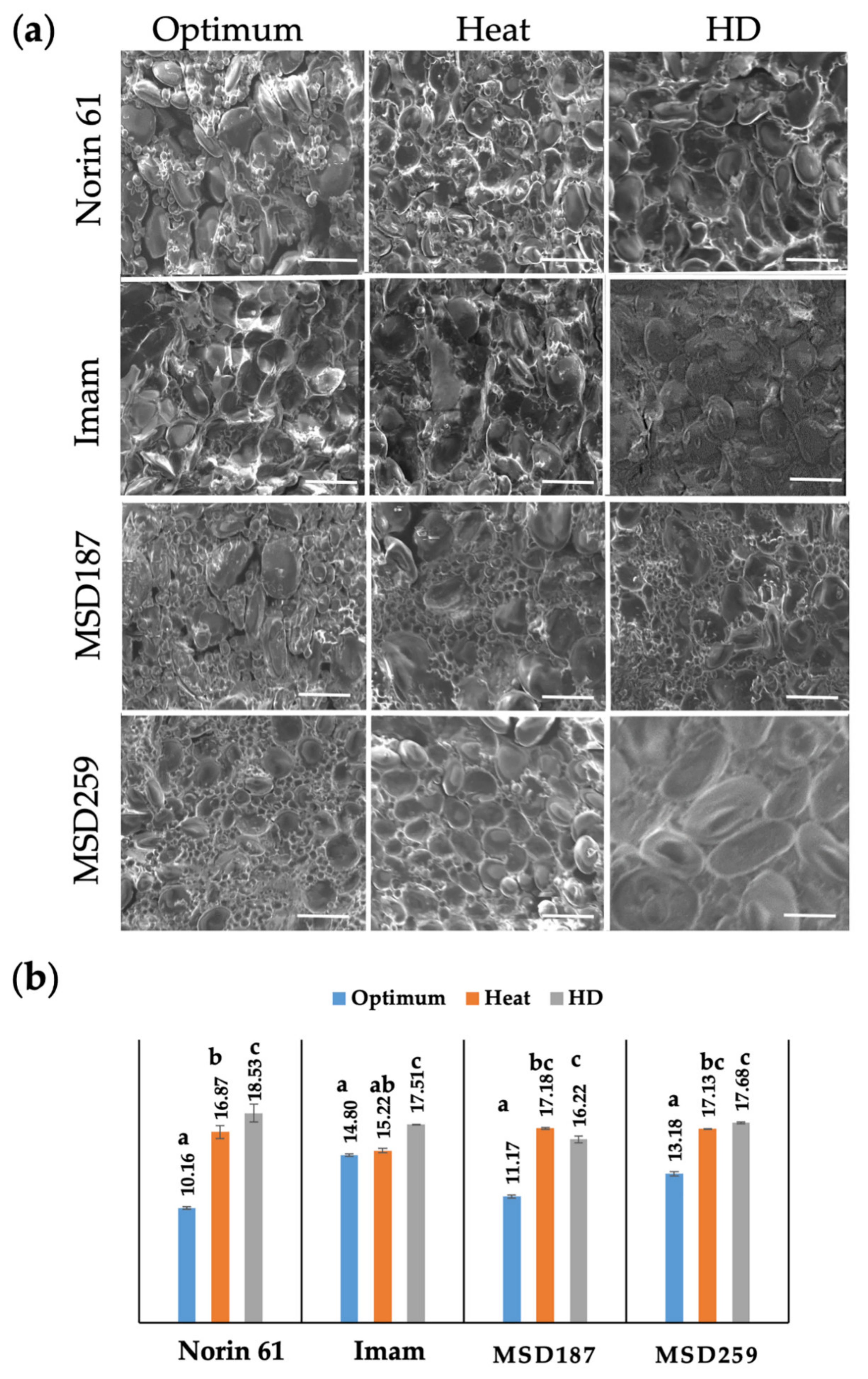
3.2. Internal Structure and Protein Content
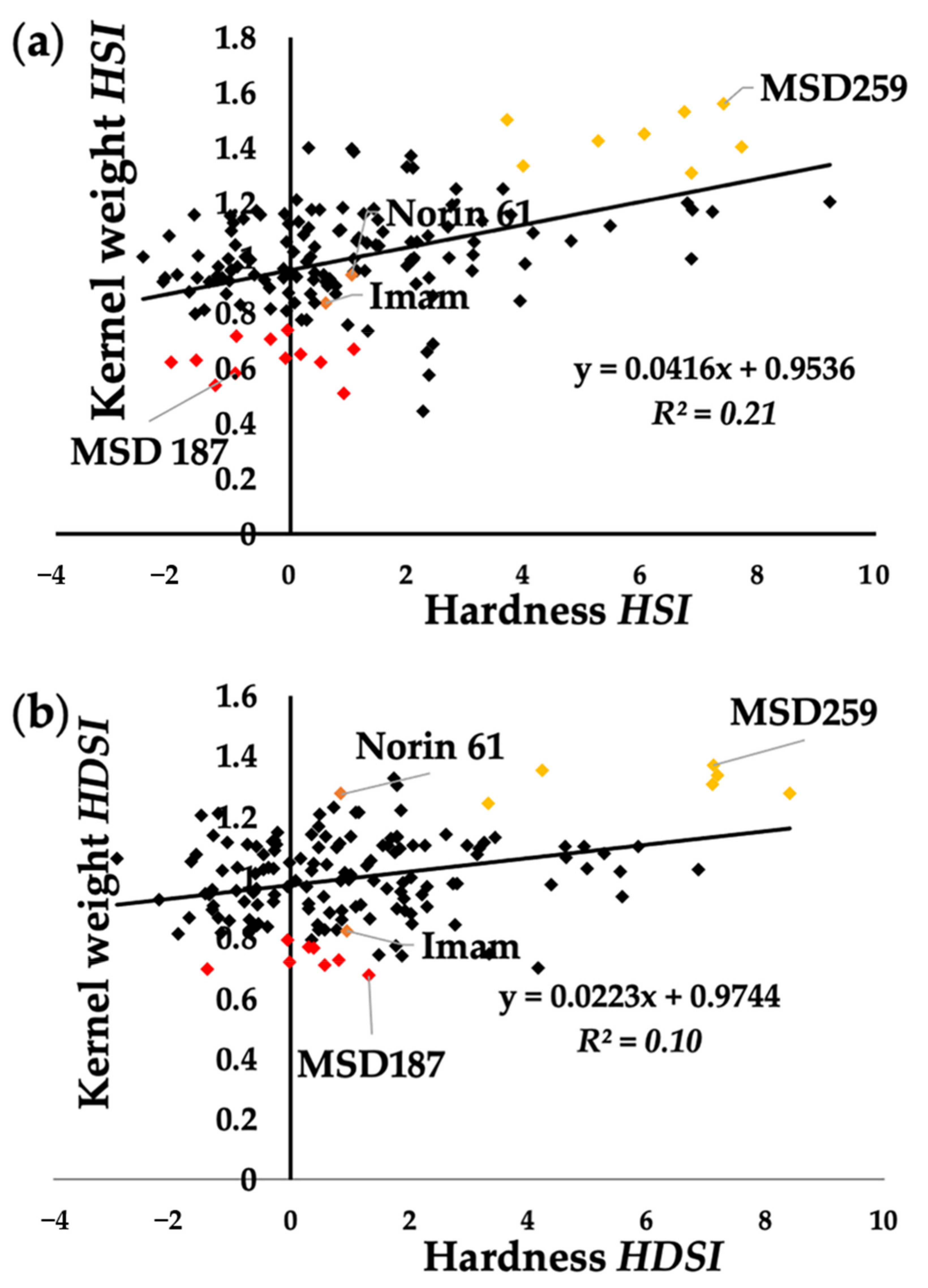
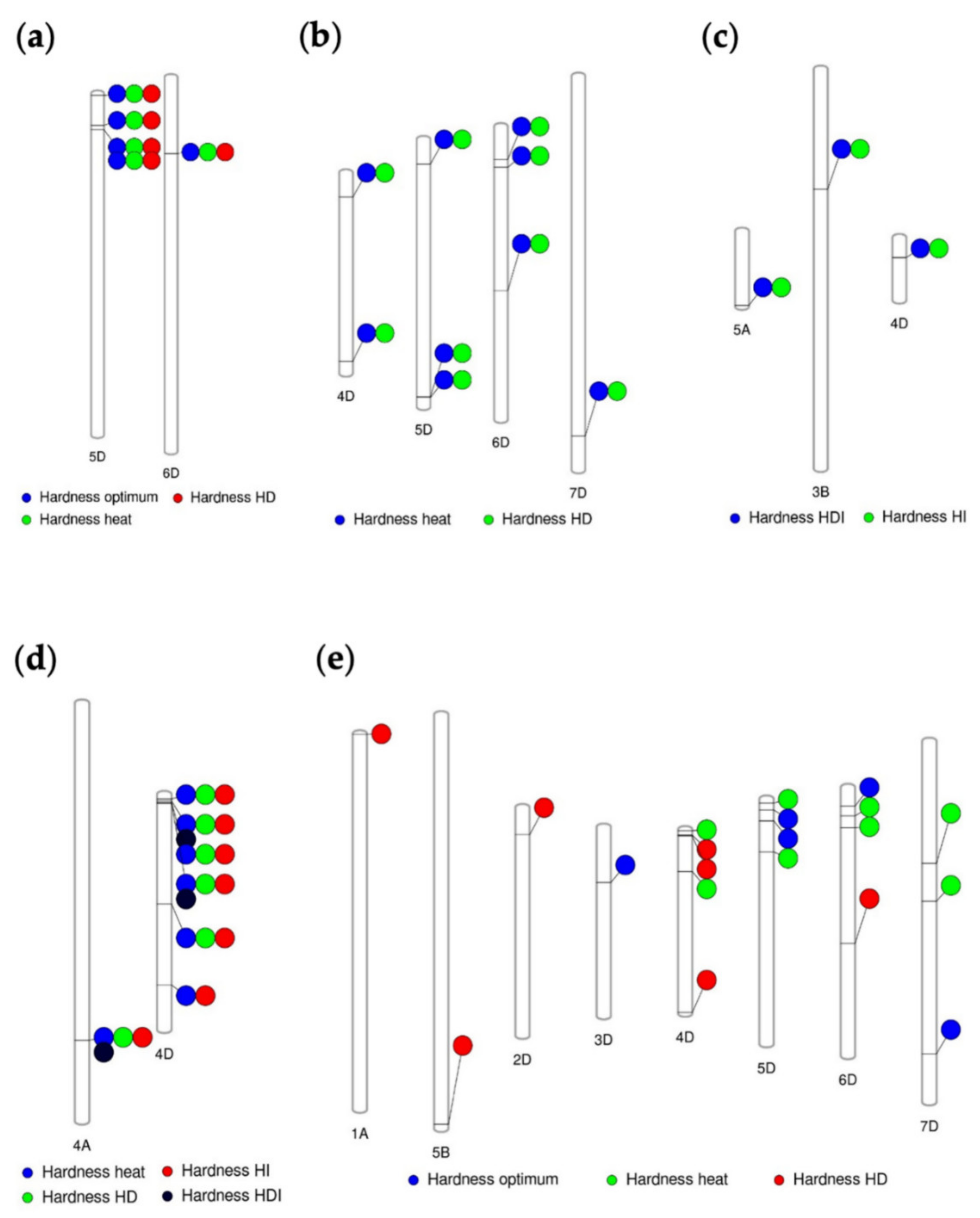
3.3. Marker Traits Association of Hardness and Hardness Indexes under Optimum and Stress Conditions
3.4. Common and Specific MTAs Associated with Kernel Hardness under Optimum and Stress Conditions
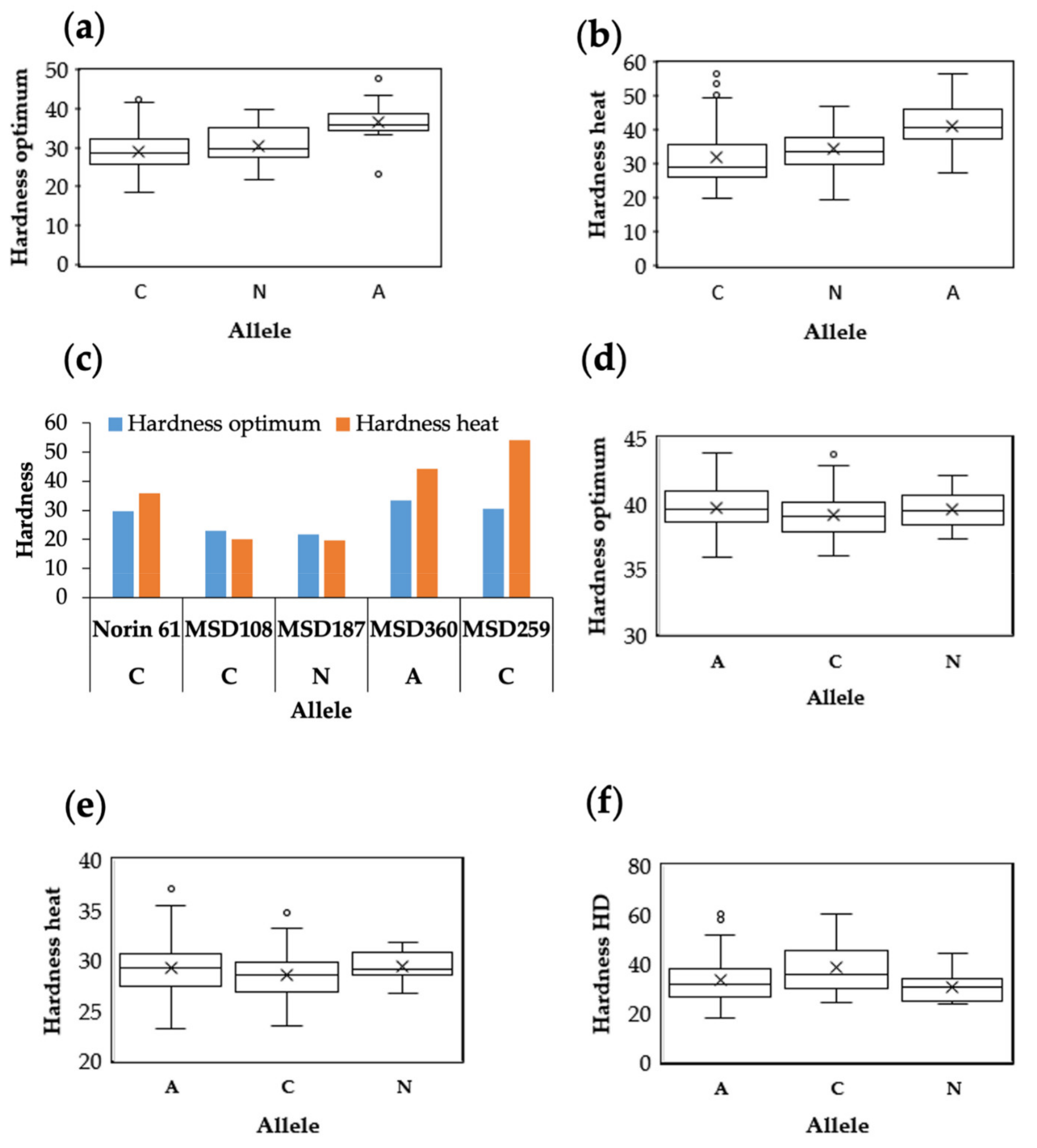
3.5. Candidate Genes for Hardness and Gene Expression
4. Discussion
4.1. Phenotypic Variation and Diversity for Hardness
4.2. Marker Traits Association for Hardness under Optimum and Stress Conditions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muqaddasi, Q.H.; Brassac, J.; Ebmeyer, E.; Kollers, S.; Korzun, V.; Argillier, O.; Stiewe, G.; Plieske, J.; Ganal, M.W.; Röder, M.S. Prospects of GWAS and predictive breeding for European winter wheat’s grain protein content, grain starch content, and grain hardness. Sci. Rep. 2020, 10, 1–17. [Google Scholar] [CrossRef]
- Giroux, M.J.; Morris, C.F. Wheat grain hardness results from highly conserved mutations in the friabilin components puroindoline a and b. Proc. Natl. Acad. Sci. USA 1998, 95, 6262–6266. [Google Scholar] [CrossRef] [Green Version]
- Bhave, M.; Morris, C.F. Molecular genetics of puroindolines and related genes: Allelic diversity in wheat and other grasses. Plant Mol. Biol. 2008, 66, 205–219. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Leonard, J.M.; von Zitzewitz, J.; Peterson, C.J.; Ross, A.S.; Riera-Lizarazu, O. Marker–trait association analysis of kernel hardness and related agronomic traits in a core collection of wheat lines. Molec. Breeding. 2014, 34, 177–184. [Google Scholar] [CrossRef]
- Kiseleva, A.A.; Leonova, I.N.; Pshenichnikova, T.A.; Salina, E.A. Dissection of novel candidate genes for grain texture in Russian wheat varieties. Plant Mol. Biol. 2020, 104, 219–233. [Google Scholar] [CrossRef]
- IPCC. Climate Change Synthesis Report Contribution of Working Groups I. II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; IPCC: Geneva, Switzerland, 2014; p. 151. [Google Scholar]
- Zandalinas, S.I.; Mittler, R.; Balfagón, D.; Arbona, V.; Gómez-Cadenas, A. Plant adaptations to the combination of drought and high temperatures. Physiol. Plant. 2018, 162, 2–12. [Google Scholar] [CrossRef] [Green Version]
- Ashraf, M. Stress-induced changes in wheat grain composition and quality. Crit. Rev. Food Sci. Nutr. 2014, 54, 1576–1583. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Liu, T.; Ning, T.; Liu, K.; Duan, X.; Wang, X.; Wang, Q.; An, Y.; Guan, X.; Tian, J.-C.; et al. Genetic dissection of wheat kernel hardness using conditional QTL mapping of kernel size and protein-related traits. Plant Mol. Biol. Report. 2018, 36, 1–12. [Google Scholar] [CrossRef]
- Szabó, B.P.; Gyimes, E.; Véha, A.; Horváth, Z.H. Flour quality and kernel hardness connection in winter wheat. Acta Univ. Sapientiae Aliment. 2016, 9, 33–40. [Google Scholar] [CrossRef] [Green Version]
- Triboï, E.; Martre, P.; Triboï-Blondel, A.M. Environmentally-induced changes in protein composition in developing grains of wheat are related to changes in total protein content. J. Exp. Bot. 2003, 54, 1731–1742. [Google Scholar] [CrossRef]
- Pasha, I.; Anjum, F.M.; Morris, C.F. Grain hardness: A major determinant of wheat quality. Food Sci. Technol. Int. 2010, 16, 511–522. [Google Scholar] [CrossRef]
- Bordes, J.; Ravel, C.; Le Gouis, J.; Lapierre, A.; Charmet, G.; Balfourier, F. Use of a global wheat core collection for association analysis of flour and dough quality traits. J. Cereal Sci. 2011, 54, 137–147. [Google Scholar] [CrossRef]
- Wang, G.; Leonard, J.M.; Ross, A.S.; Peterson, C.J.; Zemetra, R.S.; Campbell, K.G.; Riera-Lizarazu, O. Identification of genetic factors controlling kernel hardness and related traits in a recombinant inbred population derived from a soft × ‘extra-soft’ wheat (Triticum aestivum L.) cross. Theor. Appl. Genet. 2012, 124, 207–221. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, F.; Zhao, C.; Lv, G.; Sun, C.; Pan, Y.; Guo, X.; Chen, F. Genome-wide association study of six quality traits reveals the association of the TaRPP13L1 gene with flour colour in Chinese bread wheat. Plant Biotechnol. J. 2019, 17, 2106–2122. [Google Scholar] [CrossRef] [Green Version]
- Hernández-Espinosa, N.; Mondal, S.; Autrique, E.; Gonzalez-Santoyo, H.; Crossa, J.; Huerta-Espino, J.; Singh, R.P.; Guzmán, C. Milling, processing and end-use quality traits of CIMMYT spring bread wheat germplasm under drought and heat stress. Field Crop. Res. 2018, 215, 104–112. [Google Scholar] [CrossRef]
- Tomás, D.; Viegas, W.; Silva, M. Effects of post-anthesis heat waves on the grain quality of seven European wheat varieties. Agronomy 2020, 10, 268. [Google Scholar] [CrossRef] [Green Version]
- Gorafi, Y.S.A.; Kim, J.S.; Elbashir, A.A.E.; Tsujimoto, H. A population of wheat multiple synthetic derivatives: An effective platform to explore, harness and utilize genetic diversity of Aegilops tauschii for wheat improvement. Theor. Appl. Genet. 2018, 131, 1615–1626. [Google Scholar] [CrossRef]
- Elhadi, G.M.I.; Kamal, N.M.; Gorafi, Y.S.A.; Yamasaki, Y.; Takata, K.; Tahir, I.S.A.; Itam, M.O.; Tanaka, H.; Tsujimoto, H. Exploitation of tolerance of wheat kernel weight and shape-related traits from Aegilops tauschii under heat and combined heat-drought stresses. Int. J. Mol. Sci. 2021, 22, 1830. [Google Scholar] [CrossRef]
- Saito, H.; Tamura, M.; Ogawa, Y. Starch digestibility of various Japanese commercial noodles made from different starch sources. Food Chem. 2019, 283, 390–396. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Kuehl, R.O.; Ray, I.M.; Hui, R.; Soleri, D. Evaluation of simple methods for estimating broad-sense heritability in stands of randomly planted genotypes. Crop Sci. 1998, 38, 1125–1129. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Borrill, P.; Ramirez-Gonzalez, R.; Uauy, C. expVIP: A customizable RNA-seq data analysis and visualization platform. Plant Physiol. 2016, 170, 2172–2186. [Google Scholar] [CrossRef] [Green Version]
- Gedye, K.R.; Morris, C.F.; Bettge, A.D. Determination and evaluation of the sequence and textural effects of the puroindoline a and puroindoline b genes in a population of synthetic hexaploid wheat. Theor. Appl. Genet. 2004, 109, 1597–1603. [Google Scholar] [CrossRef] [PubMed]
- Peterson, C.J.; Graybosch, R.A.; Shelton, D.R.; Baenziger, P.S. Baking quality of hard winter wheat: Response of cultivars to environment in the Great Plains. Euphytica 1997, 100, 157–162. [Google Scholar] [CrossRef]
- Ozturk, A.; Aydin, F. Effect of Water Stress at Various Growth stages on some quality characteristics of winter wheat. J. Agron. Crop Sci. 2004, 190, 93–99. [Google Scholar] [CrossRef]
- Ramya, K.T.; Jain, N.; Gandhi, N.; Arora, A.; Singh, P.K.; Singh, A.M.; Singh, G.P.; Prabhu, K.V. Assessing heat stress tolerance and genetic diversity among exotic and Indian wheat genotypes using simple sequence repeats and agro-physiological traits. Plant Genet. Resour. Characterisation Util. 2017, 15, 208–220. [Google Scholar] [CrossRef]
- Fleitas, M.C.; Mondal, S.; Gerard, G.S.; Hernández-Espinosa, N.; Singh, R.P.; Crossa, J.; Guzmán, C. Identification of CIMMYT spring bread wheat germplasm maintaining superior grain yield and quality under heat-stress. J. Cereal Sci. 2020, 93. [Google Scholar] [CrossRef]
- Prathap, V.; Tyagi, A. Correlation between expression and activity of ADP glucose pyrophosphorylase and starch synthase and their role in starch accumulation during grain filling under drought stress in rice. Plant Physiol. Biochem. 2020, 157, 239–243. [Google Scholar] [CrossRef]
- Simmonds, D.H.; Barlow, K.K.; Wrigley, C.W. Biochemical basis of grain hardness in wheat. Cereal Chem. 1973, 50, 553–562. [Google Scholar]
- Okada, M.; Ikeda, T.M.; Yoshida, K.; Takumi, S. Effect of the U genome on grain hardness in nascent synthetic hexaploids derived from interspecific hybrids between durum wheat and Aegilops umbellulata. J. Cereal Sci. 2018, 83, 153–161. [Google Scholar] [CrossRef] [Green Version]
- Peterson, C.J.; Graybosch, R.A.; Baenziger, P.S.; Grombacher, A.W. Genotype and environment effects on quality characteristics of hard red winter wheat. Crop Sci. 1992, 32, 98–103. [Google Scholar] [CrossRef]
- Deol, K.K.; Mukherjee, S.; Gao, F.; Brûlé-Babel, A.; Stasolla, C.; Ayele, B.T. Identification and characterization of the three homeologues of a new sucrose transporter in hexaploid wheat (Triticum aestivum L.). BMC Plant Biol. 2013, 13. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, S.; Liu, A.; Deol, K.K.; Kulichikhin, K.; Stasolla, C.; BrÛlé-Babel, A.; Ayele, B.T. Transcriptional coordination and abscisic acid mediated regulation of sucrose transport and sucrose-to-starch metabolism related genes during grain filling in wheat (Triticum aestivum L.). Plant Sci. 2015, 240, 143–160. [Google Scholar] [CrossRef]
- Al-Sheikh Ahmed, S.; Zhang, J.; Ma, W.; Dell, B. Contributions of TaSUTs to grain weight in wheat under drought. Plant Mol. Biol. 2018, 98, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Van Ooijen, G.; Mayr, G.; Kasiem, M.M.A.; Albrecht, M.; Cornelissen, B.J.C.; Takken, F.L.W. Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. J. Of Exp. Bot. 2008, 59, 1383–1397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belkhadir, Y.; Subramaniam, R.; Dangl, J.L. Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr. Opin. In Plant Biol. 2004, 7, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Giroux, M.J.; Sripo, T.; Gerhardt, S.; Sherwood, J. Puroindolines: Their role in grain hardness and plant defence. Biotechnol. Genet. Eng. Rev. 2003, 20, 277–290. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, P.; Khurana, P. Characterization of a novel zinc finger transcription factor (TaZnF) from wheat conferring heat stress tolerance in Arabidopsis. Cell Stress Chaperones 2018, 23, 253–267. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.; Rodríguez-Quijano, M.; Giraldo, P.; Pinto, L.; Vázquez, J.F.; Carrillo, J.M.; Igrejas, G. Effect of allelic variation at glutenin and puroindoline loci on bread-making quality: Favorable combinations occur in less toxic varieties of wheat for celiac patients. Eur. Food Res. Technol. 2017, 243, 743–752. [Google Scholar] [CrossRef]
- Uauy, C.; Distelfeld, A.; Fahima, T.; Blechl, A.; Dubcovsky, J. A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 2006, 314, 1298–1301. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, M.; Wan, Y.; Tosi, P.; Leverington, M.; Snape, J.; Mitchell, R.A.C.; Shewry, P.R. Identification and genetic mapping of variant forms of puroindoline b expressed in developing wheat grain. J. Cereal Sci. 2008, 48, 722–728. [Google Scholar] [CrossRef]
- Wang, F.; Fu, L.; Kong, W.; Ye, Y.; Chen, X.; Zhou, Q.; Chen, B. Constitutive expression of StAATP, a potato plastidic ATP/ADP transporter gene, increases starch content in transgenic Arabidopsis. Biotechnol. Biotechnol. Equip. 2017, 312, 250–258. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Xiong, S.; Wei, Y.; Meng, X.; Wang, X.; Ma, X. The role of glutamine synthetase isozymes in enhancing nitrogen use efficiency of N-efficient winter wheat. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borrell, B.A.; Hammer, G.; Oosterom, E.V.A.N. Stay-green: A consequence of the balance between supply and demand for nitrogen during grain filling? Ann. Appl. Biol. 2001, 3902, 91–95. [Google Scholar] [CrossRef]
- Pinto, R.S.; Reynolds, M.P.; Mathews, K.L.; McIntyre, C.L.; Olivares-Villegas, J.J.; Chapman, S.C. Heat and drought adaptive QTL in a wheat population designed to minimize confounding agronomic effects. Theor. Appl. Genet. 2010, 121, 1001–1021. [Google Scholar] [CrossRef] [Green Version]
- Muhitch, M.J. Distribution of the glutamine synthetase isozyme GSp1 in maize (Zea mays). J. Plant Physiol. 2003, 160, 601–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
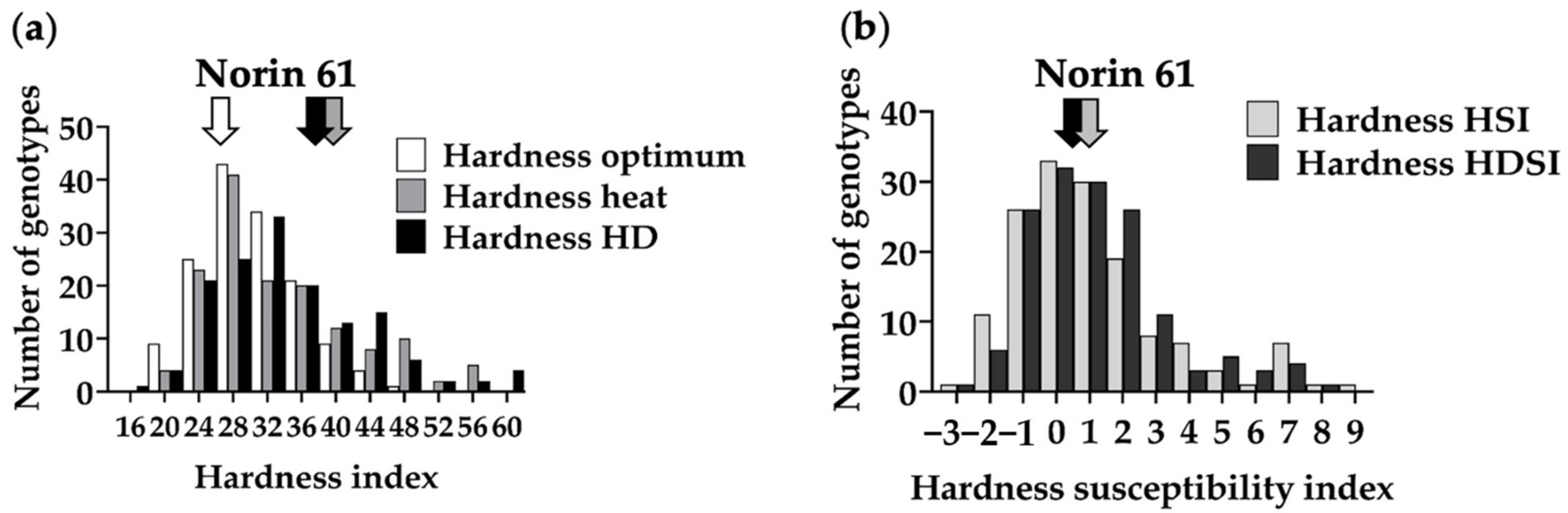

| Source of Variation | MSD Range for Hardness (Index) | Norin 61 | SED (±) | p-Value | LSD | Heritability |
|---|---|---|---|---|---|---|
| Optimum | 18.5–47.8 | 29.6 | - | *** | - | - |
| S (S1 × S2) | - | - | - | ns | - | - |
| G × S | - | - | 4.77 | *** | 9.20 | 0.90 |
| Heat | 19.6–56.9 | 35.7 | 3.60 | *** | 7.10 | - |
| HD | 25.5–60.2 | 35.2 | 6.40 | *** | 12.70 | - |
| E | - | - | - | ** | - | - |
| G × E | - | - | 3.59 | ns | 7.07 | 0.97 |
| Total Genome Contribution % | Chromosome | Marker Position | Marker | Environment | p-Value | PVE % |
|---|---|---|---|---|---|---|
| A genome (4.3%) | 1A | 441,118 | 3947128 | HD | 8.28 × 10−4 | 8.5 |
| 4A | 173,725,668 | 3025314 | Heat | 9.03 × 10−4 | 8.7 | |
| HD | 2.17 × 10−4 | 11.1 | ||||
| B genome (4.3%) | 5B | 4,332,688 | 6030814|F|0-7 | Optimum | 1.30 × 10−4 | 13.5 |
| Heat | 2.71 × 10−4 | 12.3 | ||||
| 270,359,835 | 1082888|F|0-18 | HD | 1.98 × 10−4 | 12.9 | ||
| D genome (91.4%) | 1D | 59,530,872 | 7351912|F|0-19 | Optimum | 2.32 × 10−4 | 15.2 |
| 2D | 17,717,364 | 3021443|F|0-24 | HD | 9.06 × 10−4 | 8.3 | |
| 3D | 36,431,533 | 1082930 | Optimum | 8.81 × 10−4 | 8.7 | |
| 4D | 799,855 | 1078655 | Heat | 1.22 × 10−4 | 13.0 | |
| 2,196,458 | 1062681|F|0-26 | Heat | 2.71 × 10−6 | 20.5 | ||
| HD | 2.77 × 10−5 | 17.3 | ||||
| 3,354,073 | 1665831 | Heat | 8.53 × 10−4 | 8.7 | ||
| HD | 7.77 × 10−4 | 8.5 | ||||
| 3,486,593 | 1126556 | HD | 4.85 × 10−4 | 9.8 | ||
| 4,063,428 | 4440031 | HD | 5.44 × 10−4 | 10.5 | ||
| 4,135,716 | 3946288 | Heat | 4.03 × 10−5 | 15.3 | ||
| HD | 6.79 × 10−4 | 10.2 | ||||
| 4,676,059 | 5332499 | Heat | 3.39 × 10−4 | 11.5 | ||
| HD | 9.14 × 10−5 | 12.6 | ||||
| 14,681,664 | 3574405 | Heat | 1.69 × 10−5 | 15.6 | ||
| HD | 6.69 × 10−5 | 13.1 | ||||
| 27,756,435 | 1001438|F|0-46 | Heat | 7.18 × 10−6 | 18.9 | ||
| 56,518,479 | 998809|F|0-7 | Heat | 7.30 × 10−5 | 14.4 | ||
| HD | 5.85 × 10−4 | 11.2 | ||||
| 98,321,462 | 1043872|F|0-49 | Heat | 5.61 × 10−4 | 11.7 | ||
| 113,875,447 | 3950661|F|0-9 | Heat | 6.59 × 10−4 | 12.0 | ||
| HD | 1.61 × 10−4 | 14.6 | ||||
| 120,386,388 | 1218881 | HD | 7.28 × 10−4 | 10.9 | ||
| 5D | 849,701 | 1127970 | Optimum | 5.51 × 10−7 | 22.6 | |
| Heat | 4.69 × 10−6 | 18.9 | ||||
| HD | 9.94 × 10−5 | 14.5 | ||||
| 1,268,790 | 3957566 | Optimum | 7.99 × 10−5 | 14.1 | ||
| 1,465,909 | 1090404 | Optimum | 1.44 × 10−5 | 15.0 | ||
| 2,351,222 | 3532985 | Optimum | 1.33 × 10−5 | 16.1 | ||
| Heat | 1.06 × 10−4 | 12.1 | ||||
| 2,504,012 | 5573281|F|0-7 | Optimum | 8.85 × 10−6 | 18.3 | ||
| Heat | 3.46 × 10−5 | 16.2 | ||||
| 3,757,073 | 5573405 | Optimum | 5.94 × 10−5 | 13.0 | ||
| 6,987,738 | 3948152 | Optimum | 2.14 × 10−4 | 11.0 | ||
| 14,328,401 | 4540116 | Optimum | 8.32 × 10−4 | 8.3 | ||
| 14,984,317 | 3944483 | Optimum | 7.40 × 10−5 | 13.9 | ||
| Heat | 1.33 × 10−5 | 15.3 | ||||
| HD | 1.05 × 10−5 | 16.3 | ||||
| 14,984,556 | 1025407 | Optimum | 8.54 × 10−5 | 12.6 | ||
| Heat | 7.17 × 10−5 | 12.3 | ||||
| HD | 7.29 × 10−5 | 12.1 | ||||
| 17,157,888 | 3021240|F|0-28 | Optimum | 3.77 × 10−4 | 12.0 | ||
| Heat | 5.40 × 10−5 | 14.9 | ||||
| HD | 1.09 × 10−4 | 13.7 | ||||
| 17,257,369 | 2242137|F|0-66 | Heat | 4.28 × 10−4 | 12.8 | ||
| 34,818,075 | 1093560|F|0-46 | Heat | 3.47 × 10−4 | 12.3 | ||
| 155,393,575 | 3950421 | Heat | 4.14 × 10−5 | 13.6 | ||
| HD | 1.52 × 10−4 | 11.9 | ||||
| 156,123,909 | 3936784 | Heat | 4.76 × 10−5 | 13.3 | ||
| HD | 1.04 × 10−3 | 8.4 | ||||
| 6D | 12,507,971 | 1102905|F|0-25 | Optimum | 5.84 × 10−4 | 10.0 | |
| 18,973,446 | 3960692|F|0-32 | Heat | 4.25 × 10−4 | 12.0 | ||
| 19,795,263 | 1074408|F|0-12 | Heat | 1.23 × 10−4 | 13.7 | ||
| HD | 1.65 × 10−4 | 13.1 | ||||
| 24,752,141 | 5357783 | Heat | 5.89 × 10−5 | 12.8 | ||
| HD | 2.62 × 10−4 | 10.4 | ||||
| 26,576,895 | 1073587|F|0-24 | Heat | 9.59 × 10−4 | 11.2 | ||
| 35,991,349 | 2242479 | Optimum | 2.43 × 10−4 | 12.3 | ||
| Heat | 1.19 × 10−4 | 12.1 | ||||
| HD | 1.02 × 10−4 | 12.3 | ||||
| 99,107,045 | 7348732 | Heat | 8.68 × 10−5 | 11.6 | ||
| HD | 3.85 × 10−4 | 9.4 | ||||
| 102,962,817 | 5992592 | HD | 6.21 × 10−4 | 9.2 | ||
| 7D | 80,561,020 | 3222372|F|0-27 | Heat | 6.97 × 10−6 | 19.8 | |
| 105,476,798 | 3944062 | Heat | 1.63 × 10−5 | 14.4 | ||
| 206,383,743 | 1261369|F|0-13 | Optimum | 7.69 × 10−4 | 12.5 | ||
| 217,517,266 | 1065454|F|0-13 | Heat | 3.28 × 10−4 | 9.5 | ||
| HD | 3.27 × 10−4 | 9.5 |
| Marker | Chromosome | Trait (Environment) | PVE (%) | Gene | Protein | Function |
|---|---|---|---|---|---|---|
| 3947128 | 1A | HD | 8.5 | TraesCS1A02G008000 | celiac disease | Gluten sensitivity |
| 1099828 | 3B | HDI | 12.0 | TraesCS3B02G533100 | No-apical-meristem (NAM) protein domain–containing protein | Increased protein content |
| 998809|F|0-7 | 4D | Heat, HD, HI, HDI | 14.4 | TraesCS4D02G117900 | ATP/ADP carrier protein | Nitrogen-use efficiency |
| 1062681|F|0-26 | 4D | Heat, HD, HI | 17.3 | TraesCS4D02G047400 | Glutamine synthetase | Nitrogen-use efficiency |
| 1127970 | 5D | Optimum, Heat, HD | 22.6 | TraesCS5D02G004100 | Puroindoline-a | Kernel hardness |
| 3532985 | 5D | Optimum, Heat | 16.1 | TraesCS5D02G001200 | Sucrose transporter | Starch accumulation |
| 3944483 | 5D | Optimum, Heat, HD | 16.3 | TraesCS5D02G037000 | NB-ARC domain–containing protein | Biotic stress |
| 1025407 | 5D | Heat, HD | 12.6 | TraesCS5D02G036300 | Zinc finger, RING-type domain–containing protein | Abiotic stress |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elhadi, G.M.I.; Kamal, N.M.; Gorafi, Y.S.A.; Yamasaki, Y.; Ban, Y.; Kato, K.; Tahir, I.S.A.; Ishii, T.; Tanaka, H.; Tsujimoto, H. Novel Loci for Kernel Hardness Appeared as a Response to Heat and Combined Heat-Drought Conditions in Wheat Harboring Aegilops tauschii Diversity. Agronomy 2021, 11, 1061. https://doi.org/10.3390/agronomy11061061
Elhadi GMI, Kamal NM, Gorafi YSA, Yamasaki Y, Ban Y, Kato K, Tahir ISA, Ishii T, Tanaka H, Tsujimoto H. Novel Loci for Kernel Hardness Appeared as a Response to Heat and Combined Heat-Drought Conditions in Wheat Harboring Aegilops tauschii Diversity. Agronomy. 2021; 11(6):1061. https://doi.org/10.3390/agronomy11061061
Chicago/Turabian StyleElhadi, Gamila Mohamed Idris, Nasrein Mohamed Kamal, Yasir Serag Alnor Gorafi, Yuji Yamasaki, Yusuke Ban, Keita Kato, Izzat Sidahmed Ali Tahir, Takayoshi Ishii, Hiroyuki Tanaka, and Hisashi Tsujimoto. 2021. "Novel Loci for Kernel Hardness Appeared as a Response to Heat and Combined Heat-Drought Conditions in Wheat Harboring Aegilops tauschii Diversity" Agronomy 11, no. 6: 1061. https://doi.org/10.3390/agronomy11061061







