Development of Novel Blackgram (Vigna mungo (L.) Hepper) Mutants and Deciphering Genotype × Environment Interaction for Yield-Related Traits of Mutants
Abstract
:1. Introduction
2. Materials and Methods
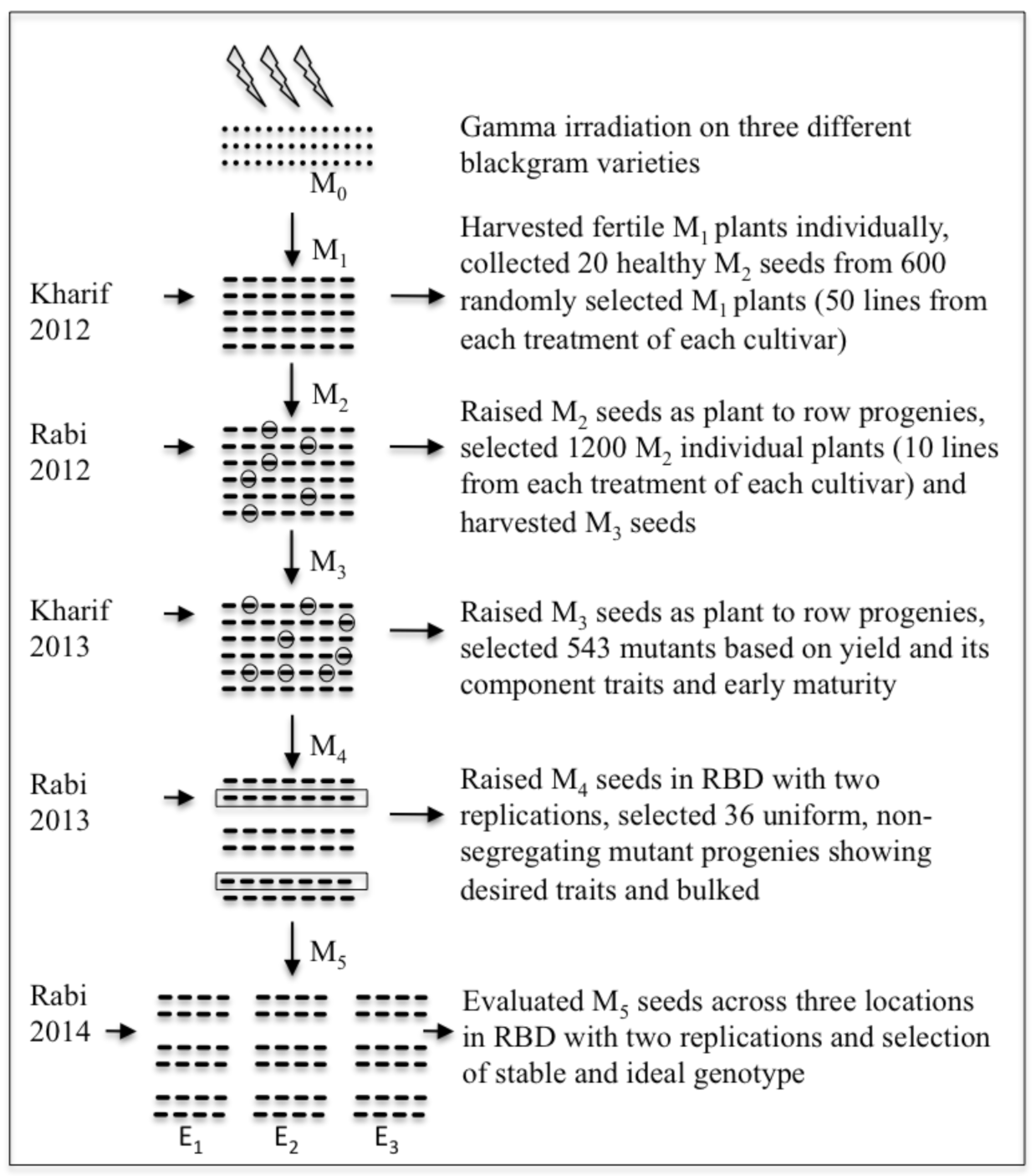
2.1. Plant Genetic Materials and Gamma Irradiation
2.2. Location, Experimental Design, and Development of Mutants
2.3. Selection Method, Data Collection, and Statistical Analysis
2.3.1. Selection Method
2.3.2. Data Collection
2.3.3. Statistical Analysis
2.4. M5 Evaluation, GEI, and Stability of Mutant Lines for Grain Yield
3. Results
3.1. Assessing the Effects of Mutagen on Yield-Related Traits in M1 and M2 Populations
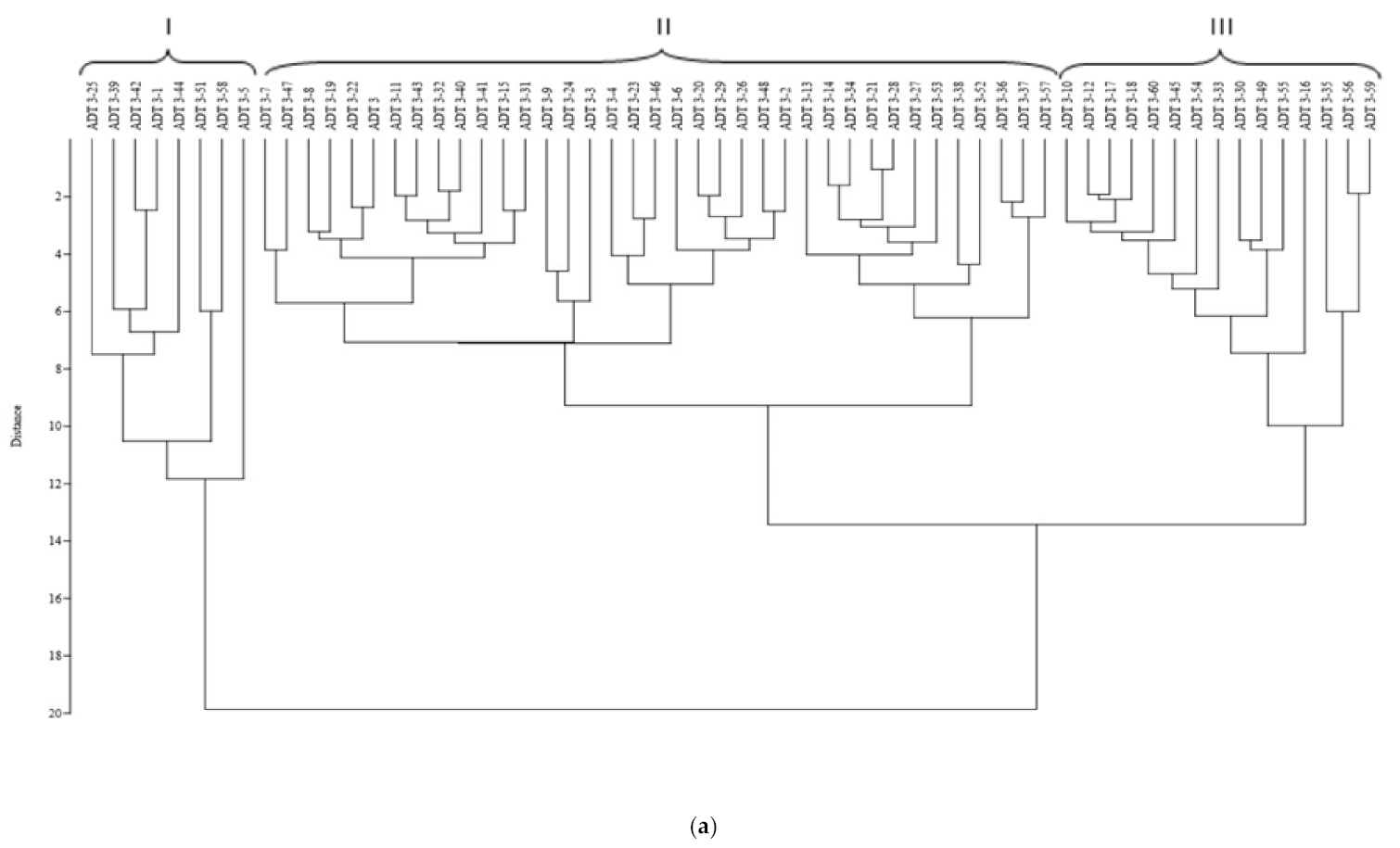
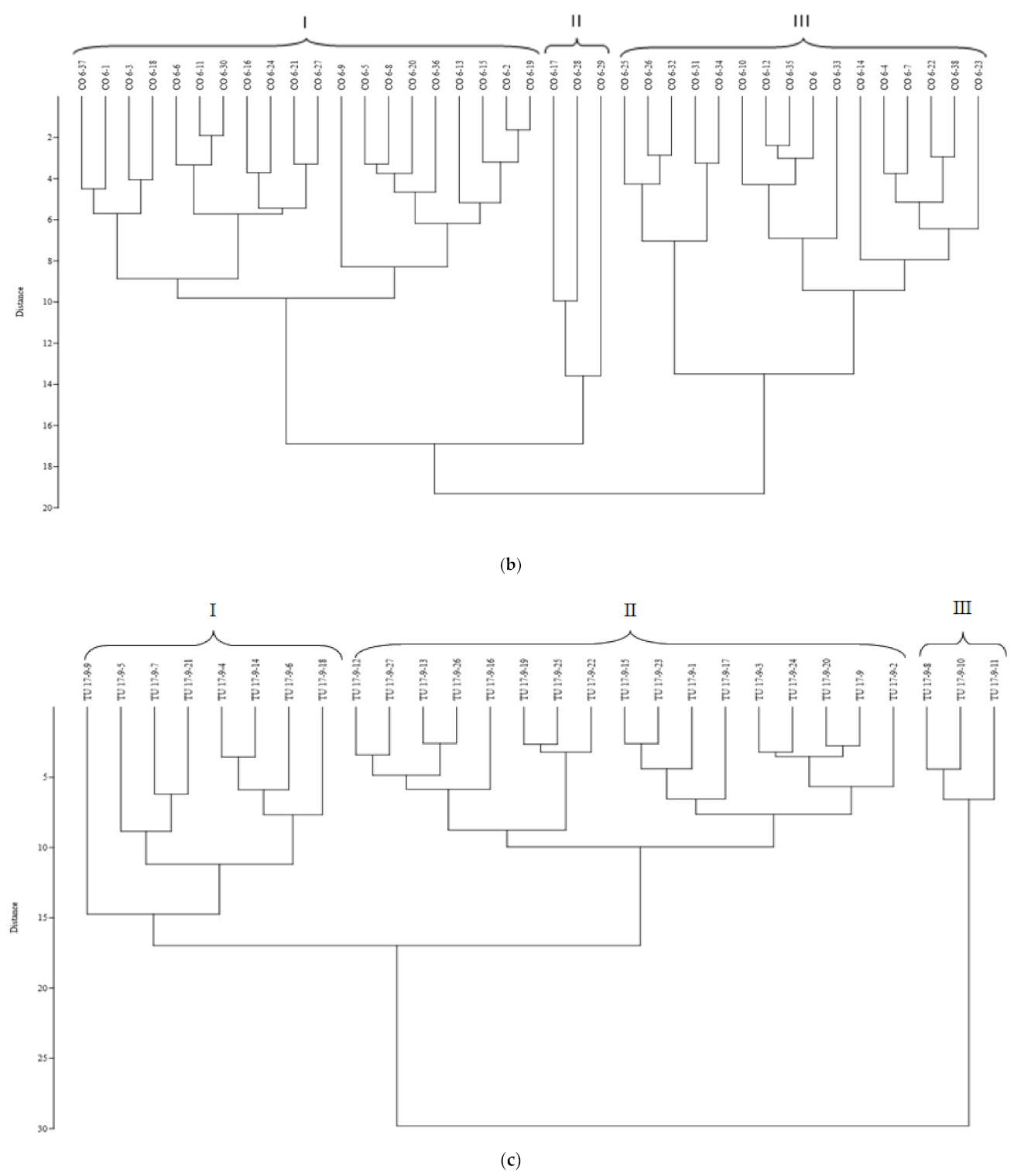
3.2. Assessing Genetic Heterogeneity on Yield-Related Traits in M3 Population
3.3. Assessing Genetic Parameters on Yield-Related Traits in the M4 Population
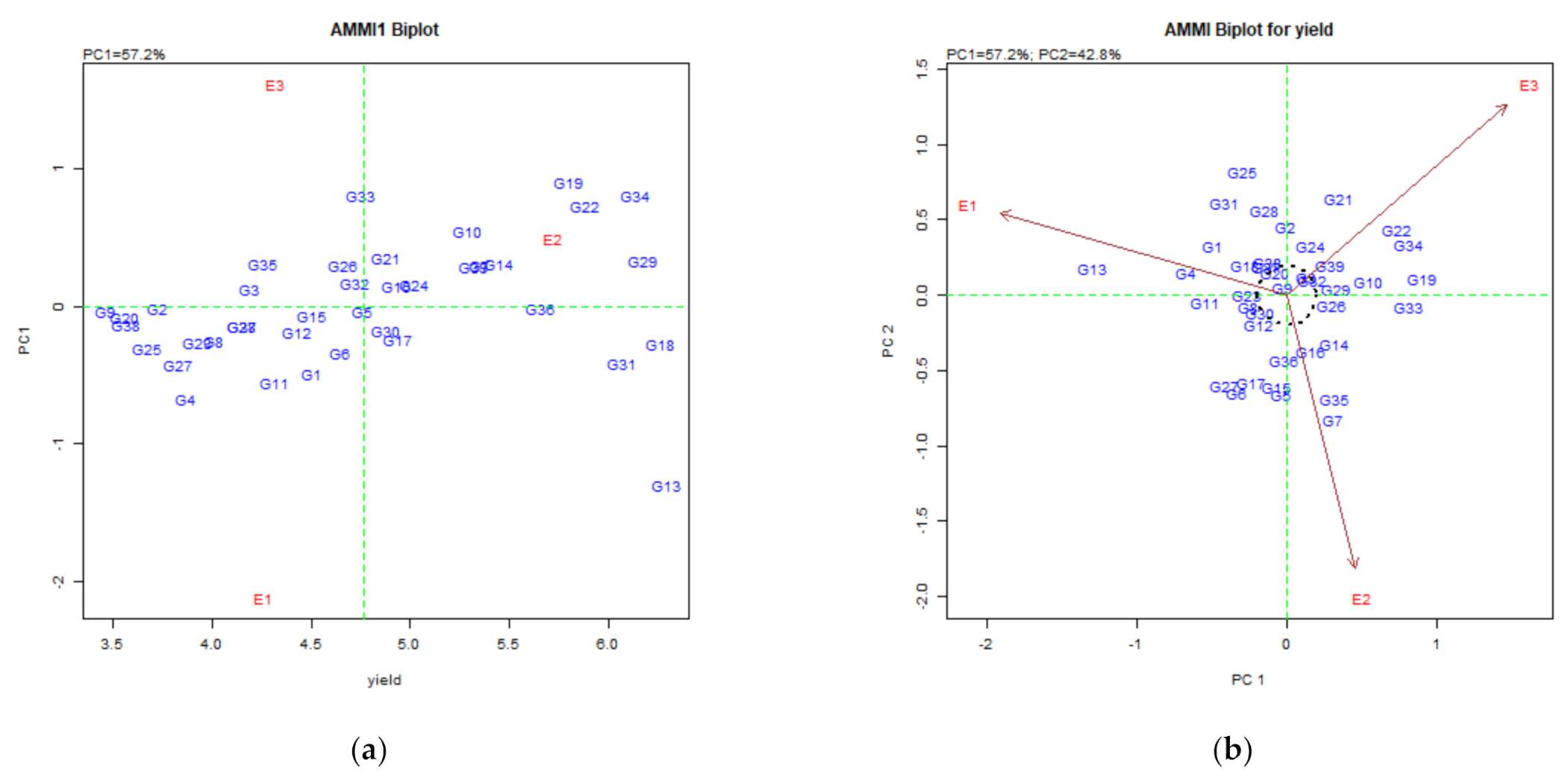
3.4. Assessing GEI and Stability of Mutant Lines for Grain Yield in M5 Population
3.4.1. ANOVA and per se Grain Yield Performance
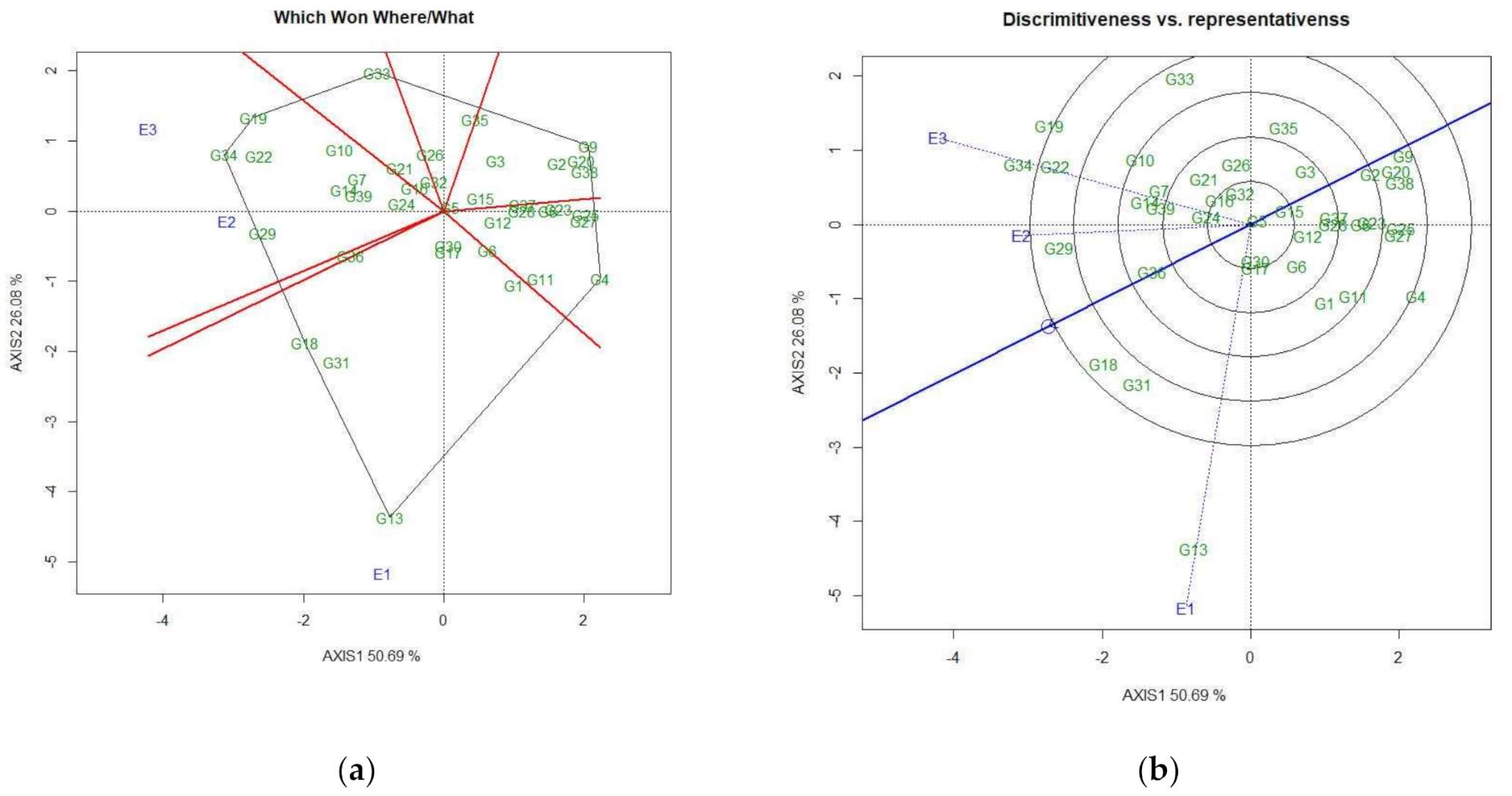
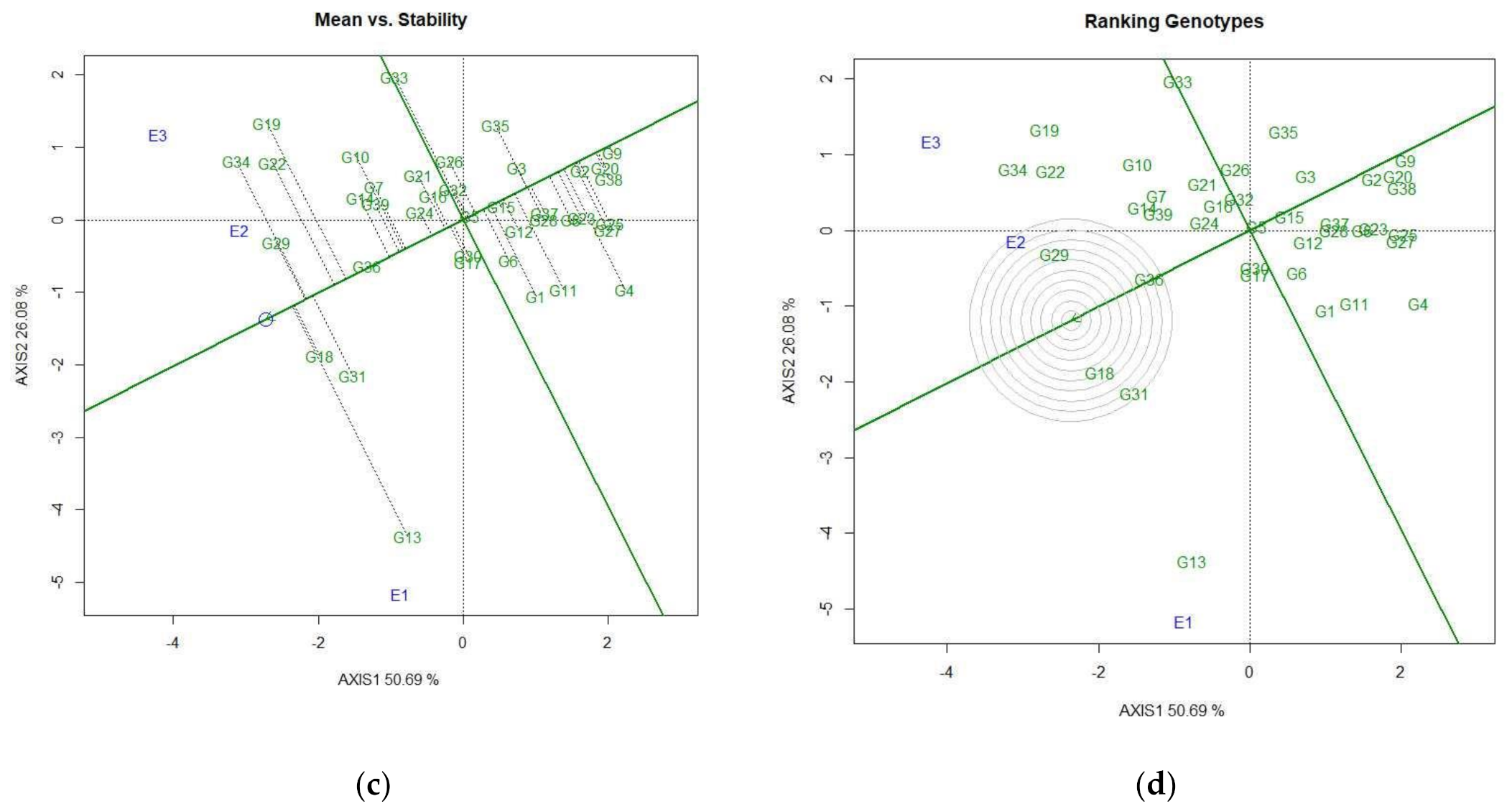
3.4.2. Identification of Genotypes for Favorable Environments
3.4.3. Identification of Genotypes for Broad-Spectrum Adaptability
3.4.4. Discovering Promising Genotypes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaewwongwal, A.; Kongjaimun, A.; Somta, P.; Chankaew, S.; Yimram, T.; Srinives, P. Genetic diversity of the black gram [Vigna mungo (L.) Hepper] gene pool as revealed by SSR markers. Breed. Sci. 2015, 65, 127–137. [Google Scholar] [CrossRef] [Green Version]
- Ozukum, C.; Sharma, M.B. Variability among urdbean (Vigna mungo L. Hepper) for yield and yield components. Int. J. Res. Innov. Appl. Sci. 2017, 2, 2454–6194. [Google Scholar]
- Government of India. Ministry of Agriculture and Farmers Welfare. Agricultural Statistics at a Glance; Directorate of Economics and Statistics, D.o.a., Cooperation and Farmers Welfare, Ed.; Controller of Publication: New Delhi, India, 2018.
- Somta, P.; Chen, J.; Yundaeng, C.; Yuan, X.; Yimram, T.; Tomooka, N.; Chen, X. Development of an SNP-based high-density linkage map and QTL analysis for bruchid (Callosobruchus maculatus F.) resistance in black gram (Vigna mungo (L.) Hepper). Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Arumuganathan, K.; Earle, E. Nuclear DNA content of some important plant species. Plant Mol. Biol. Report. 1991, 9, 208–218. [Google Scholar] [CrossRef]
- Shu, Q.Y.; Lagoda, P.J.L. Mutation techniques for gene discovery and crop improvement. Mol. Plant Breed. 2007, 5, 193–195. [Google Scholar]
- Laskar, R.A.; Khan, S. Assessment on induced genetic variability and divergence in the mutagenized lentil populations of microsperma and macrosperma cultivars developed using physical and chemical mutagenesis. PLoS ONE 2017, 12, e0184598. [Google Scholar] [CrossRef] [Green Version]
- Tulmann Neto, A.; Ando, A.; Figueira, A.; Latado, R.; Dos Santos, P.; Correa, L.; Peres, L.; Hauagge, R.; Pulcinelli, C.; Ishiy, T. Genetic improvement of crops by mutation techniques in Brazil. Plant Mutat. Rep. 2011, 2, 24–37. [Google Scholar]
- Singh, V.; Ramkrishna, K.; Arya, R.K. Induced chemical mutagenesis in cowpea [Vigna unguiculata (L.) Walp.]. Indian J. Genet. Plant Breed 2006, 66, 312–315. [Google Scholar]
- Begum, S.N.; Islam, M.M. BINAMOOG-8: Green shiny colour mutant variety of mungbean and hope for the farmer. Bangladesh J. Nucl. Agric. 2013, 25, 11–18. [Google Scholar]
- Sonu, G.; Samiullah, K. Induced mutagenesis in urdbean (Vigna mungo L. Hepper): A review. Int. J. Bot. 2010, 6, 194–206. [Google Scholar]
- Horn, L.N.; Ghebrehiwot, H.M.; Shimelis, H.A. Selection of novel cowpea genotypes derived through gamma irradiation. Front. Plant Sci. 2016, 7, 262. [Google Scholar] [CrossRef] [Green Version]
- Singh, M.; Upadhyaya, H.D.; Bisht, I.S. (Eds.) Genetic and Genomic Resources of Grain Legume Improvement; Elsevier: London, UK, 2013. [Google Scholar]
- Morita, R.; Kusaba, M.; Iida, S.; Yamaguchi, H.; Nishio, T.; Nishimura, M. Molecular characterization of mutations induced by gamma irradiation in rice. Genes Genet. Syst. 2009, 84, 361–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cecchini, E.; Mulligan, B.J.; Covey, S.N.; Milner, J.J. Characterization of gamma irradiation-induced deletion mutations at a selectable locus in Arabidopsis. Mutat. Res. Fundam. Mol. Mech. Mutagen. 1998, 401, 199–206. [Google Scholar] [CrossRef]
- Tank, H.K.; Sharma, P. The Correlation and path coefficient analysis in blackgram [Vigna mungo (L.) Hepper]. Electron. J. Plant Breed. 2019, 10, 1333–1338. [Google Scholar] [CrossRef]
- Konda, C.; Salimath, P.; Mishra, M. Genotype and environment interaction for yield and its components in blackgram (Vigna mungo (L.) Hepper). Legume Res. Int. J. 2009, 32, 195–198. [Google Scholar]
- Viotto Del Conte, M.; Carneiro, P.C.S.; Vilela de Resende, M.D.; Lopes da Silva, F.; Peternelli, L.A. Overcoming collinearity in path analysis of soybean [Glycine max (L.) Merr.] grain oil content. PLoS ONE 2020, 15, e0233290. [Google Scholar]
- Salimi, S.; Moradi, S. Effect the correlation, regression and path analysis in soybean genotypes (Glycine Max, L.) under moisture and normal condition. Int. J. Agron. Plant Prod. 2012, 3, 447–454. [Google Scholar]
- Singh, B.; Das, A.; Parihar, A.; Bhagawati, B.; Singh, D.; Pathak, K.; Dwivedi, K.; Das, N.; Keshari, N.; Midha, R. Delineation of genotype-by-environment interactions for identification and validation of resistant genotypes in mungbean to root-knot nematode (Meloidogyne incognita) using GGE biplot. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, W.; Kang, M.S.; Ma, B.; Woods, S.; Cornelius, P.L. GGE biplot vs. AMMI analysis of genotype-by-environment data. Crop. Sci. 2007, 47, 643–653. [Google Scholar] [CrossRef]
- Revanappa, S.; Kamannavar, P.; Vijaykumar, A.; Ganajaxi, M.; Gajanan, D.; Arunkumar, B.; Salimath, P. Genotype × environment interaction and stability analysis for grain yield in blackgram (Vigna mungo L.). Legume Res. Int. J. 2012, 35, 56–58. [Google Scholar]
- Rita, N.; Kigwie, S.; Noren, K.; Brojen, N.; Pramesh, K. Stability analysis of high yielding varieties of black gram (Vigna mungo L. hepper). Electron. J. Plant Breed. 2016, 22, 1–9. [Google Scholar] [CrossRef]
- Poli, Y.; Balakrishnan, D.; Desiraju, S.; Panigrahy, M.; Voleti, S.R.; Mangrauthia, S.K.; Neelamraju, S. Genotype × environment interactions of Nagina22 rice mutants for yield traits under low phosphorus, water limited and normal irrigated conditions. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Oladosu, Y.; Rafii, M.Y.; Abdullah, N.; Magaji, U.; Miah, G.; Hussin, G.; Ramli, A. Genotype× environment interaction and stability analyses of yield and yield components of established and mutant rice genotypes tested in multiple locations in Malaysia. Acta Agric. Scand. Sect. B Soil Plant Sci. 2017, 67, 590–606. [Google Scholar] [CrossRef]
- Atta, B.M.; Shah, T.M.; Abbas, G.; Haq, M.A. Genotype × environment interaction for seed yield in kabuli chickpea (Cicer arietinum L.) genotypes developed through mutation breeding. Pak. J. Bot 2009, 41, 1883–1890. [Google Scholar]
- TNAU-CPGA. Tamil Nadu Agricultural University Crop Production Guide Agriculture. Available online: Agritech.tnau.ac.in/agriculture/CropProduction/Pulses/pulses_blackgram.html (accessed on 16 August 2012).
- IBPGR. Descriptor for Vigna mungo and V. Radiata (Revised); International Board for Plant Genetic Resources: Rome, Italy, 1985. [Google Scholar]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Zobel, R.W.; Wright, M.J.; Gauch, H.G., Jr. Statistical analysis of a yield trial. Agron. J. 1988, 80, 388–393. [Google Scholar] [CrossRef]
- Crossa, J. Statistical analyses of multilocation trials. In Advances in Agronomy; Elsevier: London, UK, 1990; Volume 44, pp. 55–85. [Google Scholar]
- Yan, W.; Tinker, N.A. Biplot analysis of multi-environment trial data: Principles and applications. Can. J. Plant Sci. 2006, 86, 623–645. [Google Scholar] [CrossRef] [Green Version]
- Pacheco, A.; Vargas, M.; Alvarado, G.; Rodríguez, F.; Crossa, J.; Burgueño, J. GEA-R (genotype × environment analysis with R for windows) version 4.0. HDL 2015, 11529, V16. [Google Scholar]
- Singh, R.; Kole, C. Delineation of EMS induced genetic variability in some agronomic traits in mungbean (Vigna radiata L. Wilczek). Crop. Res. 2006, 32, 94–96. [Google Scholar]
- Deepalakshmi, A.; Kumar, C. Creation of genetic variability for different polygenic traits in blackgram {Vigna mungo (L.) Hepper} through induced mutagenesis. Legume Res. Int. J. 2004, 27, 188–192. [Google Scholar]
- Wani, M.R. Estimates of genetic variability in mutated populations and the scope of selection for yield attributes in Vigna radiata (L.) Wilczek. Egypt. J. Biol. 2006, 8. [Google Scholar]
- Al-Ghzawi, A.L.A.; Bsoul, E.; Aukour, F.; Al-Ajlouni, Z.; Al-Azzam, M.; Ajlouni, M.M. Genetic variation for quantitative traits in Jordanian lentil landraces. Adv. Environ. Bio. 2011, 5, 3676–3681. [Google Scholar]
- Amin, R.; Laskar, R.A.; Khan, S. Assessment of genetic response and character association for yield and yield components in Lentil (Lens culinaris L.) population developed through chemical mutagenesis. Cogent Food Agric. 2015, 1, 1000715. [Google Scholar] [CrossRef]
- Patial, M.; Thakur, S.; Singh, K. Comparative mutagenic effectiveness and efficiency of physical and chemical mutagen and induced variability in ricebean (Vigna umbellata Thunb, Ohwi and Ohashi). Legume Res. Int. J. 2015, 38, 30–36. [Google Scholar] [CrossRef]
- Dhole, V.; Reddy, K. Genetic analysis and variability studies in mutants induced through electron beam and gamma rays in mungbean (Vigna radiata L. Wilczek). Electron. J. Plant Breed. 2018, 9, 304–312. [Google Scholar] [CrossRef]
- Diouf, M.; Diallo, S.; Badiane, F.A.; Diack, O.; Diouf, D. Development of new cowpea (Vigna unguiculata) mutant genotypes, analysis of their agromorphological variation, genetic diversity and population structure. bioRxiv 2020. [Google Scholar] [CrossRef]
- Senapati, N.; Misra, R.C. Genetic divergence and selection indices among the micromutant lines in blackgram [Vigna mungo (L.) Hepper]. J. Crop Sci. Biotech. 2009, 12, 69–72. [Google Scholar] [CrossRef]
- Kuralarasan, V.; Vanniarajan, C.; Kanchana, S.; Veni, K.; Lavanya, S.A. Genetic divergence, heritability and genetic advance in mutant lines of urdbean [Vigna mungo (L.) Hepper]. Legume Res. Int. J. 2018, 41, 833–836. [Google Scholar]
- Das, T.R.; Baisakh, B. Genetic divergence among mutant genotypes of greengram [Vigna radiata (L.) Wilczek]. Electron. J. Plant Breed. 2019, 10, 160–166. [Google Scholar] [CrossRef]
- Gnankambary, K.; Sawadogo, N.; Diéni, Z.; Batieno, T.B.J.; Tignegré, J.B.D.S.; Sawadogo, M.; Ouédraogo, T.J. Assessment of Cowpea (Vigna unguiculata (L.) Walp.) mutant lines for drought tolerance. Int. J. Agron. 2020, 2020. [Google Scholar] [CrossRef]
- Afuape, S.; Okocha, P.; Njoku, D. Multivariate assessment of the agromorphological variability and yield components among sweet potato (Ipomoea batatas (L.) Lam) landraces. Afr. J. Plant Sci. 2011, 5, 123–132. [Google Scholar]
- Munaro, L.B.; Benin, G.; Marchioro, V.S.; de Assis Franco, F.; Silva, R.R.; da Silva, C.L.; Beche, E. Brazilian spring wheat homogeneous adaptation regions can be dissected in major megaenvironments. Crop. Sci. 2014, 54, 1374–1383. [Google Scholar] [CrossRef]
- Alam, M.A.; Sarker, Z.I.; Farhad, M.; Hakim, M.A.; Barma, N.C.D.; Hossain, M.I.; Rahman, M.M.; Islam, R. Yield stability of newly released wheat varieties in multi-environments of Bangladesh. Int. J. Plant Soil Sci. 2015, 6, 150–161. [Google Scholar] [CrossRef]
- Tonk, F.A.; Ilker, E.; Tosun, M. Evaluation of genotype × environment interactions in maize hybrids using GGE biplot analysis. Crop. Breed. Appl. Biotech. 2011, 11, 01–09. [Google Scholar] [CrossRef] [Green Version]
- Vaezi, B.; Pour-Aboughadareh, A.; Mohammadi, R.; Armion, M.; Mehraban, A.; Hossein-Pour, T.; Dorii, M. GGE biplot and AMMI analysis of barley yield performance in Iran. Cereal Res. Commun. 2017, 45, 500–511. [Google Scholar] [CrossRef] [Green Version]
- Horn, L.; Shimelis, H.; Sarsu, F.; Mwadzingeni, L.; Laing, M.D. Genotype-by-environment interaction for grain yield among novel cowpea (Vigna unguiculata L.) selections derived by gamma irradiation. Crop J. 2018, 6, 306–313. [Google Scholar] [CrossRef]
- Baraki, F.; Gebregergis, Z.; Belay, Y.; Berhe, M.; Zibelo, H. Genotype × environment interaction and yield stability analysis of mung bean (Vigna radiata (L.) Wilczek) genotypes in Northern Ethiopia. Cogent Food Agric. 2020, 6, 1729581. [Google Scholar] [CrossRef]
- Samyuktha, S.; Malarvizhi, D.; Karthikeyan, A.; Dhasarathan, M.; Hemavathy, A.; Vanniarajan, C.; Sheela, V.; Hepziba, S.; Pandiyan, M.; Senthil, N. Delineation of genotype × environment interaction for identification of stable genotypes to grain yield in mungbean. Front. Agron. 2020, 2. [Google Scholar] [CrossRef]
- Kendal, E.; Sener, O. Examination of genotype × environment interactions by GGE biplot analysis in spring durum wheat. Indian J. Genet. 2015, 75, 341–348. [Google Scholar] [CrossRef]


| cv. ADT 3 | cv. Co 6 | cv. TU 17-9 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Parameter | Mean | Min | Max | Mean | Min | Max | Mean | Min | Max |
| DF | 38.93 | 36.00 | 41.50 | 39.91 | 38.00 | 41.00 | 37.56 | 35.00 | 41.50 |
| PH | 22.37 | 14.75 | 32.84 | 26.86 | 18.00 | 36.50 | 22.12 | 14.25 | 32.00 |
| NB | 2.85 | 1.25 | 4.85 | 2.82 | 1.65 | 4.50 | 2.95 | 1.50 | 5.00 |
| NC | 11.88 | 6.00 | 22.25 | 13.31 | 4.75 | 24.25 | 12.06 | 5.50 | 20.25 |
| PC | 2.86 | 2.00 | 4.00 | 3.11 | 2.00 | 4.00 | 2.84 | 2.00 | 3.85 |
| NPP | 26.62 | 8.75 | 45.75 | 33.45 | 12.25 | 57.90 | 26.36 | 13.00 | 51.25 |
| PL | 4.89 | 4.03 | 5.79 | 4.85 | 4.00 | 5.45 | 4.78 | 4.09 | 5.35 |
| NS | 6.56 | 5.50 | 7.75 | 6.31 | 5.75 | 7.00 | 6.38 | 5.00 | 7.00 |
| SPY | 4.43 | 0.64 | 8.81 | 4.52 | 1.39 | 9.02 | 4.30 | 1.51 | 9.23 |
| Quantitative Traits | cv. ADT 3 | cv. Co 6 | cv. TU 17-9 |
|---|---|---|---|
| Grain Yield (g) | Grain Yield (g) | Grain Yield (g) | |
| Days to 50% flowering | −0.013 | 0.181 | 0.443 * |
| Plant height (cm) | 0.537 ** | 0.187 | 0.493 ** |
| Number of fertile branches | 0.234 | 0.564 ** | 0.513 ** |
| Number of clusters per plant | 0.648 ** | 0.813 ** | 0.767 ** |
| Number of pods per cluster | 0.682 ** | 0.530 ** | 0.740 ** |
| Number of pods per plant | 0.754 ** | 0.818 ** | 0.851 ** |
| Pod length (cm) | 0.096 | 0.084 | 0.197 |
| Number of seeds per pod | −0.174 | 0.085 | 0.141 |
| cv. ADT 3 Mutants | cv. Co 6 Mutants | cv. TU 17-9 Mutants | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Traits | PC 1 | PC 2 | PC 3 | PC 1 | PC 2 | PC 3 | PC 1 | PC 2 | PC 3 |
| Days to 50% flowering | 0.379 | −0.154 | 0.768 | 0.607 | −0.320 | −0.439 | 0.343 | −0.092 | 0.335 |
| Plant height (cm) | 0.879 | 0.047 | 0.230 | 0.934 | −0.095 | −0.170 | 0.444 | −0.078 | −0.024 |
| Number of branches per plant | 0.839 | 0.003 | −0.229 | 0.640 | 0.104 | 0.530 | 0.363 | −0.106 | −0.518 |
| Number of clusters per plant | 0.934 | −0.007 | 0.081 | 0.878 | −0.199 | −0.035 | 0.457 | −0.153 | −0.161 |
| Number of pods per cluster | −0.037 | 0.904 | −0.072 | 0.261 | 0.780 | −0.213 | 0.060 | 0.640 | −0.243 |
| Number of pods per plant | −0.127 | 0.648 | 0.534 | 0.235 | 0.782 | −0.144 | −0.010 | 0.681 | −0.068 |
| Pod length (cm) | 0.872 | 0.109 | −0.194 | 0.896 | −0.229 | 0.008 | 0.446 | 0.106 | −0.180 |
| Number of seeds per pod | −0.123 | 0.619 | −0.135 | 0.186 | 0.012 | 0.798 | 0.254 | 0.171 | 0.616 |
| Single plant yield (g) | 0.528 | 0.223 | −0.301 | 0.456 | 0.453 | 0.114 | 0.277 | 0.191 | 0.342 |
| Eigenvalue | 3.57 | 1.71 | 1.14 | 3.59 | 1.64 | 1.22 | 4.22 | 1.72 | 0.91 |
| % variance | 39.62 | 18.97 | 12.65 | 39.88 | 18.20 | 13.55 | 46.85 | 19.10 | 10.08 |
| % cumulative of variance | 39.62 | 58.59 | 71.24 | 39.88 | 58.08 | 71.63 | 46.85 | 65.95 | 76.03 |
| Sum of Squares | Degrees of Freedom | Mean sum of Squares | F-Value | Probability | Variation Explained (%) | |
|---|---|---|---|---|---|---|
| Environment | 107.55 | 2.00 | 53.77 | 29.98 | 0.00 | 23.58 |
| Genotype | 159.02 | 38.00 | 4.18 | 2.33 | 0.00 | 34.86 |
| Environment × Genotype | 189.57 | 76.00 | 2.49 | 1.39 | 0.05 | 41.56 |
| PC1 | 108.52 | 39.00 | 2.78 | 1.55 | 0.04 | 57.24 |
| PC2 | 81.05 | 37.00 | 2.19 | 1.22 | 0.21 | 42.76 |
| Residuals | 209.85 | 117.00 | 1.79 |
| S.No | Codes | Source | Aduthurai (E1) | Kattuthottam (E2) | Vamban (E3) | Mean | Gain in Selection (%) | PC 1 | PC 2 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | G1 | ADT 3 derived mutant | 5.23 | 4.56 | 3.71 | 4.50 | 8 | −0.38 | 0.25 |
| 2 | G2 | ADT 3 derived mutant | 3.52 | 3.77 | 3.88 | 3.72 | −10 | −0.01 | 0.35 |
| 3 | G3 | ADT 3 derived mutant | 3.49 | 4.97 | 4.10 | 4.19 | 1 | 0.09 | 0.09 |
| 4 | G4 | TU 17-9 derived mutant | 4.88 | 4.19 | 2.52 | 3.86 | −27 | −0.52 | 0.11 |
| 5 | G5 | Co 6 derived mutant | 3.94 | 7.02 | 3.32 | 4.76 | 33 | −0.03 | −0.51 |
| 6 | G6 | Co 6 derived mutant | 4.45 | 6.75 | 2.74 | 4.65 | 30 | −0.26 | −0.50 |
| 7 | G7 | ADT 3 derived mutant | 3.70 | 8.12 | 4.22 | 5.35 | 29 | 0.23 | −0.64 |
| 8 | G8 | Co 6 derived mutant | 3.99 | 4.99 | 3.04 | 4.01 | 12 | −0.20 | −0.06 |
| 9 | G9 | TU 17-9 derived mutant | 3.05 | 4.29 | 3.04 | 3.46 | −35 | −0.02 | 0.04 |
| 10 | G10 | ADT 3 derived mutant | 3.69 | 6.33 | 5.86 | 5.29 | 27 | 0.42 | 0.07 |
| 11 | G11 | ADT 3 derived mutant | 4.94 | 5.10 | 2.90 | 4.32 | 4 | −0.42 | −0.04 |
| 12 | G12 | ADT 3 derived mutant | 4.20 | 5.69 | 3.40 | 4.43 | 7 | −0.15 | −0.15 |
| 13 | G13 | ADT 3 derived mutant | 8.64 | 6.26 | 3.99 | 6.30 | 52 | −1.00 | 0.13 |
| 14 | G14 | ADT 3 derived mutant | 4.08 | 7.22 | 5.05 | 5.45 | 31 | 0.24 | −0.25 |
| 15 | G15 | ADT 3 derived mutant | 3.77 | 6.66 | 3.09 | 4.50 | 8 | −0.05 | −0.47 |
| 16 | G16 | ADT 3 derived mutant | 3.87 | 6.71 | 4.21 | 4.93 | 19 | 0.12 | −0.29 |
| 17 | G17 | ADT 3 derived mutant | 4.59 | 6.93 | 3.28 | 4.94 | 19 | −0.19 | −0.44 |
| 18 | G18 | ADT 3 derived mutant | 6.46 | 6.69 | 5.66 | 6.27 | 51 | −0.21 | 0.15 |
| 19 | G19 | ADT 3 derived mutant | 3.46 | 6.98 | 6.97 | 5.80 | 40 | 0.69 | 0.09 |
| 20 | G20 | ADT 3 derived mutant | 3.31 | 4.17 | 3.19 | 3.56 | −14 | −0.06 | 0.12 |
| 21 | G21 | ADT 3 derived mutant | 4.01 | 4.73 | 5.89 | 4.88 | 18 | 0.27 | 0.49 |
| 22 | G22 | ADT 3 derived mutant | 4.09 | 6.32 | 7.22 | 5.88 | 42 | 0.56 | 0.34 |
| 23 | G23 | ADT 3 derived mutant | 3.98 | 4.74 | 3.05 | 3.92 | −6 | −0.20 | 0.01 |
| 24 | G24 | ADT 3 derived mutant | 4.38 | 5.40 | 5.28 | 5.02 | 21 | 0.12 | 0.25 |
| 25 | G25 | TU 17-9 derived mutant | 4.29 | 2.83 | 3.88 | 3.67 | −31 | −0.23 | 0.63 |
| 26 | G26 | Co 6 derived mutant | 3.48 | 5.90 | 4.59 | 4.66 | 31 | 0.23 | −0.05 |
| 27 | G27 | Co 6 derived mutant | 3.85 | 5.78 | 1.86 | 3.83 | 7 | −0.32 | −0.46 |
| 28 | G28 | Co 6 derived mutant | 4.29 | 3.90 | 4.25 | 4.15 | 16 | −0.11 | 0.43 |
| 29 | G29 | Co 6 derived mutant | 5.00 | 7.22 | 6.32 | 6.18 | 73 | 0.25 | 0.03 |
| 30 | G30 | Co 6 derived mutant | 4.68 | 5.97 | 3.98 | 4.88 | 37 | −0.14 | −0.09 |
| 31 | G31 | TU 17-9 derived mutant | 6.81 | 5.58 | 5.81 | 6.07 | 14 | −0.32 | 0.47 |
| 32 | G32 | TU 17-9 derived mutant | 3.91 | 5.56 | 4.68 | 4.72 | −11 | 0.13 | 0.08 |
| 33 | G33 | TU 17-9 derived mutant | 2.49 | 6.25 | 5.50 | 4.75 | −11 | 0.62 | −0.06 |
| 34 | G34 | TU 17-9 derived mutant | 4.11 | 6.82 | 7.48 | 6.14 | 15 | 0.62 | 0.26 |
| 35 | G35 | ADT 3 derived mutant | 2.67 | 6.74 | 3.35 | 4.25 | 2 | 0.24 | −0.53 |
| 36 | G36 | ADT 3 derived mutant | 4.93 | 7.46 | 4.57 | 5.66 | 36 | −0.01 | −0.33 |
| Parents/checks | |||||||||
| 37 | G37 | ADT 3 | 4.05 | 4.66 | 3.73 | 4.15 | −0.11 | 0.14 | |
| 38 | G38 | Co 6 | 3.47 | 4.01 | 3.21 | 3.57 | −0.10 | 0.17 | |
| 39 | G39 | TU 17-9 | 4.32 | 6.02 | 5.62 | 5.32 | 0.22 | 0.15 | |
| Mean | 4.26 | 5.73 | 4.32 | 4.77 | |||||
| Range | 2.49–8.64 | 2.83–8.12 | 1.86–7.48 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhasarathan, M.; Geetha, S.; Karthikeyan, A.; Sassikumar, D.; Meenakshiganesan, N. Development of Novel Blackgram (Vigna mungo (L.) Hepper) Mutants and Deciphering Genotype × Environment Interaction for Yield-Related Traits of Mutants. Agronomy 2021, 11, 1287. https://doi.org/10.3390/agronomy11071287
Dhasarathan M, Geetha S, Karthikeyan A, Sassikumar D, Meenakshiganesan N. Development of Novel Blackgram (Vigna mungo (L.) Hepper) Mutants and Deciphering Genotype × Environment Interaction for Yield-Related Traits of Mutants. Agronomy. 2021; 11(7):1287. https://doi.org/10.3390/agronomy11071287
Chicago/Turabian StyleDhasarathan, Manickam, Seshadri Geetha, Adhimoolam Karthikeyan, Datchinamoorthy Sassikumar, and Narayanapillai Meenakshiganesan. 2021. "Development of Novel Blackgram (Vigna mungo (L.) Hepper) Mutants and Deciphering Genotype × Environment Interaction for Yield-Related Traits of Mutants" Agronomy 11, no. 7: 1287. https://doi.org/10.3390/agronomy11071287
APA StyleDhasarathan, M., Geetha, S., Karthikeyan, A., Sassikumar, D., & Meenakshiganesan, N. (2021). Development of Novel Blackgram (Vigna mungo (L.) Hepper) Mutants and Deciphering Genotype × Environment Interaction for Yield-Related Traits of Mutants. Agronomy, 11(7), 1287. https://doi.org/10.3390/agronomy11071287







