Characterization of Different Arundo donax L. Clones from the Mediterranean Region
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Measurements
2.3. Statistical Analysis
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lewandowski, I.; Scurlock, J.M.; Lindvall, E.; Christou, M. The development and current status of perennial rhizomatous grasses as energy crops in the US and Europe. Biomass Bioenergy 2003, 25, 335–361. [Google Scholar] [CrossRef]
- Sánchez, E.; Scordia, D.; Lino, G.; Arias, C.; Cosentino, S.L.; Nogués, S. Salinity and Water Stress Effects on Biomass Production in Different Arundo donax L. Clones. BioEnergy Res. 2015, 8, 1461–1479. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, E.; Gil, S.; Azcón-Bieto, J.; Nogués, S. The response of Arundo donax L. (C 3) and Panicum virgatum (C 4) to different stresses. Biomass Bioenergy 2016, 85, 335–345. [Google Scholar] [CrossRef]
- Sánchez, E.; Lino, G.; Arias, C.; Serrat, X.; Nogués, S. Photosynthesis, resource acquisition and growth responses of two biomass crops subjected to water stress. J. Plant. Sci. 2018, 6, 68–86. [Google Scholar]
- De Stefano, R.; Cappetta, E.; Guida, G.; Mistretta, C.; Caruso, G.; Giorio, P.; Albrizio, R.; Tucci, M. Screening of giant reed (Arundo donax L.) ecotypes for biomass production under salt stress. Plant. Biosyst. Int. J. Deal. Asp. Plant. Biol. 2017, 152, 911–917. [Google Scholar] [CrossRef]
- Webster, R.J.; Driever, S.; Kromdijk, J.; McGrath, J.; Leakey, A.; Siebke, K.; Demetriades-Shah, T.; Bonnage, S.; Peloe, T.; Lawson, T.; et al. High C3 photosynthetic capacity and high intrinsic water use efficiency underlies the high productivity of the bioenergy grass Arundo donax. Sci. Rep. 2016, 6, 20694. [Google Scholar] [CrossRef] [PubMed]
- Boland, J.M. The Importance of Layering in the Rapid Spread of Arundo donax (Giant Reed). Madroño 2006, 53, 303–312. [Google Scholar] [CrossRef]
- Hunter, A.W.S. A Karyo-Systematic Investigation in the Gramineae. Can. J. Res. 1934, 11, 213–241. [Google Scholar] [CrossRef] [Green Version]
- Bucci, A.; Cassani, E.; Landoni, M.; Cantaluppi, E.; Pilu, R. Analysis of chromosome number and speculations on the origin of Arundo donax L. (Giant Reed). Cytol. Genet. 2013, 47, 237–241. [Google Scholar] [CrossRef] [Green Version]
- Hoagland, D.R.; Arnon, D.I. The Water-Culture Method for Growing Plants without Soil, 2nd ed.; Circular. California Agricultural Experiment Station; University of California: Berkeley, CA, USA, 1950; p. 32. [Google Scholar]
- Haddadchi, A.; Gross, C.; Fatemi, M. The expansion of sterile Arundo donax (Poaceae) in southeastern Australia is accompanied by genotypic variation. Aquat. Bot. 2013, 104, 153–161. [Google Scholar] [CrossRef]
- Vittoria, A. Natura e neutralizzazione dell’avvelenamento colchicinico in Graminacee con riferimenti alle fitotossico-si, fisiologiche e patologiche. Acta Med. Vet. 1958, 4, 1–108. [Google Scholar]
- Mariani, C.; Cabrini, R.; Danin, A.; Piffanelli, P.; Fricano, A.; Gomarasca, S.; DiCandilo, M.; Grassi, F.; Soave, C. Origin, diffusion and reproduction of the giant reed (Arundo donax L.): A promising weedy energy crop. Ann. Appl. Biol. 2010, 157, 191–202. [Google Scholar] [CrossRef]
- Avdulov, N. Karyo-Systematische Untersuchung der Familie Gramineem. Bull. Appl. Bot. Genet. Plant. Breed. 1931, 44, 428. [Google Scholar]
- Christopher, I.; Abraham, A. Studies on the cytology and phylogeny of South Indian grasses. I. Bambu soideae, Oryzoideae, Arundinoideae and Festucoideae. Cytologia 1971, 36, 579–594. [Google Scholar] [CrossRef] [Green Version]
- Nackley, L.L.; Vogt, K.A.; Kim, S.-H. Arundo donax water use and photosynthetic responses to drought and elevated CO2. Agric. Water Manag. 2014, 136, 13–22. [Google Scholar] [CrossRef]
- Haworth, M.; Cosentino, S.L.; Marino, G.; Brunetti, C.; Scordia, D.; Testa, G.; Riggi, E.; Avola, G.; Loreto, F.; Centritto, M. Physiological responses of Arundo donaxecotypes to drought: A common garden study. GCB Bioenergy 2017, 9, 132–143. [Google Scholar] [CrossRef] [Green Version]
- Wullschleger, S. Biochemical Limitations to Carbon Assimilation in C3 Plants—A Retrospective Analysis of theA/CiCurves from 109 Species. J. Exp. Bot. 1993, 44, 907–920. [Google Scholar] [CrossRef]
- Cosentino, S.L.; Copani, V.; D’Agosta, G.M.; Sanzone, E.; Mantineo, M. First results on evaluation of Arundo donax L. clones collected in Southern Italy. Ind. Crop. Prod. 2006, 23, 212–222. [Google Scholar] [CrossRef]
- Amaducci, S.; Perego, A. Field evaluation of Arundo donax clones for bioenergy production. Ind. Crop. Prod. 2015, 75, 122–128. [Google Scholar] [CrossRef]
- Hardion, L.; Verlaque, R.; Saltonstall, K.; Leriche, A.; Vila, B. Origin of the invasive Arundo donax (Poaceae): A Trans-Asian Expedition in Herbaria. Ann. Bot. 2014, 114, 455–462. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, R.; Liow, P.-S.; Spencer, D.F.; Jasieniuk, M. Molecular evidence for a single genetic clone of invasive Arundo donax in the United States. Aquat. Bot. 2008, 88, 113–120. [Google Scholar] [CrossRef]
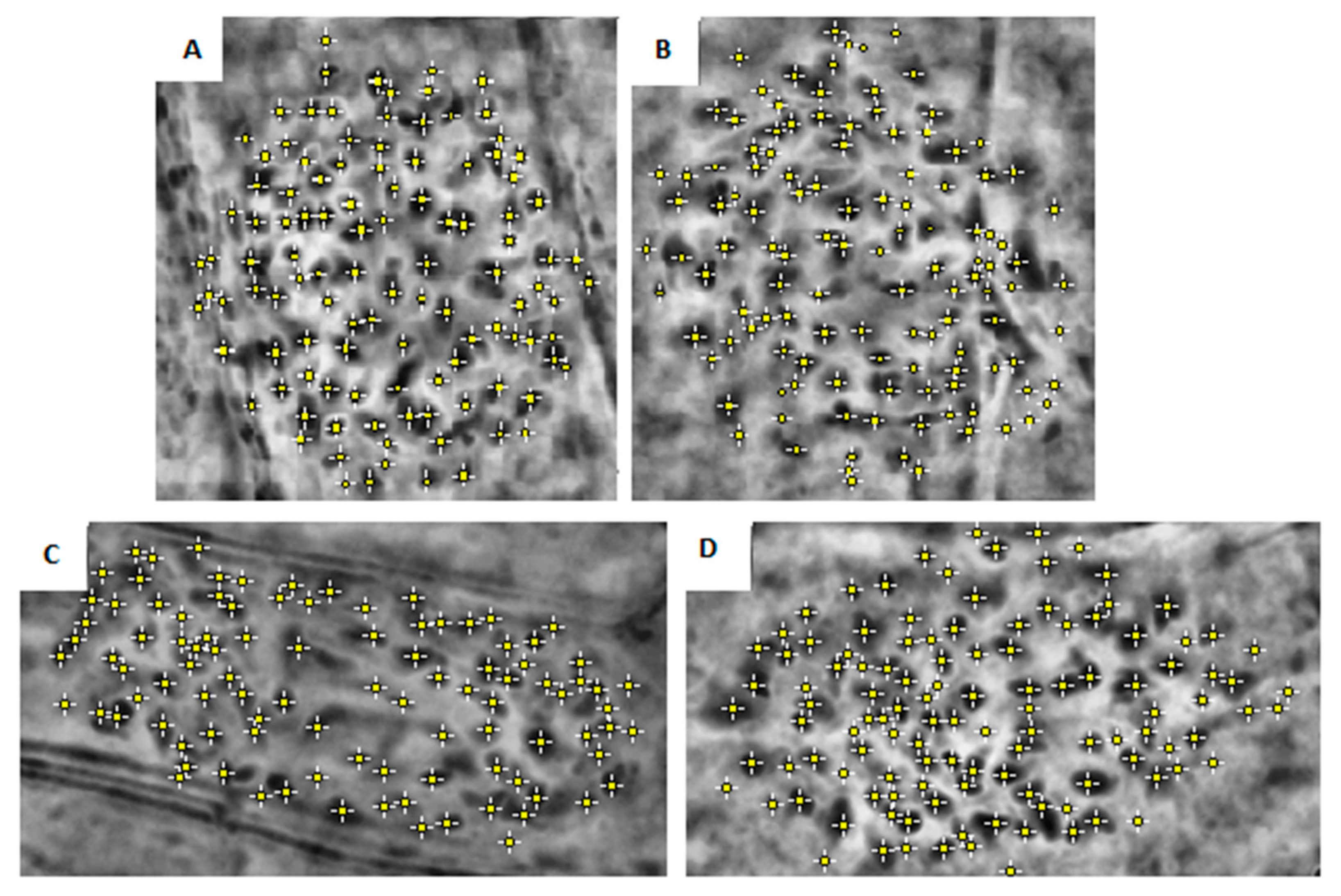
| Clone | Chromosome Numbers Mean ± SE | ||
|---|---|---|---|
| Fondachello | 122 | ±1.51 | a |
| Martinensis | 116 | ±2.05 | b |
| Piccoplant | 110 | ±0.8 | c |
| Granadensis | 98 | ±10.88 | d |
| Piccoplant | Fondachello | Martinensis | Granadensis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Asat | 23.1 | ±0.3 | n.s | 22.1 | ±0.5 | n.s | 22.8 | ±0.9 | n.s | 26.2 | ±2.3 | n.s |
| gs | 0.30 | ±0.04 | n.s | 0.35 | ±0.01 | n.s | 0.35 | ±0.05 | n.s | 0.29 | ±0.05 | n.s |
| Vcmax | 113.2 | ±3.1 | n.s | 108.2 | ±5.3 | n.s | 106.7 | ±1.5 | n.s | 120.8 | ±3.8 | n.s |
| Jmax | 243.3 | ±6.0 | n.s | 229.5 | ±7.5 | n.s | 226.9 | ±24.7 | n.s | 261.0 | ±13.5 | n.s |
| T | 5.46 | ±0.79 | n.s | 5.99 | ±0.67 | n.s | 8.43 | ±0.94 | n.s | 6.64 | ±1.02 | n.s |
| WUEi | 4.43 | ±0.64 | n.s | 2.76 | ±0.28 | n.s | 3.79 | ±0.49 | n.s | 4.02 | ±0.28 | n.s |
| Fv/Fm | 0.795 | ±0.002 | a | 0.781 | ±0.007 | ab | 0.772 | ±0.001 | b | 0.784 | ±0.002 | ab |
| Fv’/Fm’ | 0.497 | ±0.004 | n.s | 0.505 | ±0.008 | n.s | 0.501 | ±0.003 | n.s | 0.518 | ±0.017 | n.s |
| NPQ | 1.507 | ±0.015 | n.s | 1.372 | ±0.108 | n.s | 1.397 | ±0.018 | n.s | 1.337 | ±0.096 | n.s |
| RWC | 97.64 | ±1.05 | ab | 98.48 | ±0.76 | a | 97.73 | ±0.84 | Ab | 93.40 | ±1.62 | b |
| SPAD | 44.5 | ±0.9 | b | 43.8 | ±0.7 | b | 46.9 | ±0.3 | Ab | 48.6 | ±0.7 | a |
| Piccoplant | Fondachello | Martinensis | Granadensis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H | 75.8 | ±7.2 | n.s. | 61.3 | ±10.4 | n.s. | 57.3 | ±3.6 | n.s. | 47.1 | ±4.9 | n.s. |
| LA | 0.24 | ±0.03 | a | 0.06 | ±0.02 | b | 0.14 | ±0.02 | ab | 0.20 | ±0.02 | a |
| SDW | 26.8 | ±3.4 | n.s. | 16.4 | ±2.5 | n.s. | 16.8 | ±3.6 | n.s. | 17.8 | ±6.1 | n.s. |
| S/R | 4.3 | ±0.2 | n.s. | 4.6 | ±0.5 | n.s. | 3.3 | ±0.3 | n.s. | 4.1 | ±0.8 | n.s. |
| LAI | 21.2 | ±2.6 | a | 5.7 | ±1.6 | b | 12.7 | ±2.1 | ab | 15.7 | ±3.4 | ab |
| LMA | 46.0 | ±1.2 | n.s. | 39.1 | ±8.1 | n.s. | 51.4 | ±5.1 | n.s. | 49.0 | ±5.3 | n.s. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez, E.; Lino, G.; Serrat, X.; Nogués, S. Characterization of Different Arundo donax L. Clones from the Mediterranean Region. Agronomy 2021, 11, 1347. https://doi.org/10.3390/agronomy11071347
Sánchez E, Lino G, Serrat X, Nogués S. Characterization of Different Arundo donax L. Clones from the Mediterranean Region. Agronomy. 2021; 11(7):1347. https://doi.org/10.3390/agronomy11071347
Chicago/Turabian StyleSánchez, Elena, Gladys Lino, Xavier Serrat, and Salvador Nogués. 2021. "Characterization of Different Arundo donax L. Clones from the Mediterranean Region" Agronomy 11, no. 7: 1347. https://doi.org/10.3390/agronomy11071347
APA StyleSánchez, E., Lino, G., Serrat, X., & Nogués, S. (2021). Characterization of Different Arundo donax L. Clones from the Mediterranean Region. Agronomy, 11(7), 1347. https://doi.org/10.3390/agronomy11071347






