Identification and Characterization of SPL Transcription Factor Family Reveals Organization and Chilling-Responsive Patterns in Cabbage (Brassica oleracea var. capitata L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of BoSPL Genes in B. oleracea
2.2. Characterization and Phylogenetic Analysis of BoSPL Proteins
2.3. Chromosomal Distribution
2.4. Cis-Acting Element Analysis
2.5. Organs/Tissues Expression Analyses of BoSPLs
2.6. Chilling Stress, Sample Collection and RNA-Seq Analysis
3. Results
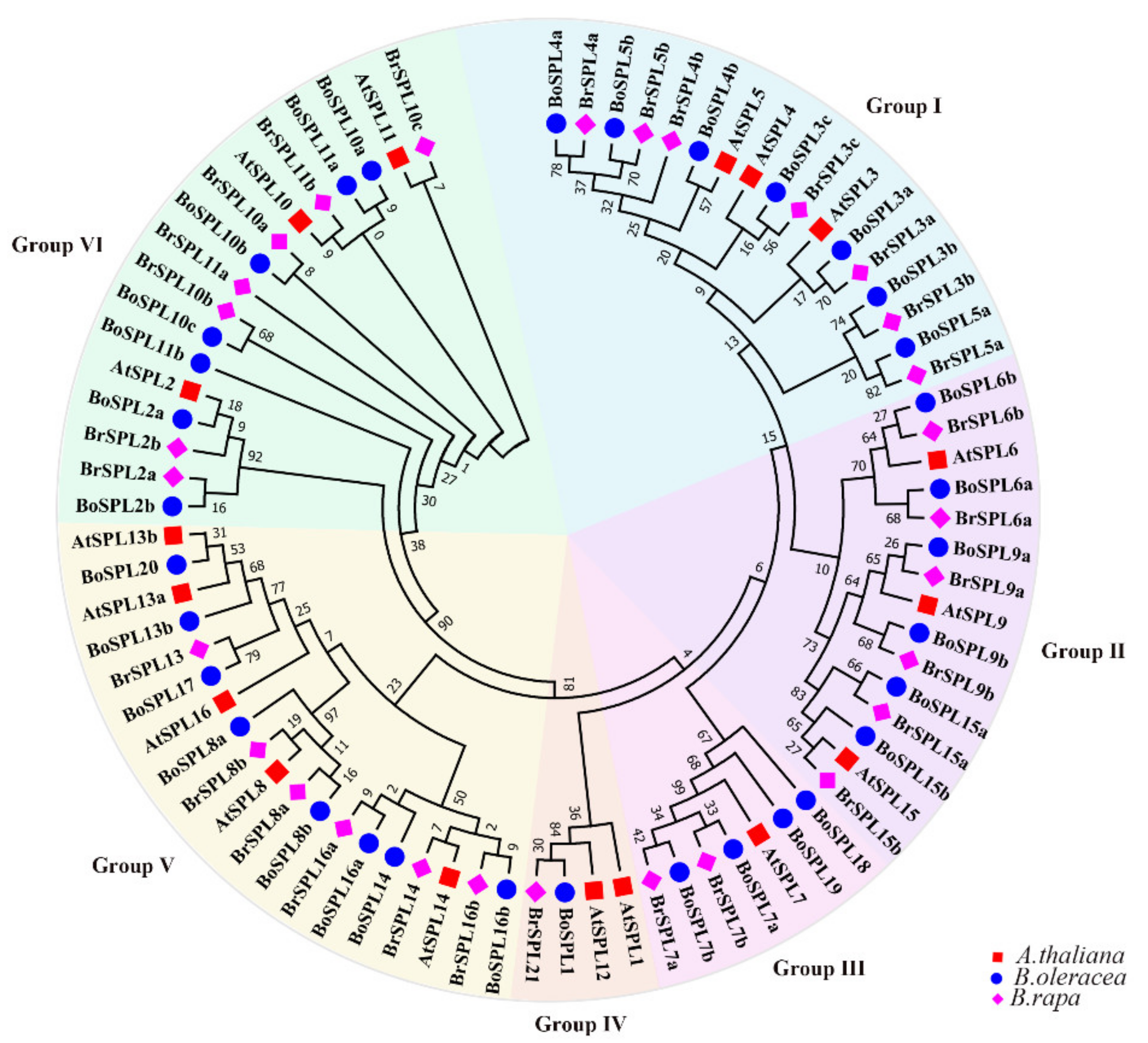
3.1. Identification and Phylogenetic Analysis of SPL Family Genes in B. oleracea
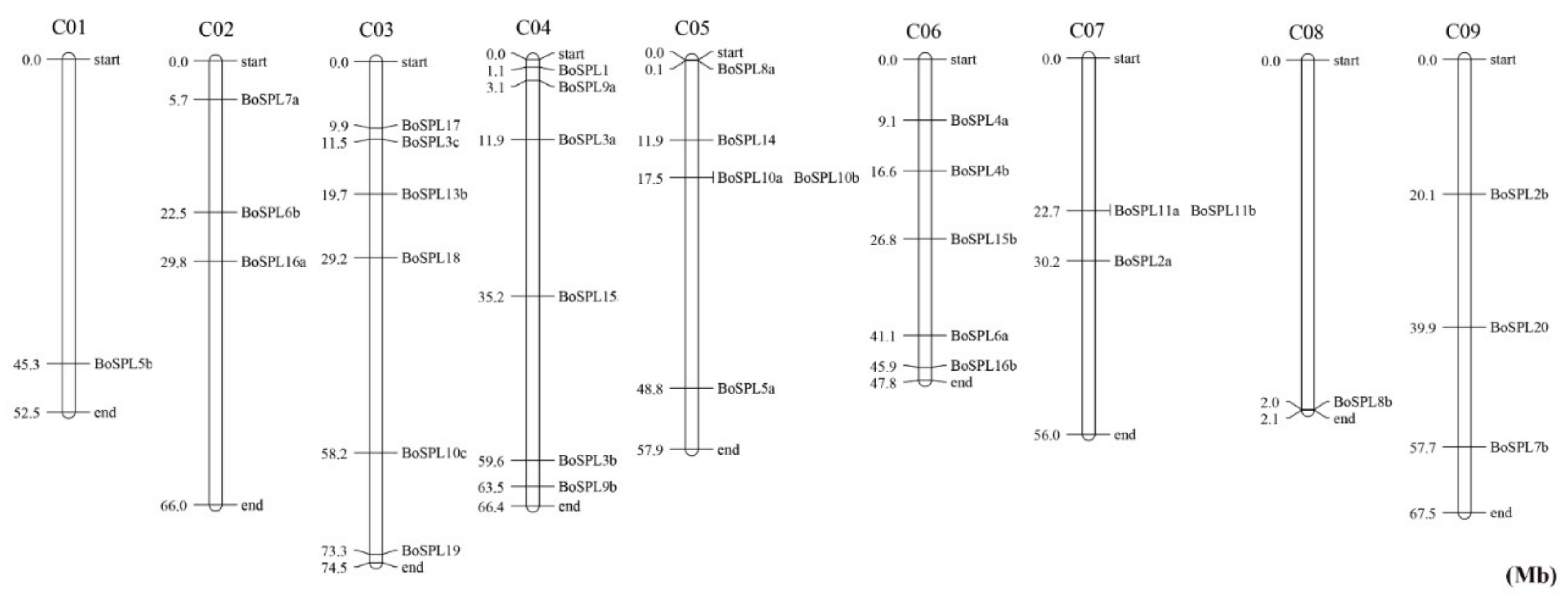
3.2. Chromosomal Distribution of BoSPL Genes
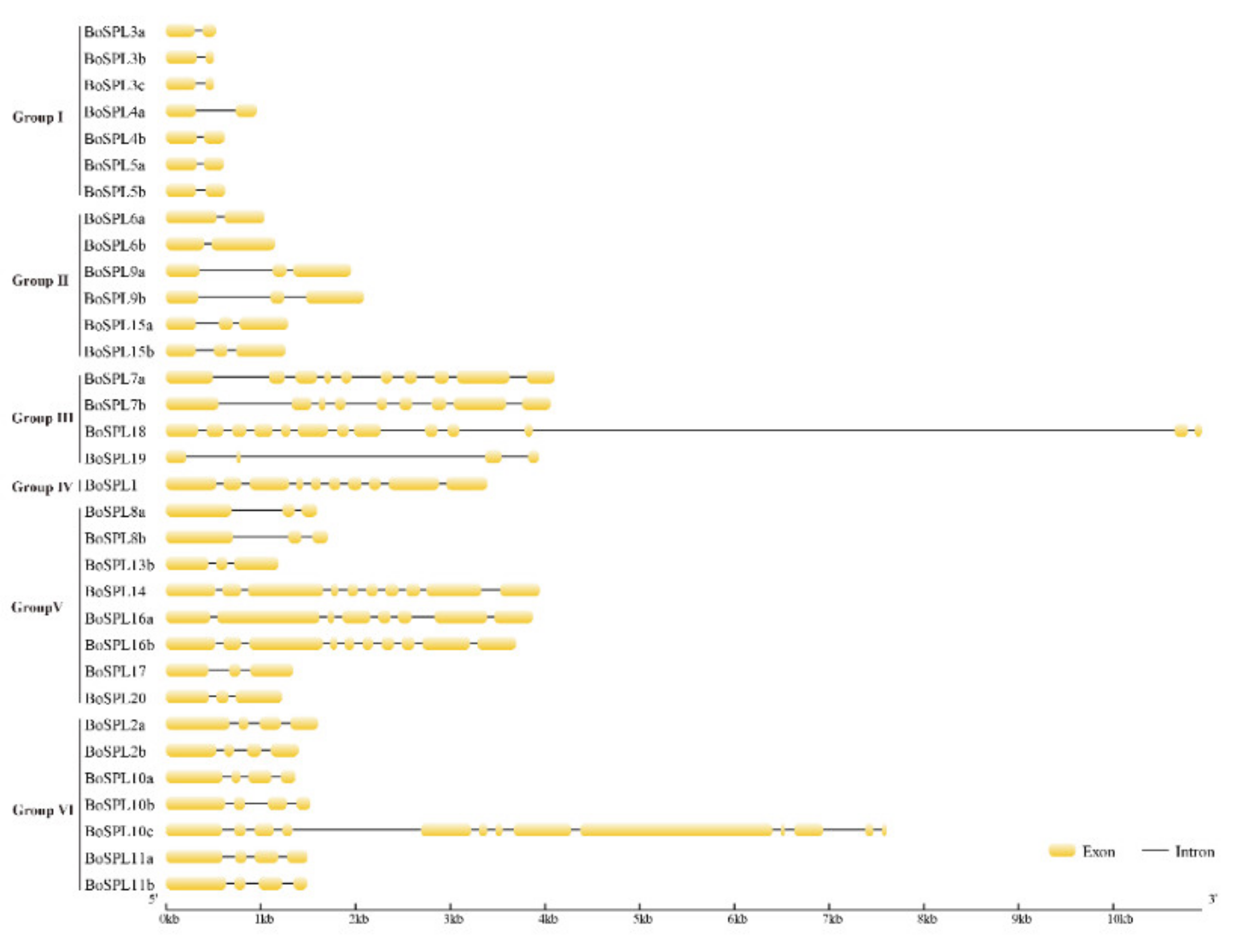
3.3. Gene Structure and Conserved Protein Motifs of BoSPL Genes
3.4. Analysis of Putative Promoter Regions in BoSPL Genes
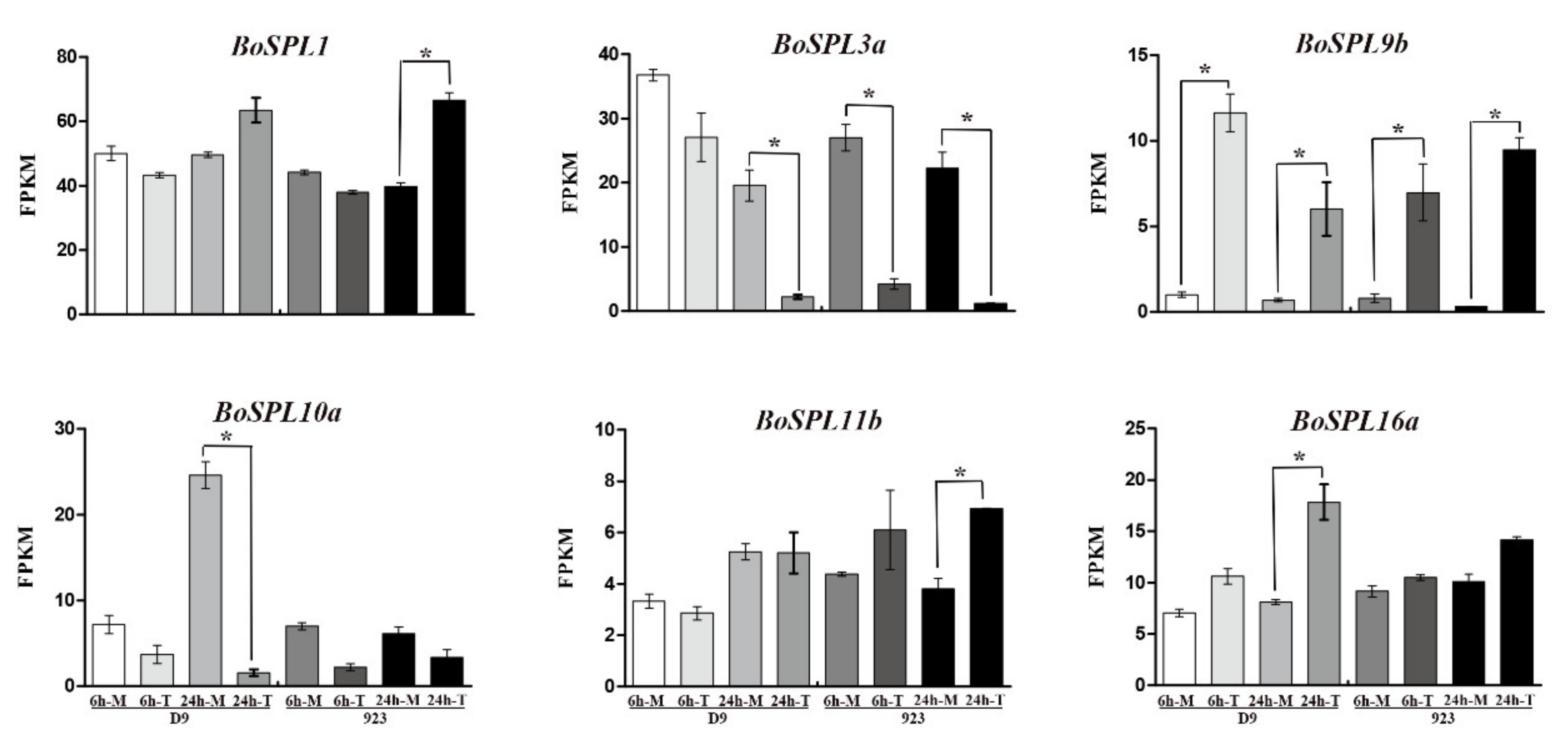
3.5. Expression Profile of BoSPLs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, X.; Zhang, Z.; Liu, D.; Kai, Z.; Li, A.; Long, M. SQUAMOSA promoter-binding protein-like transcription factors: Star players for plant growth and development. J. Integr. Plant Biol. 2010, 52, 946–951. [Google Scholar] [CrossRef] [PubMed]
- Gou, J.; Fu, C.; Liu, S.; Tang, C.; Debnath, S.; Flanagan, A.; Ge, Y.; Tang, Y.; Jiang, Q.; Larson, P.; et al. The miR156-SPL4 module predominantly regulates aerial axillary bud formation and controls shoot architecture. New Phytol. 2017, 216, 829–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gou, J.; Felippes, F.; Liu, C.; Weigel, D.; Wang, J. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 2011, 23, 1512–1522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, S.; Galvão, V.; Zhang, Y.; Horrer, D.; Zhang, T.; Hao, Y. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA promoter binding-like transcription factors. Plant Cell 2012, 24, 3320–3332. [Google Scholar] [CrossRef] [Green Version]
- Cui, L.; Shan, J.; Shi, M.; Gao, J.; Lin, H. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2014, 80, 1108–1117. [Google Scholar] [CrossRef]
- Klein, J.; Saedler, H.; Huijser, P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 1996, 250, 7–16. [Google Scholar]
- Cardon, G.; Höhmann, S.; Klein, J.; Nettesheim, K.; Saedler, H.; Huijser, P. Molecular characterisation of the Arabidopsis SBP-box genes. Gene 1999, 237, 91–104. [Google Scholar] [CrossRef]
- Xie, K.; Wu, C.; Xiong, L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding like transcription factors and microRNA156 in rice. Plant Physiol. 2006, 142, 280–293. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Wang, X.; Gu, S.; Hu, Z.; Xu, H.; Xu, C. Comparative study of SBP-box gene family in Arabidopsis and rice. Gene 2008, 407, 1–11. [Google Scholar] [CrossRef]
- Tan, H.; Song, X.; Duan, W.; Wang, Y.; Hou, X. Genome-wide analysis of the SBP-box gene family in Chinese cabbage (Brassica rapa ssp. pekinensis). Genome 2015, 58, 463–477. [Google Scholar] [CrossRef]
- Salinas, M.; Xing, S.; Höhmann, S.; Berndtgen, R.; Huijser, P. Genomic organization, phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato. Planta 2012, 235, 1171–1184. [Google Scholar] [CrossRef]
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Tateno, M.; Yamasaki, T.; Yabuki, T.; Aoki, M.; Seki, E.; Matsuda, T.; Nunokawa, E. A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J. Mol. Biol. 2004, 337, 49–63. [Google Scholar] [CrossRef]
- Matthew, W.; Brenda, J.; Lee, P. Prediction of plant microRNA targets. Cell 2002, 110, 513–520. [Google Scholar]
- Jeyakumar, J.; Ali, A.; Wang, W.M.; Thiruvengadam, M. Characterizing the Role of the miR156-SPL Network in Plant Development and Stress Response. Plants 2020, 9, 1206. [Google Scholar] [CrossRef] [PubMed]
- Stief, A.; Altmann, S.; Hoffmann, K.; Pant, B.D.; Scheible, W.R.; Bäurle, I. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 2014, 26, 1792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gandikota, M.; Birkenbihl, R.; Höhmann, S.; Cardon, G.; Saedler, H.; Huijser, P. The miRNA156/157 recognition element in the 3′UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 2007, 49, 683–693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, R.; Goel, R.; Kumari, S.; Kumari, S.; Dahuja, A. Genomic organization, phylogenetic comparison, and expression profiles of the SPL family genes and their regulation in soybean. Dev. Genes Evol. 2017, 227, 101–119. [Google Scholar] [CrossRef]
- Shao, Y.; Zhou, H.; Wu, Y.; Zhang, H.; Lin, J.; Jiang, X.; He, Q.; Zhu, J.; Li, Y.; Yu, H.; et al. OsSPL3, a SBP-domain protein, regulates crown root development in rice. Plant Cell 2019, 31, 1257–1275. [Google Scholar] [CrossRef] [Green Version]
- Cao, R.; Guo, L.; Ma, M.; Zhang, W.; Liu, X.; Zhao, H. Identification and Functional characterization of squamosa promoter bindingprotein-like Gene TaSPL16 in Wheat (Triticum aestivum L.). Front. Plant Sci. 2019, 10, 212. [Google Scholar] [CrossRef]
- Chao, L.; Liu, Y.; Chen, D. Arabidopsis transcription factors SPL1 and SPL12 confer plant thermotolerance at reproductive stage. Mol. Plant 2017, 10, 735–748. [Google Scholar] [CrossRef]
- Hou, H.; Li, J.; Gao, M. Genomic organization, phylogenetic comparison and differential expression of the SBP-box family genes in grape. PLoS ONE 2013, 8, e59358. [Google Scholar] [CrossRef]
- Ning, K.; Chen, S.; Huang, H. Molecular characterization and expression analysis of the SPL gene family with BpSPL9 transgenic lines found to confer tolerance to abiotic stress in Betula platyphylla Suk. Plant Cell Tiss. Organ Cult. 2017, 130, 469–481. [Google Scholar] [CrossRef]
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.J.; Lee, J.H.; Kim, W.; Jung, H.S.; Huijser, P.; Ahn, J.H. The microRNA156-squamosa promoter binding protein-like3 module regulates ambient temperature-responsive flowering via flowering locust in Arabidopsis. Plant Physiol. 2012, 159, 461–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergonzi, S.; Albani, M.C.; van Themaat, E.V.L.; Nordstrom, K.J.V.; Wang, R.H.; Schneeberger, K.; Coupland, G. Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science 2013, 340, 1094–1097. [Google Scholar] [CrossRef]
- Cai, X.; Wu, J.; Liang, J.; Lin, R.; Zhang, K.; Cheng, F.; Wang, X. Improved Brassica oleracea JZS assembly reveals significant changing of LTR-RT dynamics in different morphotypes. Theor. Appl. Genet. 2020, 133, 3187–3199. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, H.; Wang, J. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, F.; Wu, J.; Fang, L.; Wang, X. Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Front. Plant Sci. 2012, 3, 198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. BioRxiv 2018, 289660. [Google Scholar] [CrossRef]
- Lescot, M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Saeed, A.; Bhagabati, N.; Braisted, J.; Wei, L.; Sharov, V.; Howe, E. TM4 microarray software suite. Methods Enzymol. 2006, 411, 134–193. [Google Scholar]
- Le, D.; Nishiyama, R.; Watanabe, Y.; Vankova, R.; Tanaka, M.; Seki, M. Identification and expression analysis of cytokinin metabolic genes in soybean under normal and drought conditions in relation to cytokinin levels. PLoS ONE 2012, 7, e42411. [Google Scholar] [CrossRef] [Green Version]
- Walther, D.; Brunnemann, R.; Selbig, J. The regulatory code for transcriptional response diversity and its relation to genome structural properties in A. thaliana. PLoS Genet. 2007, 3, e11. [Google Scholar] [CrossRef]
- Pan, F.; Wang, Y.; Liu, H.; Wu, M.; Chu, W.; Chen, D. Genome-wide identification and expression analysis of SBP-like transcription factor genes in Moso Bamboo (Phyllostachys edulis). BMC Genom. 2017, 18, 486. [Google Scholar] [CrossRef] [PubMed]
- Ilhan, E. Genome-wide characterization and analysis of SBP transcription factor family in common bean (Phaseolus vulgaris L.). Appl. Ecol. Environ. Res. 2018, 16, 5467–5480. [Google Scholar] [CrossRef]
- Chen, G.; Li, J.; Liu, Y.; Zhang, Q.; Gao, Y.; Fang, K.; Cao, Q.; Qin, L.; Xing, Y. Roles of the GA-mediated SPL Gene Family and miR156 in the floral development of Chinese Chestnut (Castanea mollissima). Int. J. Mol. Sci. 2019, 20, 1577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Z.; Yu, H.; Xiong, G.; Wang, J.; Jiao, Y.; Liu, G.; Jing, Y.; Meng, X.; Hu, X.; Qian, Q.; et al. Genome wide binding analysis of the transcription activator IDEAL PLANT ARCHITECTURE1 reveals a complex network regulating rice plant architecture. Plant Cell 2013, 25, 3743–3759. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhou, L.; Shi, H.; Chern, M.; Yu, H.; Yi, H.; He, M.; He, M.; Yin, J.; Zhu, X.; et al. A single transcription factor promotes both yield and immunity in rice. Science 2018, 361, 1026–1028. [Google Scholar] [CrossRef] [Green Version]
- Cai, C.; Guo, W.; Zhang, B. Genome-wide identification and characterization of SPL transcription factor family and their evolution and expression profiling analysis in cotton. Sci. Rep. 2018, 8, 762. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, R.; Bregitzer, P.; Singh, J. Genome-wide analysis of the SPL/miR156 module and its interaction with the AP2/miR172 unit in barley. Sci. Rep. 2018, 8, 7085. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef] [Green Version]
- Moore, R.; Purugganan, M. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 15682–15687. [Google Scholar] [CrossRef] [Green Version]
- Preston, J.C.; Hileman, L.C. Functional evolution in the plant squamosa-promoter binding protein-like (spl) gene family. Front. Plant Sci. 2013, 4, 80. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Luo, J.; Chen, J.; Zhang, L.S.; Lim, J.K.; Wang, Z.J. Selection pressure causes differentiation of the SPL gene family in the Juglandaceae. Mol. Genet. Genom. 2019, 294, 1037–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaufmann, K.; Wellmer, F.; Muino, J.M.; Ferrier, T.; Wuest, S.; Kumar, V.; Serrano-Mislata, A.; Madueño, F.; Krajewski, P.; Meyerowitz, E.M.; et al. Orchestration of Floral Initiation by APETALA1. Science 2010, 328, 85–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Lin, E.; Huang, H.; Niu, M.; Tong, Z.; Zhang, J. Molecular characterization of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) gene family in Betula luminifera. Front. Plant Sci. 2018, 9, 608. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Jin, J.; He, Y.; Lu, B.; Li, D.; Chai, W. Genome-wide identification and analysis of the SBP-box family genes under Phytophthora capsici stress in pepper (Capsicum annuum L.). Front. Plant Sci. 2016, 7, 504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Park, J.J.; Kim, S.L.; Yim, J.; An, G. Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol. Biol. 2007, 65, 487–499. [Google Scholar] [CrossRef]
- Xie, J. Study on the Florescence Biology of Carya Illinoinensis. Master’s Thesis, Nanjing Forestry University, Nanjing, China, 2013. [Google Scholar]
- Stone, J.M.; Liang, X.; Nekl, E.R.; Stiers, J.J. Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J. 2005, 41, 744–754. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, S.; Grande, A.V.; Bujdoso, N.; Saedler, H.; Huijser, P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 2008, 67, 183–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyun, Y.; Vincent, C.; Tilmes, V.; Bergonzi, S.; Kiefer, C.; Richter, R.; Rafael, M.; Edouard, S.; George, C. A regulatory circuit conferring varied flowering response to cold in annual and perennial plants. Science 2019, 363, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Lian, H.; Wang, J. Plant developmental transitions: The role of microRNAs and sugars. Curr. Opin. Plant Biol. 2015, 27, 1–7. [Google Scholar] [CrossRef] [PubMed]



| Gene Name | PL (aa) | MW (kD) | pI | Instability Index | Aliphatic Index | GRAVY | Subcellular Localization Prediction |
|---|---|---|---|---|---|---|---|
| BoSPL1 | 888 | 98.89 | 5.80 | 52.76 | 78.52 | −0.44 | Nucleus |
| BoSPL2a | 429 | 47.73 | 7.99 | 57.43 | 56.15 | −0.70 | Nucleus |
| BoSPL2b | 361 | 40.55 | 8.83 | 51.95 | 59.97 | −0.72 | Nucleus |
| BoSPL3a | 147 | 17.01 | 7.05 | 105.69 | 34.63 | −1.38 | Nucleus |
| BoSPL3b | 141 | 16.48 | 6.25 | 108.70 | 31.21 | −1.52 | Nucleus |
| BoSPL3c | 135 | 15.77 | 8.17 | 111.81 | 28.96 | −1.52 | Nucleus |
| BoSPL4a | 179 | 20.42 | 9.59 | 51.65 | 49.55 | −1.19 | Nucleus |
| BoSPL4b | 183 | 21.10 | 9.26 | 73.77 | 46.28 | −1.24 | Nucleus |
| BoSPL5a | 179 | 20.71 | 9.45 | 67.67 | 50.17 | −1.17 | Nucleus |
| BoSPL5b | 176 | 20.48 | 9.60 | 52.39 | 44.94 | −1.22 | Nucleus |
| BoSPL6a | 318 | 36.03 | 8.33 | 60.86 | 57.04 | −0.81 | Nucleus |
| BoSPL6b | 358 | 40.58 | 8.84 | 60.63 | 59.61 | −0.70 | Nucleus |
| BoSPL7a | 781 | 87.38 | 6.71 | 50.64 | 78.35 | −0.41 | Nucleus |
| BoSPL7b | 731 | 81.55 | 6.00 | 51.84 | 78.89 | −0.31 | Nucleus |
| BoSPL8a | 328 | 36.54 | 8.85 | 56.37 | 48.17 | −0.81 | Nucleus |
| BoSPL8b | 335 | 37.18 | 9.01 | 55.77 | 51.31 | −0.77 | Nucleus |
| BoSPL9a | 370 | 40.39 | 8.62 | 55.79 | 45.84 | −0.86 | Nucleus |
| BoSPL9b | 365 | 40.14 | 7.67 | 60.33 | 48.60 | −0.84 | Nucleus |
| BoSPL10a | 366 | 40.71 | 8.89 | 56.22 | 50.82 | −0.82 | Nucleus |
| BoSPL10b | 364 | 40.72 | 9.01 | 48.61 | 58.87 | −0.71 | Nucleus |
| BoSPL10c | 1610 | 178.85 | 6.04 | 59.18 | 58.22 | −0.88 | Nucleus |
| BoSPL11a | 394 | 44.44 | 8.51 | 51.25 | 61.85 | −0.67 | Nucleus |
| BoSPL11b | 384 | 43.16 | 8.50 | 49.40 | 55.31 | −0.81 | Nucleus |
| BoSPL13b | 348 | 38.28 | 7.64 | 61.89 | 52.41 | −0.67 | Nucleus |
| BoSPL14 | 1030 | 113.98 | 8.73 | 62.81 | 74.03 | −0.50 | Nucleus |
| BoSPL15a | 324 | 36.13 | 9.14 | 55.22 | 57.50 | −0.64 | Nucleus |
| BoSPL15b | 325 | 36.47 | 9.19 | 55.99 | 57.63 | −0.72 | Nucleus |
| BoSPL16a | 1053 | 116.61 | 8.55 | 54.39 | 77.30 | −0.38 | Nucleus |
| BoSPL16b | 989 | 109.02 | 8.80 | 55.53 | 75.01 | −0.47 | Nucleus |
| BoSPL17 | 341 | 37.48 | 8.29 | 74.14 | 54.57 | −0.66 | Nucleus |
| BoSPL18 | 743 | 84.89 | 6.47 | 51.73 | 78.51 | −0.64 | Nucleus |
| BoSPL19 | 181 | 20.15 | 9.67 | 46.11 | 68.40 | −0.57 | Cytoplasm |
| BoSPL20 | 359 | 39.02 | 8.45 | 74.21 | 59.50 | −0.52 | Nucleus |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shan, X.; Zhang, W.; Huang, J.; Yu, F.; Qin, W.; Li, J.; Wang, S.; Dai, Z. Identification and Characterization of SPL Transcription Factor Family Reveals Organization and Chilling-Responsive Patterns in Cabbage (Brassica oleracea var. capitata L.). Agronomy 2021, 11, 1445. https://doi.org/10.3390/agronomy11071445
Shan X, Zhang W, Huang J, Yu F, Qin W, Li J, Wang S, Dai Z. Identification and Characterization of SPL Transcription Factor Family Reveals Organization and Chilling-Responsive Patterns in Cabbage (Brassica oleracea var. capitata L.). Agronomy. 2021; 11(7):1445. https://doi.org/10.3390/agronomy11071445
Chicago/Turabian StyleShan, Xi, Wei Zhang, Jianxin Huang, Fangwei Yu, Wenbin Qin, Jianbin Li, Shenyun Wang, and Zhongliang Dai. 2021. "Identification and Characterization of SPL Transcription Factor Family Reveals Organization and Chilling-Responsive Patterns in Cabbage (Brassica oleracea var. capitata L.)" Agronomy 11, no. 7: 1445. https://doi.org/10.3390/agronomy11071445
APA StyleShan, X., Zhang, W., Huang, J., Yu, F., Qin, W., Li, J., Wang, S., & Dai, Z. (2021). Identification and Characterization of SPL Transcription Factor Family Reveals Organization and Chilling-Responsive Patterns in Cabbage (Brassica oleracea var. capitata L.). Agronomy, 11(7), 1445. https://doi.org/10.3390/agronomy11071445





