Automatic Phenotyping of Tomatoes in Production Greenhouses Using Robotics and Computer Vision: From Theory to Practice
Abstract
:1. Introduction
1.1. Related Work
1.2. Goals of this Paper
- To describe the development of an integrated phenotypic system from an acquisition system (robot, commercially available cameras), a set of high-performance segmentation algorithms for tomato and ribbon detection, and an adaptation of a well-known image registration algorithm.
- To evaluate the potential of this integrated phenotypic system in a realistic production environment
- To outline the challenges that this system faces in such a complex environment.
2. Materials and Methods
2.1. Hardware
2.2. Data
2.3. Image Analysis Pipeline
- Tomato detection by Mask-Region-based Convolutional Neural Networks (MaskRCNN)
- Ribbon detection Faster Region-based Convolutional Neural Networks (FasterRCNN)
- Image registration (both vertically and horizontally, using a Discrete Fourier Transform (DFT)-based registration)
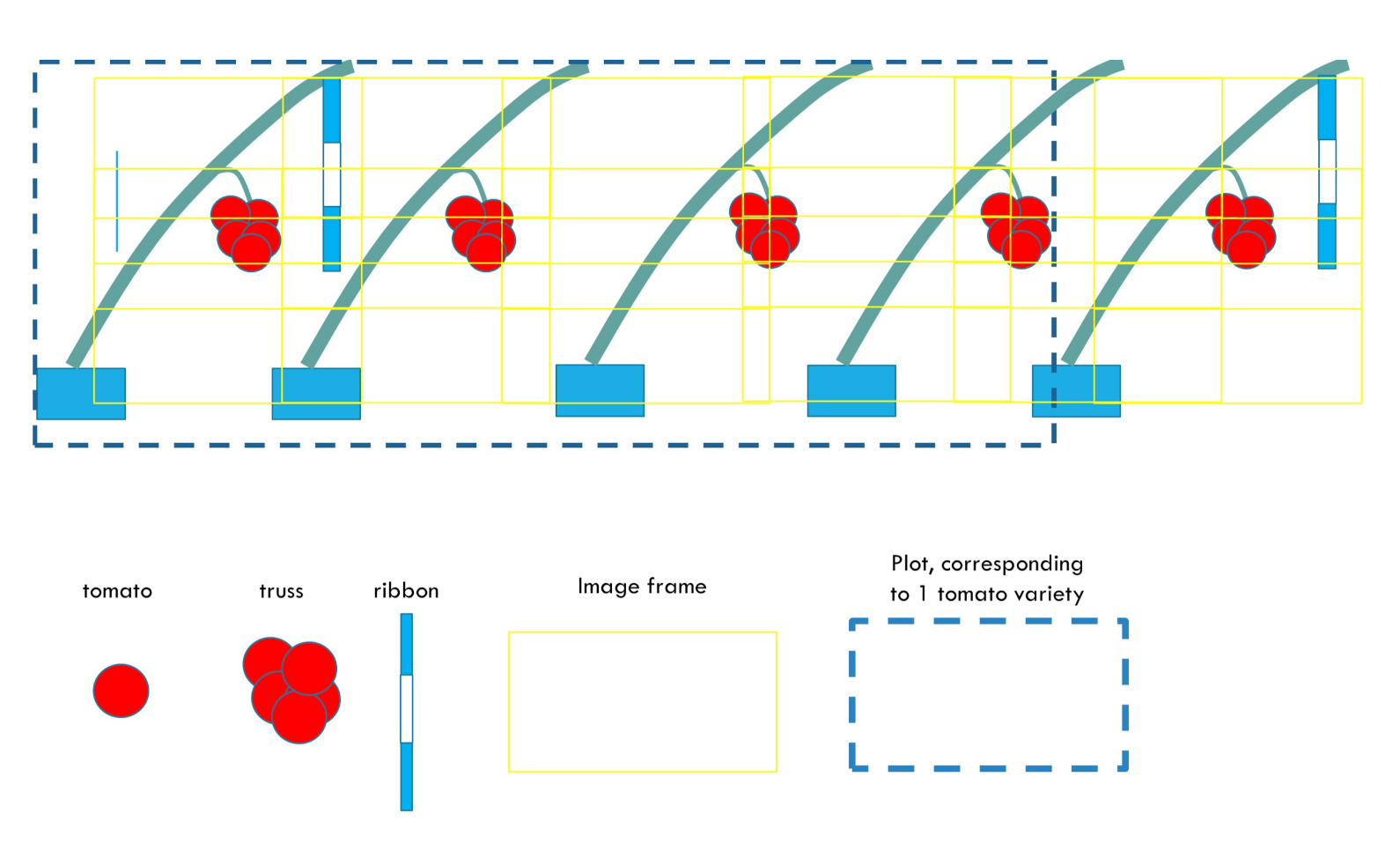
- Creation of a unified reference frame corresponding to a full row
- Position tomatoes and ribbons within this reference frame and assign tomatoes to plots
- Truss detection (Connected-component analysis);
- Identification of harvested trusses by comparison between pre- and post-harvest data.
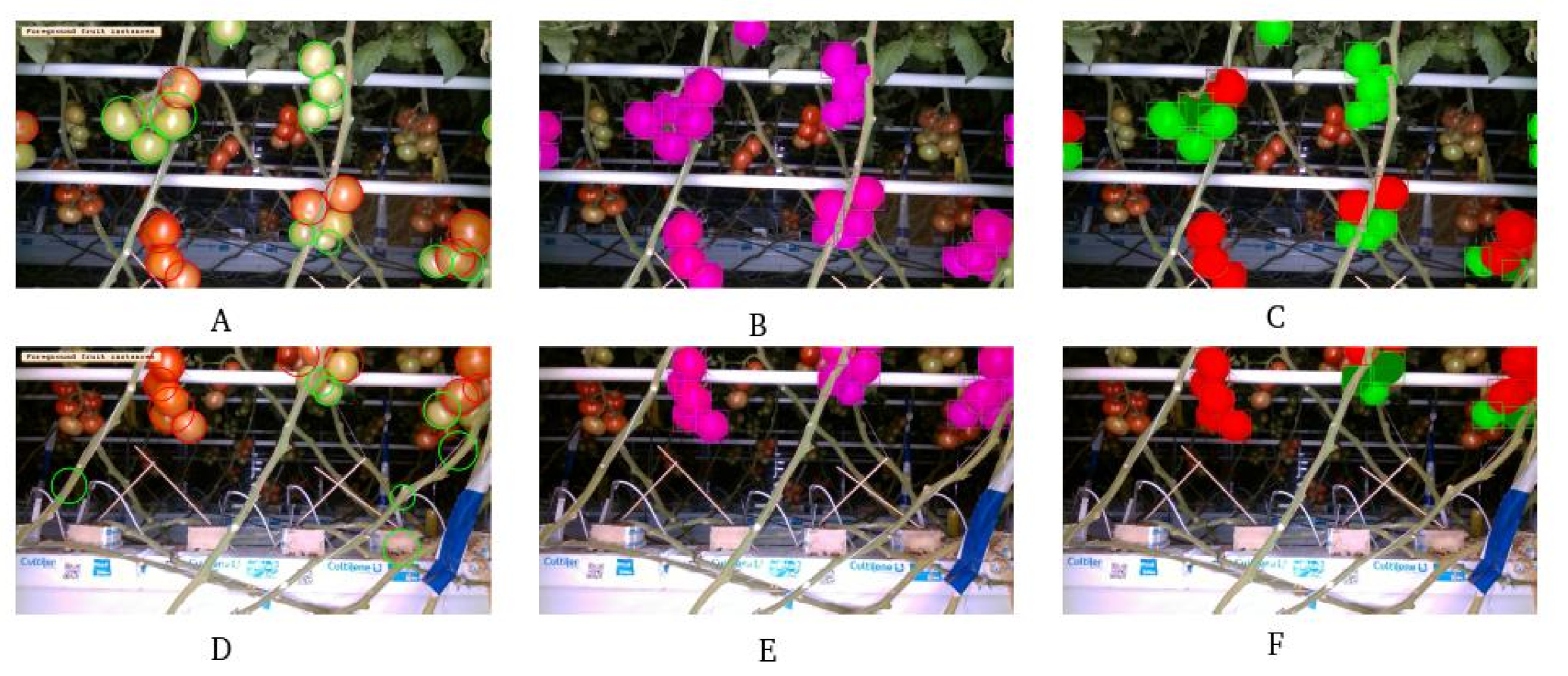
2.3.1. Fruit Detection
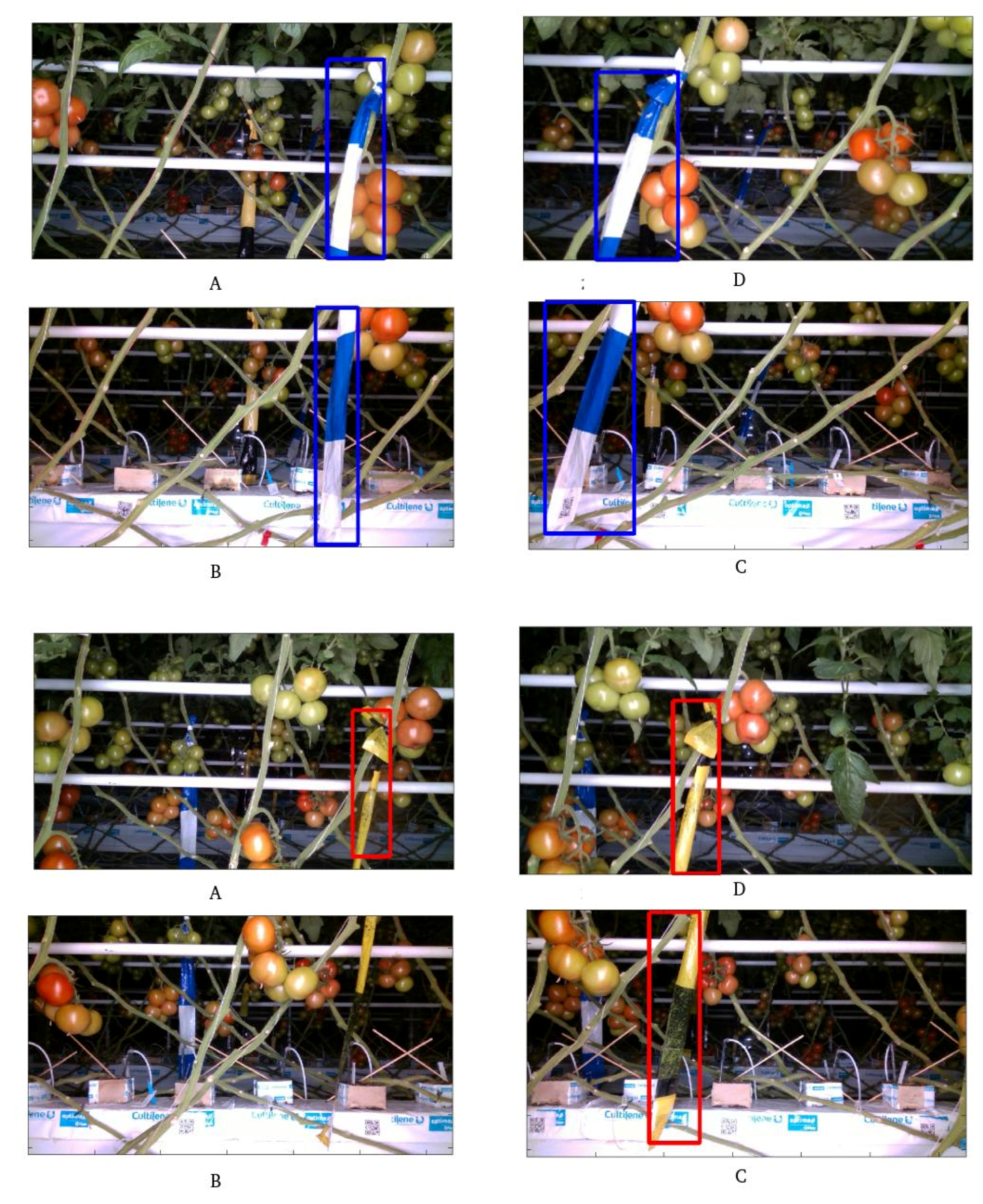
2.3.2. Ribbon Detection
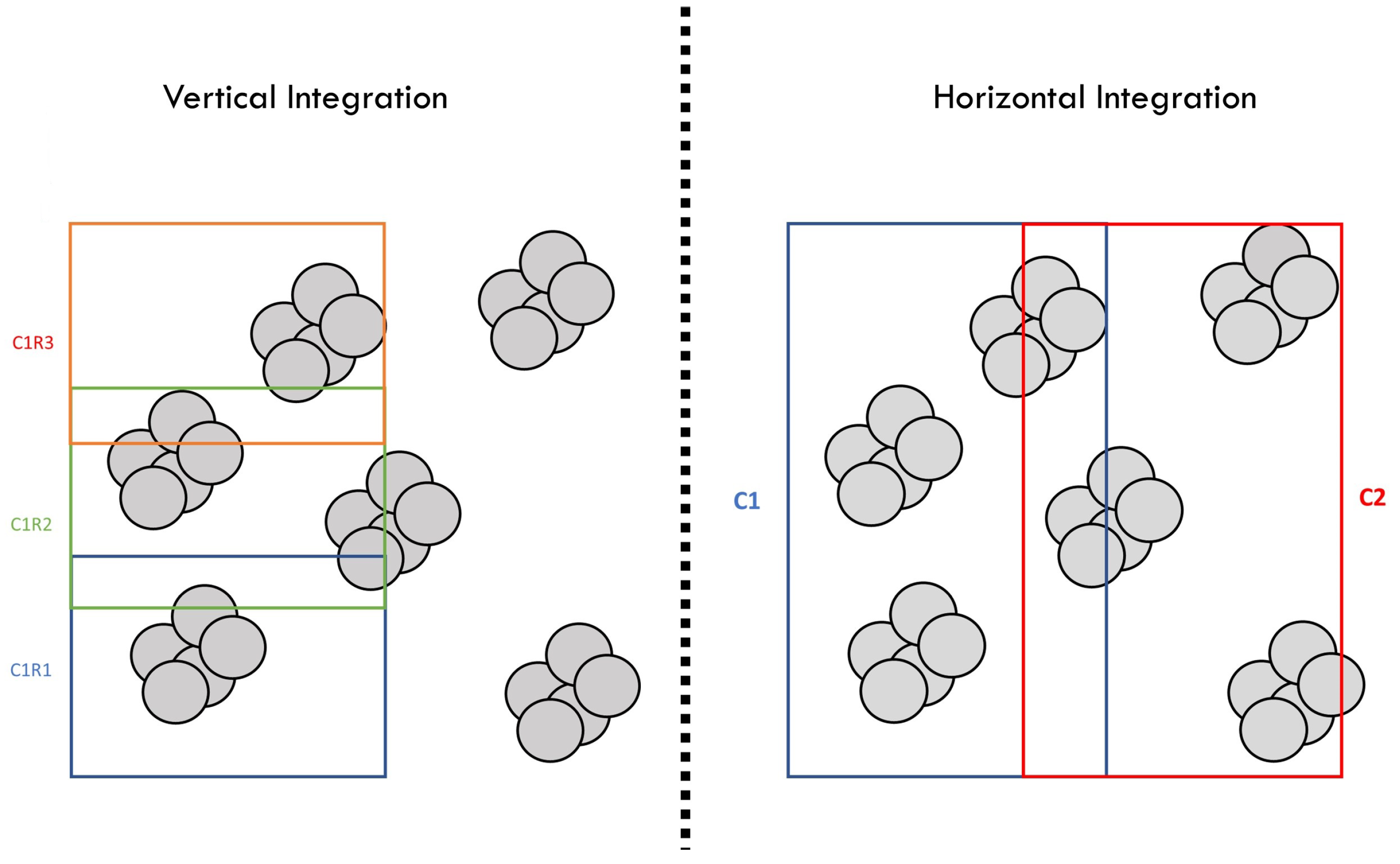
2.4. Image Registration
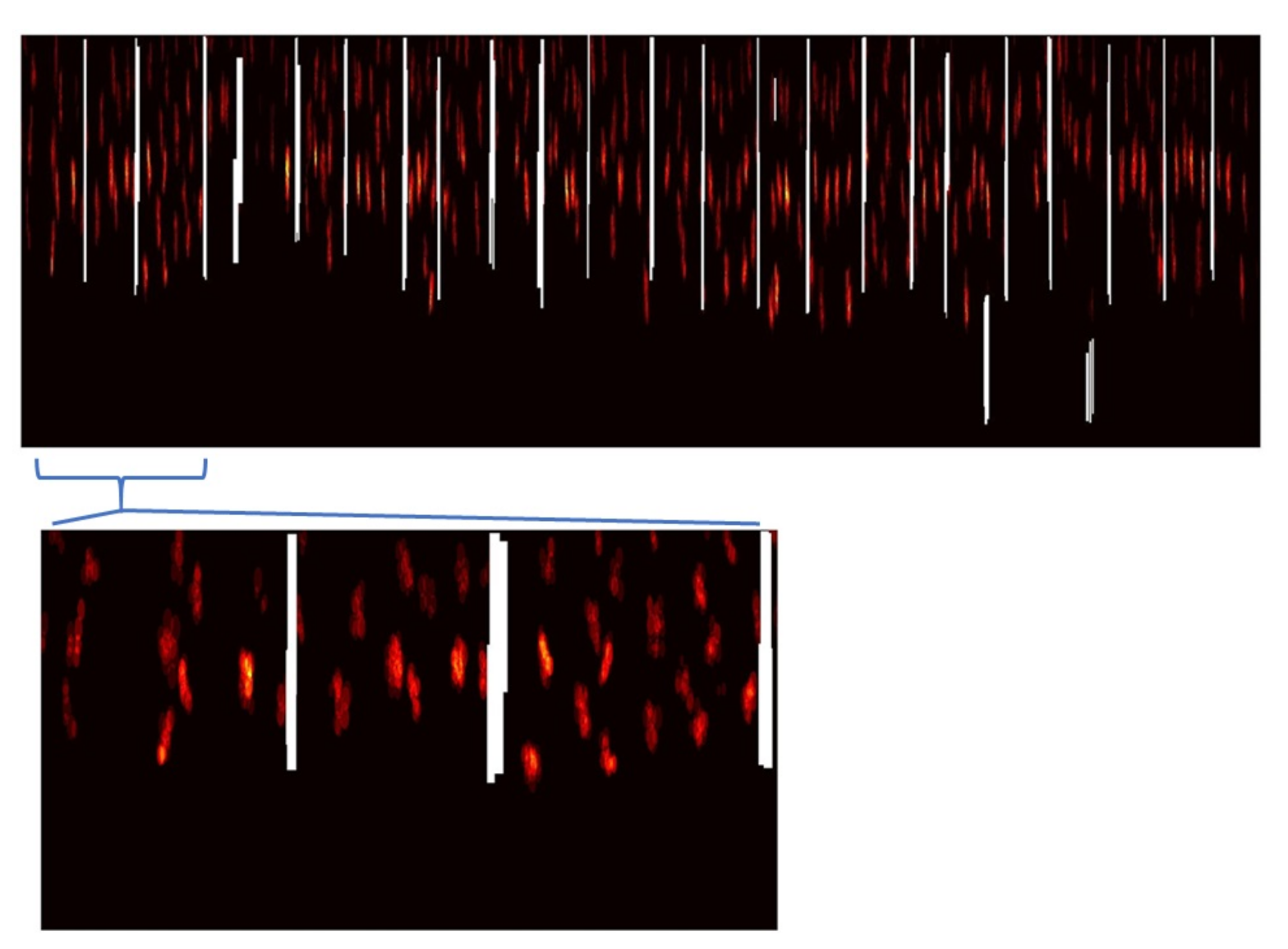
2.5. Truss Detection
2.6. Detection of Harvested Trusses
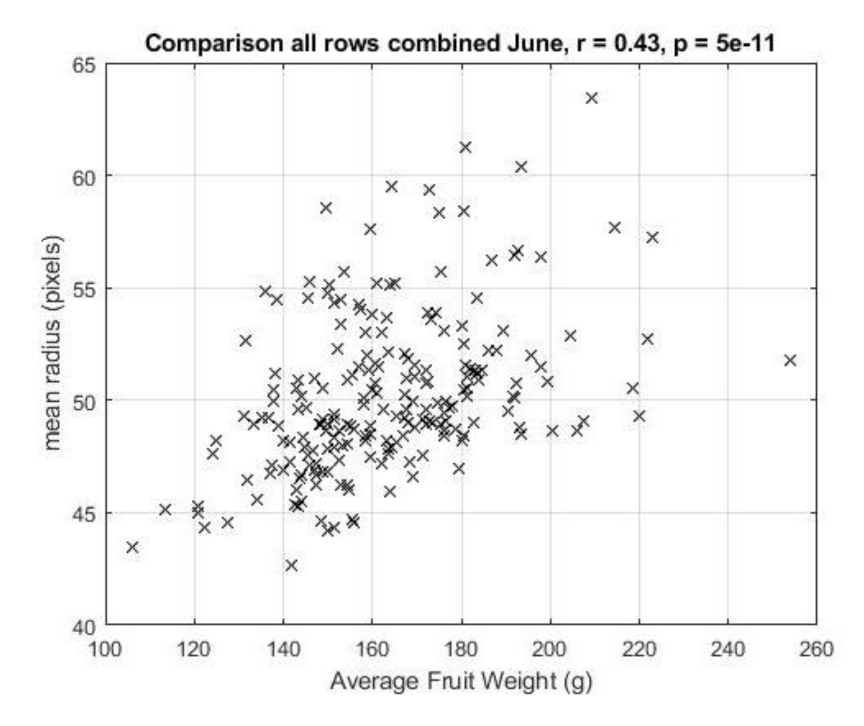
2.7. Comparison of Plot-Level Average Tomato Radius to Harvest Yield
3. Results
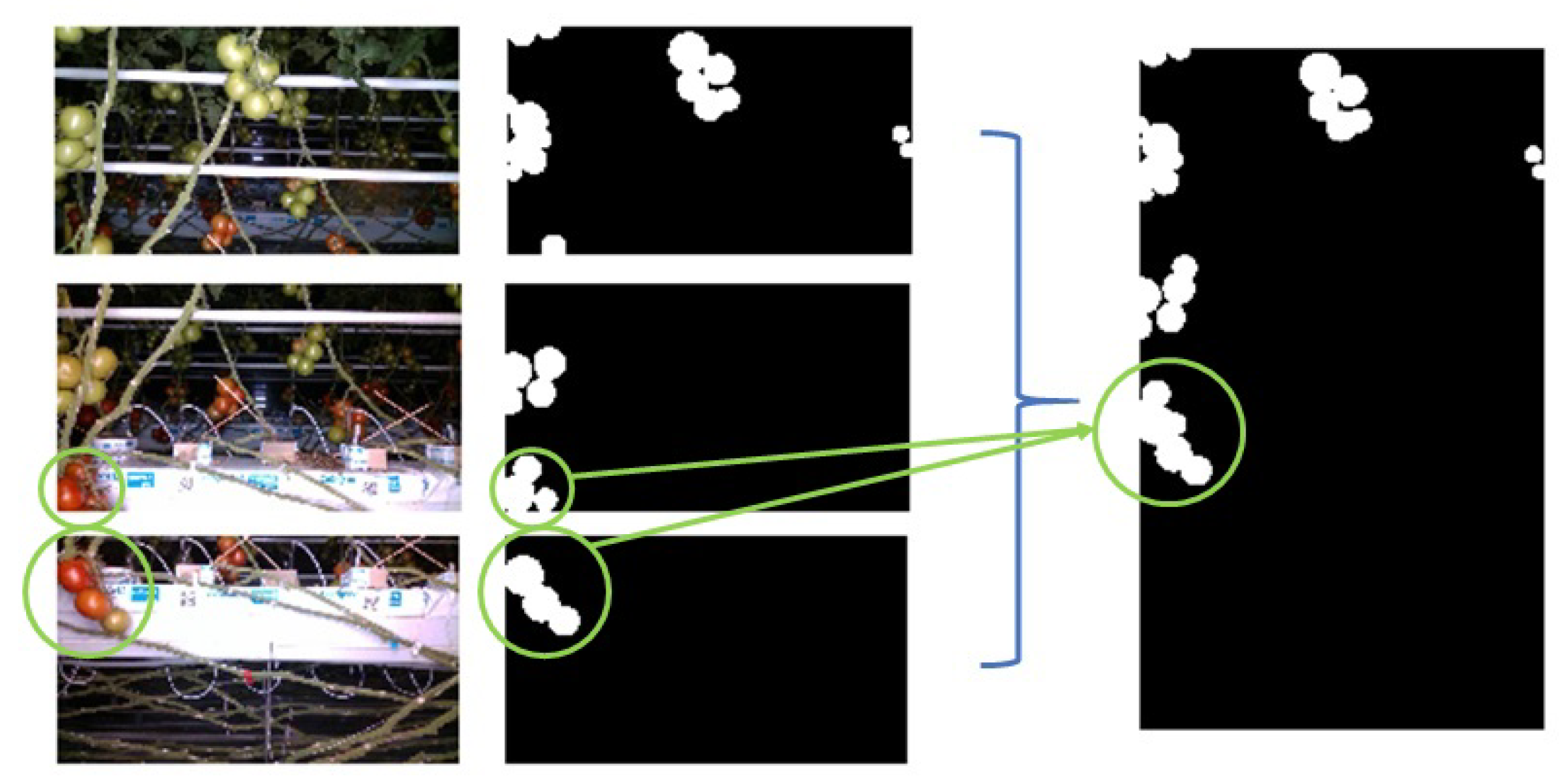
3.1. Fruit Segmentation
3.2. Ribbon Segmentation
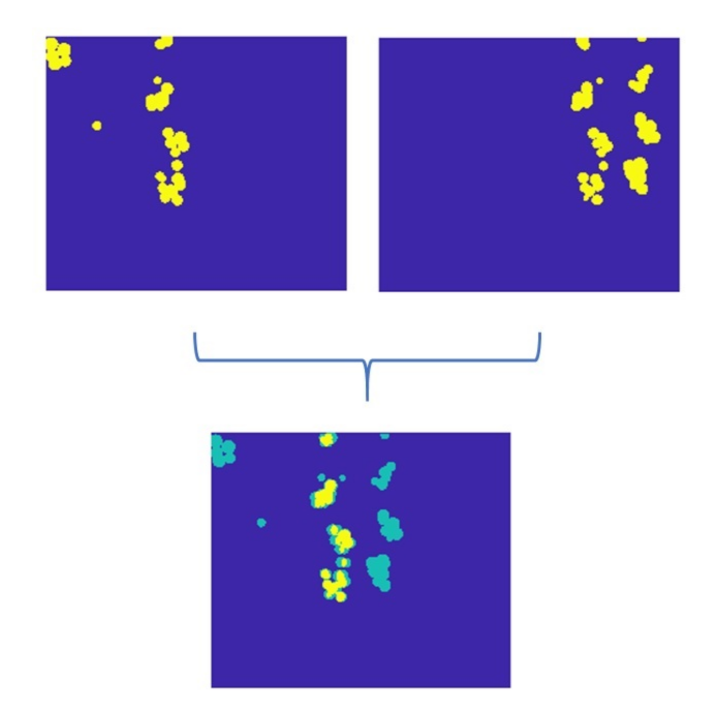
3.3. Image Registration
3.4. Overall Performance
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Furbank, R.T.; Tester, M. Phenomics—Technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 2011, 16, 635–644. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Q.; Huang, D. A review of imaging techniques for plant phenotyping. Sensors 2014, 14, 20078–20111. [Google Scholar] [CrossRef]
- Minervini, M.; Scharr, H.; Tsaftaris, S.A. Image Analysis: The New Bottleneck in Plant Phenotyping [Applications Corner]. IEEE Signal Process. Mag. 2015, 32, 126–131. [Google Scholar] [CrossRef] [Green Version]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436. [Google Scholar] [CrossRef] [PubMed]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef] [Green Version]
- Das Choudhury, S.; Samal, A.; Awada, T. Leveraging image analysis for high-throughput plant phenotyping. Front. Plant Sci. 2019, 10, 508. [Google Scholar] [CrossRef] [PubMed]
- Abade, A.S.; Ferreira, P.A.; Vidal, F.d.B. Plant Diseases recognition on images using Convolutional Neural Networks: A Systematic Review. arXiv 2020, arXiv:2009.04365. [Google Scholar]
- Burud, I.; Lange, G.; Lillemo, M.; Bleken, E.; Grimstad, L.; From, P.J. Exploring robots and UAVs as phenotyping tools in plant breeding. IFAC-PapersOnLine 2017, 50, 11479–11484. [Google Scholar] [CrossRef]
- Johansen, K.; Morton, M.J.; Malbeteau, Y.; Aragon, B.J.L.; AlMashharawi, S.; Ziliani, M.; Angel, Y.; Fiene, G.; Negrao, S.; Mousa, M.A.; et al. Predicting Biomass and Yield in a Tomato Phenotyping Experiment using UAV Imagery and Machine Learning. Front. Artif. Intell. 2020, 3, 28. [Google Scholar] [CrossRef]
- Granier, C.; Aguirrezabal, L.; Chenu, K.; Cookson, S.J.; Dauzat, M.; Hamard, P.; Thioux, J.J.; Rolland, G.; Bouchier-Combaud, S.; Lebaudy, A.; et al. PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol. 2006, 169, 623–635. [Google Scholar] [CrossRef]
- Walter, A.; Scharr, H.; Gilmer, F.; Zierer, R.; Nagel, K.A.; Ernst, M.; Wiese, A.; Virnich, O.; Christ, M.M.; Uhlig, B.; et al. Dynamics of seedling growth acclimation towards altered light conditions can be quantified via GROWSCREEN: A setup and procedure designed for rapid optical phenotyping of different plant species. New Phytol. 2007, 174, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Reuzeau, C. TraitMill (TM): A high throughput functional genomics platform for the phenotypic analysis of cereals. In In Vitro Cellular & Developmental Biology-Animal; Springer: New York, NY, USA, 2007; Volume 43, p. S4. [Google Scholar]
- Tang, Y.C.; Wang, C.; Luo, L.; Zou, X. Recognition and localization methods for vision-based fruit picking robots: A review. Front. Plant Sci. 2020, 11, 510. [Google Scholar] [CrossRef] [PubMed]
- Kootstra, G.; Wang, X.; Blok, P.M.; Hemming, J.; van Henten, E. Selective Harvesting Robotics: Current Research, Trends, and Future Directions. Curr. Robot. Rep. 2021, 1–10. [Google Scholar] [CrossRef]
- Hemming, J.; Bac, C.W.; Van Tuijl, B.; Barth, R.; Bontsema, J.; Pekkeriet, E. A robot for harvesting sweet-pepper in greenhouses. In Proceedings of the International Conference of Agricultural Engineering, Lausanne, Switzerland, 6–10 October 2014. [Google Scholar]
- Bac, C.W.; Hemming, J.; van Tuijl, B.; Barth, R.; Wais, E.; van Henten, E.J. Performance Evaluation of a Harvesting Robot for Sweet Pepper. J. Field Robot. 2017, 34, 1123–1139. [Google Scholar] [CrossRef]
- Ringdahl, O.; Kurtser, P.; Barth, R.; Edan, Y. Operational Flow of an Autonomous Sweetpepper Harvesting Robot. BO-25.06-002-003-PPO/PRI, EU-2015-03, 1409-035 EU. 2016. Available online: http://edepot.wur.nl/401245 (accessed on 6 August 2021).
- Barth, R.; IJsselmuiden, J.; Hemming, J.; Van Henten, E.J. Optimising Realism of Synthetic Agricultural Images Using Cycle Generative Adversarial Networks; Wageningen University & Research: Wageningen, The Netherlands, 2017. [Google Scholar]
- Mao, S.; Li, Y.; Ma, Y.; Zhang, B.; Zhou, J.; Wang, K. Automatic cucumber recognition algorithm for harvesting robots in the natural environment using deep learning and multi-feature fusion. Comput. Electron. Agric. 2020, 170, 105254. [Google Scholar] [CrossRef]
- Oberti, R.; Marchi, M.; Tirelli, P.; Calcante, A.; Iriti, M.; Tona, E.; Hočevar, M.; Baur, J.; Pfaff, J.; Schütz, C.; et al. Selective spraying of grapevines for disease control using a modular agricultural robot. Biosyst. Eng. 2016, 146, 203–215. [Google Scholar] [CrossRef]
- Paulin, S.; Botterill, T.; Lin, J.; Chen, X.; Green, R. A comparison of sampling-based path planners for a grape vine pruning robot arm. In Proceedings of the 2015 6th International Conference on Automation, Robotics and Applications (ICARA), Queenstown, New Zealand, 17–19 February 2015; IEEE: New York, NY, USA, 2015; pp. 98–103. [Google Scholar]
- Kaljaca, D.; Vroegindeweij, B.; Henten, E.J.V. Coverage trajectory planning for a bush trimming robot arm. J. Field Robot. 2019, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Cuevas-Velasquez, H.; Gallego, A.J.; Tylecek, R.; Hemming, J.; van Tuijl, B.; Mencarelli, A.; Fisher, R.B. Real-time Stereo Visual Servoing for Rose Pruning with Robotic Arm. In Proceedings of the 2020 International Conference on Robotics and Automation, Paris, France, 31 May–31 August 2020. [Google Scholar]
- Ruckelshausen, A.; Biber, P.; Dorna, M.; Gremmes, H.; Klose, R.; Linz, A.; Rahe, F.; Resch, R.; Thiel, M.; Trautz, D.; et al. BoniRob–an autonomous field robot platform for individual plant phenotyping. Precis. Agric. 2009, 9, 1. [Google Scholar]
- van der Heijden, G.; Song, Y.; Horgan, G.; Polder, G.; Dieleman, A.; Bink, M.; Palloix, A.; van Eeuwijk, F.; Glasbey, C. SPICY: Towards automated phenotyping of large pepper plants in the greenhouse. Funct. Plant Biol. 2012, 39, 870–877. [Google Scholar] [CrossRef]
- Zhou, J.; Chen, H.; Zhou, J.; Fu, X.; Ye, H.; Nguyen, H.T. Development of an automated phenotyping platform for quantifying soybean dynamic responses to salinity stress in greenhouse environment. Comput. Electron. Agric. 2018, 151, 319–330. [Google Scholar] [CrossRef]
- Shah, D.; Tang, L.; Gai, J.; Putta-Venkata, R. Development of a Mobile Robotic Phenotyping System for Growth Chamber-based Studies of Genotype x Environment Interactions. IFAC-PapersOnLine 2016, 49, 248–253. [Google Scholar] [CrossRef]
- Zhang, J.; Gong, L.; Liu, C.; Huang, Y.; Zhang, D.; Yuan, Z. Field Phenotyping Robot Design and Validation for the Crop Breeding. IFAC-PapersOnLine 2016, 49, 281–286. [Google Scholar] [CrossRef]
- Virlet, N.; Sabermanesh, K.; Sadeghi-Tehran, P.; Hawkesford, M.J. Field Scanalyzer: An automated robotic field phenotyping platform for detailed crop monitoring. Funct. Plant Biol. 2016, 44, 143–153. [Google Scholar] [CrossRef] [Green Version]
- Boogaard, F.P.; Rongen, K.S.; Kootstra, G.W. Robust node detection and tracking in fruit-vegetable crops using deep learning and multi-view imaging. Biosyst. Eng. 2020, 192, 117–132. [Google Scholar] [CrossRef]
- Bargoti, S.; Underwood, J.P. Image segmentation for fruit detection and yield estimation in apple orchards. J. Field Robot. 2017, 34, 1039–1060. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Chen, S.W.; Liu, C.; Shivakumar, S.S.; Das, J.; Taylor, C.J.; Underwood, J.; Kumar, V. Monocular camera based fruit counting and mapping with semantic data association. IEEE Robot. Autom. Lett. 2019, 4, 2296–2303. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Rathod, V.; Sun, C.; Zhu, M.; Korattikara, A.; Fathi, A.; Fischer, I.; Wojna, Z.; Song, Y.; Guadarrama, S.; et al. Speed/accuracy trade-offs for modern convolutional object detectors. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 7310–7311. [Google Scholar]
- Mu, Y.; Chen, T.S.; Ninomiya, S.; Guo, W. Intact Detection of Highly Occluded Immature Tomatoes on Plants Using Deep Learning Techniques. Sensors 2020, 20, 2984. [Google Scholar] [CrossRef] [PubMed]
- Koller, M.; Upadhyaya, S. Prediction of processing tomato yield using a crop growth model and remotely sensed aerial images. Trans. ASAE 2005, 48, 2335–2341. [Google Scholar] [CrossRef]
- Ashapure, A.; Oh, S.; Marconi, T.G.; Chang, A.; Jung, J.; Landivar, J.; Enciso, J. Unmanned aerial system based tomato yield estimation using machine learning. In Autonomous Air and Ground Sensing Systems for Agricultural Optimization and Phenotyping IV; International Society for Optics and Photonics: Bellingham, WA, USA, 2019; Volume 11008, p. 110080O. [Google Scholar]
- Darrigues, A.; Hall, J.; van der Knaap, E.; Francis, D.M.; Dujmovic, N.; Gray, S. Tomato analyzer-color test: A new tool for efficient digital phenotyping. J. Am. Soc. Hortic. Sci. 2008, 133, 579–586. [Google Scholar] [CrossRef] [Green Version]
- Stein, M.; Bargoti, S.; Underwood, J. Image Based Mango Fruit Detection, Localisation and Yield Estimation Using Multiple View Geometry. Sensors 2016, 16, 1915. [Google Scholar] [CrossRef] [PubMed]
- Schonberger, J.L.; Frahm, J.M. Structure-from-motion revisited. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 4104–4113. [Google Scholar]
- Fujinaga, T.; Yasukawa, S.; Li, B.; Ishii, K. Image mosaicing using multi-modal images for generation of tomato growth state map. J. Robot. Mechatron. 2018, 30, 187–197. [Google Scholar] [CrossRef]
- Gan, H.; Lee, W.S.; Alchanatis, V. A photogrammetry-based image registration method for multi-camera systems–With applications in images of a tree crop. Biosyst. Eng. 2018, 174, 89–106. [Google Scholar] [CrossRef]
- Liu, X.; Chen, S.W.; Aditya, S.; Sivakumar, N.; Dcunha, S.; Qu, C.; Taylor, C.J.; Das, J.; Kumar, V. Robust fruit counting: Combining deep learning, tracking, and structure from motion. In Proceedings of the 2018 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS), Madrid, Spain, 1–5 October 2018; IEEE: New York, NY, USA, 2018; pp. 1045–1052. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, S.; Masuzawa, H.; Miura, J.; Oishi, S. 3D Semantic Mapping in Greenhouses for Agricultural Mobile Robots with Robust Object Recognition Using Robots’ Trajectory. In Proceedings of the 2018 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Miyazaki, Japan, 7–10 October 2018; pp. 357–362. [Google Scholar]
- Gené-Mola, J.; Sanz-Cortiella, R.; Rosell-Polo, J.R.; Morros, J.R.; Ruiz-Hidalgo, J.; Vilaplana, V.; Gregorio, E. Fruit detection and 3D location using instance segmentation neural networks and structure-from-motion photogrammetry. Comput. Electron. Agric. 2020, 169, 105165. [Google Scholar] [CrossRef]
- Afonso, M.; Fonteijn, H.; Fiorentin, F.S.; Lensink, D.; Mooij, M.; Faber, N.; Polder, G.; Wehrens, R. Tomato Fruit Detection and Counting in Greenhouses Using Deep Learning. Front. Plant Sci. 2020, 11, 1759. [Google Scholar] [CrossRef]
- Afonso, M.; Mencarelli, A.; Polder, G.; Wehrens, R.; Lensink, D.; Faber, N. Detection of tomato flowers from greenhouse images using colorspace transformations. In Proceedings of the EPIA Conference on Artificial Intelligence, Vila Real, Portugal, 3–6 September 2019; pp. 146–155. [Google Scholar]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; IEEE: New York, NY, USA, 2017; pp. 2980–2988. [Google Scholar]
- Xie, S.; Girshick, R.; Dollár, P.; Tu, Z.; He, K. Aggregated residual transformations for deep neural networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; IEEE: New York, NY, USA, 2017; pp. 5987–5995. [Google Scholar]
- Girshick, R. Fast R-CNN. In Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015; pp. 1440–1448. [Google Scholar] [CrossRef]
- Guizar-Sicairos, M.; Thurman, S.T.; Fienup, J.R. Efficient subpixel image registration algorithms. Opt. Lett. 2008, 33, 156–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fonteijn, H.; Afonso, M.; Lensink, D.; Mooij, M.; Faber, N.; Vroegop, A.; Polder, G.; Wehrens, R. Automatic Phenotyping of Tomatoes in Production Greenhouses Using Robotics and Computer Vision: From Theory to Practice. Agronomy 2021, 11, 1599. https://doi.org/10.3390/agronomy11081599
Fonteijn H, Afonso M, Lensink D, Mooij M, Faber N, Vroegop A, Polder G, Wehrens R. Automatic Phenotyping of Tomatoes in Production Greenhouses Using Robotics and Computer Vision: From Theory to Practice. Agronomy. 2021; 11(8):1599. https://doi.org/10.3390/agronomy11081599
Chicago/Turabian StyleFonteijn, Hubert, Manya Afonso, Dick Lensink, Marcel Mooij, Nanne Faber, Arjan Vroegop, Gerrit Polder, and Ron Wehrens. 2021. "Automatic Phenotyping of Tomatoes in Production Greenhouses Using Robotics and Computer Vision: From Theory to Practice" Agronomy 11, no. 8: 1599. https://doi.org/10.3390/agronomy11081599






