Resistance Response of the Recently Discovered Species Nicotiana mutabilis to Potato virus Y (PVY) and Tomato spotted wilt virus (TSWV) Compared to Other Sources of Resistance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
- Nicotiana alata, section Alatae;
- Nicotiana forgetiana, section Alatae;
- Nicotiana tabacum, section Nicotiana. In our research, this species was represented by two cultivars:
- −
- −
- Burley 21—belongs to the light air-cured type of tobacco. Susceptible to all strains of PVY and to TSWV.
2.2. Resistance Tests
- −
- temperature, 24 °C during the day/16 °C at night;
- −
- light and darkness duration, 16 h light/8 h darkness.
- IUNG 21—mild isolate that does not break va resistance in the VAM cultivar. It is not detectable by monoclonal antibodies directed against the necrotic serotype (MoAbs anti YN IgG 112511, Bioreba) [21];
- IUNG 20—severe isolate that breaks the va resistance of cultivar VAM. Detectable by two types of Bioreba antibodies (MoAbs anti Y IgG 112511 and MoAbs anti YN IgG 112512) [21].
2.3. Serological Tests
- −
- lower part included four lower leaves;
- −
- middle part included four leaves above the lower part;
- −
- upper part included the four youngest leaves on the plant.
2.4. Molecular Tests
3. Results
3.1. Resistance Response to PVY
3.2. Resistance Response to TSWV
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Doroszewska, T.; Depta, A.; Czubacka, A. Album of Nicotiana Species; Institute of Soil Science and Plant Cultivation: Puławy, Poland, 2009. [Google Scholar]
- Kostoff, D. The origin of the tetraploid Nicotiana from Bathurst. Cur. Sci. 1939, 8, 110–111. [Google Scholar]
- Goodspeed, T.H. The genus Nicotiana; Chronica Botanica: Waltham, MA, USA, 1954. [Google Scholar]
- Burbidge, N.T. The Australian species of Nicotiana L. (Solanaceae). Aust. J. Bot. 1960, 8, 342–380. [Google Scholar] [CrossRef]
- Merxmüller, H.; Buttler, L.P. Nicotiana in der Afrikanischen Namib-Ein Pflanzen geographisches und phylogenetisches Ratsel. Itl. Bot. 1975, 12, 91–104. [Google Scholar]
- Knapp, S.; Chase, M.; Clarkson, J.J. Nomenclatural changes and a new sectional classification in Nicotiana (Solanaceae). Taxon 2004, 53, 73–82. [Google Scholar] [CrossRef]
- Lewis, R.S.; Nicholson, J.S. Aspects of the evolution of Nicotiana tabacum L. and the status of the United States Nicotiana Germplasm Collection. Genet. Resour. Crop Evol. 2007, 54, 727–740. [Google Scholar] [CrossRef]
- Stehmann, J.R.; Semir, J.; Ippolito, A. Nicotiana mutabilis (Solanaceae), a new species from southern Brazil. Kew Bull. 2002, 57, 639–646. [Google Scholar] [CrossRef]
- Doroszewska, T.; Berbeć, A. Methodology of Tobacco Integrated Pest Management; Institute of Soil Science and Plant Cultivation: Puławy, Poland, 2015. (In Polish) [Google Scholar]
- Doroszewska, T. Wide Hybridization and Genetic Transformation in Breeding for Resistance to Potato virus Y (PVY) in Tobacco (Nicotiana tabacum L.); Institute of Soil Science and Plant Cultivation: Puławy, Poland, 2004. [Google Scholar]
- Scholthof, K.B.K.; Adkins, S.; Czosnek, H.; Palukaitis, P.; Jacquot, E.; Hohn, T.; Hohn, B.; Saunders, K.; Candresse, T.; Ahlquist, P.; et al. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 2011, 12, 938–954. [Google Scholar] [CrossRef]
- Simmonds, P.; Aiewsakun, P. Virus classification—Where do you draw the line? Arch. Virol. 2018, 163, 2037–2046. [Google Scholar] [CrossRef] [Green Version]
- Gorbalenya, A.E.; Krupovic, M.; Mushegian, A.; Kropinski, A.M.; Siddell, S.G.; Varsani, A.; Adams, M.J.; Davison, A.J.; Dutilh, B.E.; Harrach, B.; et al. The new scope of virus taxonomy: Partitioning the virosphere into 15 hierarchical ranks. Nat. Microbiol. 2020, 5, 668–674. [Google Scholar]
- Robaglia, C.; Durand-Tardif, M.; Tronchet, M.; Boudazin, G.; Astier-Manifacier, S.; Casse-Delbart, F. Nucleotide sequence of Potato virus Y (N strain) genomic RNA. J. Gen. Virol. 1989, 70, 935–947. [Google Scholar] [CrossRef]
- Thole, V.; Dalmay, T.; Burgyan, J.; Balazs, E. Cloning and sequencing of Potato virus Y (Hungarian isolate) genomic RNA. Gene 1993, 123, 149–156. [Google Scholar] [CrossRef]
- Green, K.J.; Brown, C.J.; Gray, S.M.; Karasev, A.V. Phylogenetic study of recombinant strains of Potato virus Y. Virology 2017, 507, 40–52. [Google Scholar] [CrossRef]
- Chrzanowska, M. Differentiation of Potato virus Y (PVY) isolates. Phytopathol. Pol. 1994, 8, 15–20. [Google Scholar]
- Le Romancer, M.; Kerlan, C.; Nedellec, M. Biological characterization of various geographical isolates of Potato virus Y inducing superficial necrosis on potato tubers. Plant Pathol. 1994, 43, 138–144. [Google Scholar] [CrossRef]
- Tribodet, M.; Glais, L.; Kerlan, C.; Jacquot, E. Characterization of Potato virus Y (PVY) molecular determinants involved in the vein necrosis symptom induced by PVYN isolates in infected Nicotiana tabacum cv. Xanthi. J. Gen. Virol. 2005, 86, 2101–2105. [Google Scholar] [CrossRef]
- Crosslin, J.M. PVY: An Old Enemy and A Continuing Challenge. Am. J. Potato Res. 2013, 90, 2–6. [Google Scholar] [CrossRef]
- Gugerli, P.; Fries, P. Characterization of monoclonal antibodies to Potato virus Y and their use for virus detection. J. Gen. Virol. 1983, 64, 2471–2477. [Google Scholar] [CrossRef]
- Drake, J.W. Rates of spontaneous mutation among RNA viruses. Proc. Natl. Acad. Sci. USA 1993, 90, 4171–4175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Przybyś, M.; Doroszewska, T.; Berbeć, A. Point mutation in the viral genome-linked protein (VPg) of Potato virus Y probably correspond with ability to overcome resistance of tobacco. J. Food Agric. Environ. 2013, 11, 986–989. [Google Scholar]
- Dluge, K.L.; Song, Z.; Wang, B.; Steede, W.T.; Xiao, B.; Liu, Y.; Dewey, R.E. Characterization of Nicotiana tabacum genotypes possessing deletion mutations that affect potyvirus resistance and the production of trichome exudates. BMC Genom. 2018, 19, 484. [Google Scholar] [CrossRef] [PubMed]
- Ruyi, R.; Qiang, Z.; Futai, N.; Qiu, J.; Wiuqing, W.; Jicheng, W. Breeding for PVY resistance in tobacco LJ911 using CRISPR/Cas9 technology. Crop Breed. Appl. Biotechnol. 2021, 21, e31682116. [Google Scholar] [CrossRef]
- Julio, E.; Cotucheau, J.; Decorps, C.; Volpatti, R.; Sentenac, C.; Candresse, T.; Dorlhac de Borne, F. A eucariotic translation initiation factor 4E (elF4E) is responsible for the “va” tobacco recessive resistance to potyviruses. Plant. Mol. Biol. Rep. 2015, 33, 609–623. [Google Scholar] [CrossRef]
- Koelle, G. Versucherzur Vererbung der Krankheitsresistenz bei Tabak. 2 Mitt. Eine Rippen-braune-resistante Virgin A Mutante nach Anwendung kunstlicher Mutations auslosung durch Rontgenstrahlen. Table Forsch. 1958, 24, 83–84. [Google Scholar]
- Michel, V.; Julio, E.; Candresse, T.; Cotucheau, J.; Decorps, C.; Volpatti, R.; Moury, B.; Glais, L.; Dorlhac de Borne, F.; Decroocq, V.; et al. NtTPN: A RPP8-like R gene required for Potato virus Y-induced veinal necrosis in tobacco. Plant J. 2018, 95, 700–714. [Google Scholar] [CrossRef]
- Verrier, J.L.; Doroszewska, T. Tobacco Virus Collaborative Study (1996–2011) VIR Technical Report. 2018. Available online: https://www.coresta.org/documents/search?f%5B0%5D=im_field_technical_document_type%3A36193&page=5 (accessed on 1 August 2018).
- Sievert, R.C. Sources of resistance to Potato virus Y in the genus Nicotiana. Tob. Sci. 1972, 106, 92–94. [Google Scholar]
- Doroszewska, T.; Depta, A. Resistance of wild Nicotiana species to different PVY isolates. Phytopathologia 2011, 59, 9–24. [Google Scholar]
- Doroszewska, T.; Chrzanowska, M. Characterization of the main PVY resistance sources to different PVY strains. Inf. Bull. Coresta 2001, 60, 21–27. [Google Scholar]
- Wersman, E.A. Varied Roles of the Haploid Sporophyte in Plant Improvement. In Plant Breeding in the 1990s; Stalker, H.T., Murphy, J.P., Raleigh, N.C., Eds.; CABI Publ.: Wallinfort, UK, 1992; pp. 461–484. [Google Scholar]
- Lewis, R.S. Transfer of resistance to Potato virus Y (PVY) from Nicotiana africana to Nicotiana tabacum: Possible influence of tissue culture on the rate of introgression. Theor. Appl. Genet. 2005, 110, 678–687. [Google Scholar] [CrossRef]
- Lewis, R.S. Evaluation of Nicotiana tabacum genotypes possessing Nicotiana africana-derived genetic tolerance to Potato Virus Y. Crop Sci. 2007, 47, 1975–1984. [Google Scholar] [CrossRef]
- Doroszewska, T. Transfer of tolerance to different Potato virus Y (PVY) isolates from Nicotiana Africana Merxm. to Nicotiana tabacum L. Plant Breed. 2010, 129, 76–81. [Google Scholar] [CrossRef]
- Korbecka-Glinka, G.; Czubacka, A.; Przybyś, M.; Doroszewska, T. Resistance vs. tolerance to Potato virus Y in tobacco—Comparing effectiveness using virus isolates from Central Europe. Breed. Sci. 2017, 67, 459–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korbecka-Glinka, G.; Czubacka, A.; Depta, A.; Doroszewska, T. Inheritance of Potato virus Y tolerance introgressed from Nicotiana africana to cultivated tobacco. Pol. J. Agron. 2017, 31, 39–44. [Google Scholar]
- Parella, G.; Gognalons, P.; Gebre-Sellassie, K.; Vovlas, C.; Marchoux, G. An update of the host range of Tomato spotted wilt virus. J. Plant Pathol. 2003, 85, 227–264. [Google Scholar]
- Francki, R.I.B.; Fauquet, C.M.; Knudson, D.D.; Brown, F. Fifth report of the International Committee on Taxonomy of Viruses. Arch. Virol. 1991, 2, 1–450. [Google Scholar]
- Wijkamp, I.; Van Lent, J.; Kormelink, R.; Goldbach, R.; Peters, D. Multiplication of Tomato spotted wilt virus in its insect vector, Frankliniella occidentalis. J. Gen.Virol. 1993, 74, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.R. Plant viruses transmitted by thrips. Eur. J. Plant Pathol. 2005, 113, 119–157. [Google Scholar] [CrossRef]
- Mumford, R.A.; Barker, I.; Wood, K.R. The biology of the tospoviruses. Ann. Appl. Biol. 1996, 128, 159–183. [Google Scholar] [CrossRef]
- Doroszewska, T.; Berbeć, A.; Czarnecka, D.; Kawka, M. Diseases and Pests of Tobacco; Institute of Soil Science and Plant Cultivation: Puławy, Poland, 2013. [Google Scholar]
- Ivancheva-Gabrovska, T. Sources of resistance to Tomato spotted wilt virus and Thrips tabaci Lind. Spec. CORESTA Bull Symp. Sofia 1978, 96. [Google Scholar]
- Jankowski, F. Sources of resistance to TSWV (Lycopersicum virus 3) among uncultivated species of Nicotiana genus. Biul CLPT 1980, 1–2, 3–8. [Google Scholar]
- Kovalenko, A.G.; Rud, E.A.; Strelyaeva, N.I.; Oleshchenko, L.T. Responses of tobacco varieties, wild species and interspecies hybrids on artificial infection with Tomato spotted wilt virus. Mikrobiol. Zhurnal. 1987, 49, 85–89. [Google Scholar]
- Palakarcheva, M.; Yancheva, A. Genetic sources of resistance to tomato bronziness pathogen on tobacco in wild species of the genus Nicotiana. Genet Breed 1989, 22, 473–479. (In Bulgarian) [Google Scholar]
- Laskowska, D.; Doroszewska, T.; Depta, A.; Kursa, K.; Olszak-Przybyś, H.; Czubacka, A. A survey of Nicotiana germplasm for resistance to Tomato spotted wilt virus (TSWV). Euphytica 2013, 193, 207–219. [Google Scholar] [CrossRef] [Green Version]
- Moon, H.; Nicholson, J.S. AFLP and SCAR markers linked to Tomato Spotted Wilt Virus resistance in tobacco. Crop Sci. 2007, 47, 1887–1894. [Google Scholar] [CrossRef]
- Koelle, G. Genetische Analyse einer Y-virus (Rippen-braune) resistenten Mutante der Tabaksorte Virgin A. Zuchter 1961, 31, 71–72. [Google Scholar]
- Chrzanowska, M.; Doroszewska, T. Comparison between PVY isolates obtained from potato and tobacco plants in Poland. Phytopathol. Pol. 1997, 13, 63–71. [Google Scholar]
- Doroszewska, T.; Verrier, J.L. Sub-Group Collaborative Study on Potato Virus Y; Annual Subgroup Report; CORESTA CD-ROM Version. 20; Paris, France, 2004. [Google Scholar]
- Verrier, J.L. PVY Collaborative Experiment. Results; Altadis, Insitute du Tabac: Bergerac, France, 2001. [Google Scholar]
- Tsakiridis, J.P.; Gooding, G.V. Tomato spotted wilt virus in Greese. Phytopathol. Mediterr. 1972, 11, 42–47. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure from small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Sierro, N.; Battey, J.N.D.; Ouadi, S.; Bakaher, N.; Bovet, L.; Willing, A.; Geopfert, S.; Peitsch, M.C.; Ivanov, N.V. The tobacco genome sequence and its comparison with those of tomato and potato. Nat. Commun. 2014, 5, 3833. [Google Scholar] [CrossRef]
- Cardin, L.; Moury, B. First Report of Potato virus Y in Nicotiana mutabilis in France. Plant Dis. 2008, 92, 312. [Google Scholar] [CrossRef]
- Gajos, Z. Inheritance of resistance to Tomato spotted wilt virus in interspecies hybrids of Nicotiana tabacum × Nicotiana alata Link. Zesz. Prob. Post. Nauk Rol. 1981, 244, 117–126. [Google Scholar]
- Gajos, Z. Polalta, the first Polish tobacco variety resistant to Tomato spotted wilt virus was released for regional experimentation and propagation. Wiad. Tytoniowe. 1987, 31, 11–17. [Google Scholar]
- Gajos, Z. Virginia ZG-4 (Wiktoria)—A new tobacco variety resistant to Tomato spotted wilt virus (TSWV) and black root rot (Thielaviopsisbasicola Ferr.). Biul. CLPT 1993, 1–4, 5–19. [Google Scholar]
- Yancheva, A.A. Possibility for transferring combined resistance to Tomato spotted wilt virus and Thielaviopsis basicola to intercultivar tobacco hybrids. Genet. Breed. 1990, 23, 194–199. (In Bulgarian) [Google Scholar]
- Berbeć, A. Cytological study on Nicotiana tabacum L. cv. Nadwiślański Mały (2× and 4×) ×Nicotiana alata Link et Otto hybrids. Genet. Pol. 1987, 28, 251–261. [Google Scholar]
- Laskowska, D.; Berbeć, A. Cytology and fertility of viable hybrids of Nicotiana tabacum L. cv. TB-566 with N. alata Link et Otto. J. Appl. Genet. 2005, 46, 11–18. [Google Scholar]
- Laskowska, D.; Berbeć, A. Resistance to Tomato spotted wilt virus (TSWV) in Nicotiana alata and N. sanderae and in hybrids between N. tabacum and N. alata. Plant Breed. Seed Sci. 2006, 54, 91–100. [Google Scholar]
- Laskowska, D.; Berbeć, A. TSWV resistance in DH lines of tobacco (Nicotiana tabacum L.) obtained from a hybrid between ‘Polalta’ and ‘Wiślica’. Plant Breed. 2010, 129, 731–733. [Google Scholar] [CrossRef]

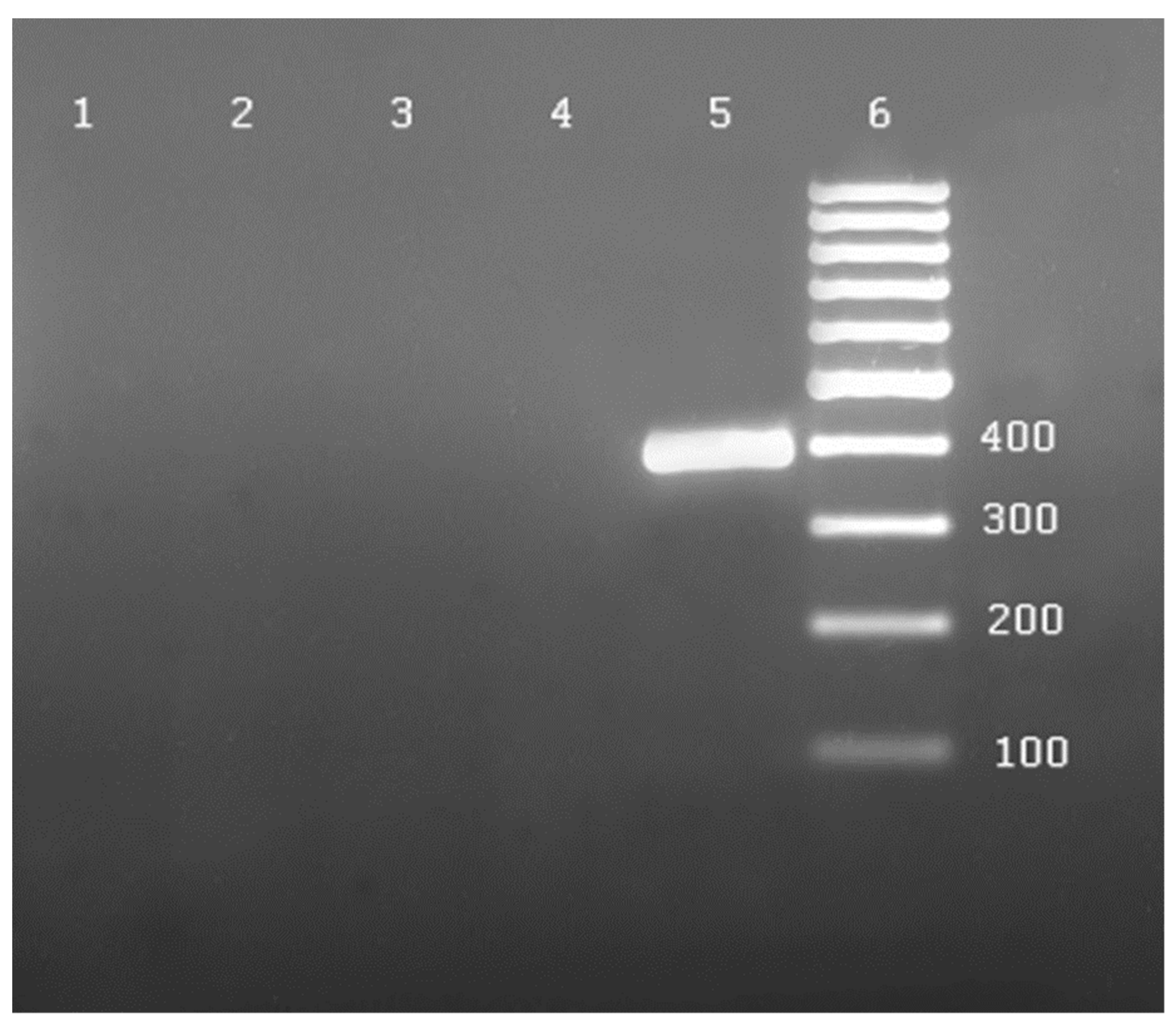





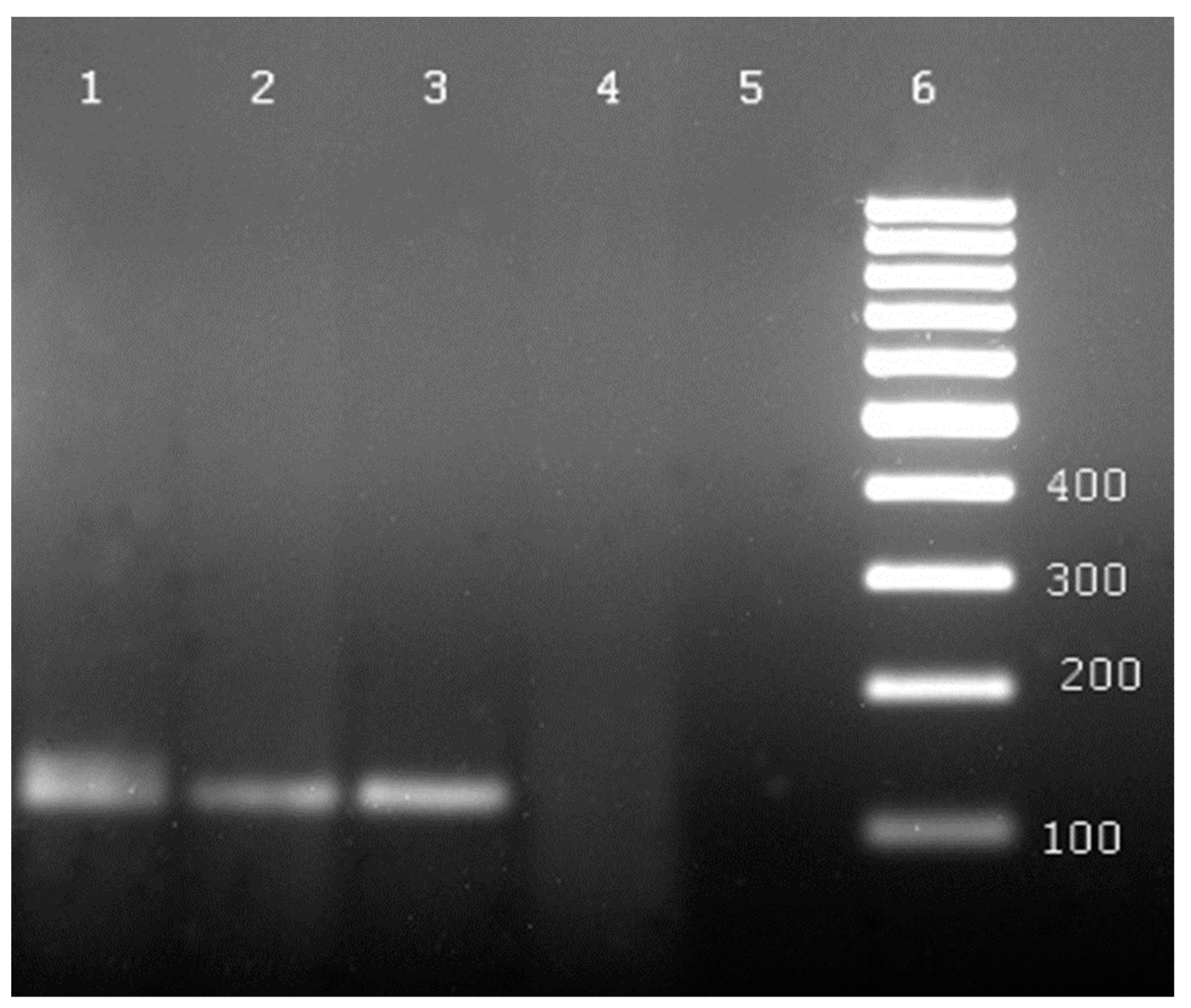
| No. | Species | Number of Plants Tested with: | ||
|---|---|---|---|---|
| PVY IUNG 21 | PVY IUNG 20 | TSWV | ||
| 1 | N. mutabilis | 20 | 20 | 40 |
| 2 | N. alata | 5 | 5 | 10 |
| 3 | N. forgetiana | 5 | 5 | 10 |
| 4 | N. tabacum cv. VAM | 5 | 5 | 10 |
| 5 | N. tabacum cv. Burley 21 | 5 | 5 | 10 |
| Species | Presence of Marker Nsyl-elF4E1 | PVY Isolate | |||
|---|---|---|---|---|---|
| IUNG 21 (Mild) | IUNG 20 (Severe) | ||||
| Symptoms | DAS-ELISA Results | Symptoms | DAS-ELISA Results | ||
| Nicotiana mutabilis | − | VC | + | VC | + |
| Nicotiana alata | − | VC | + | VC | + |
| Nicotiana forgetiana | − | VC, WL | + | VC, WL, MS | + |
| Nicotiana tabacum cv. VAM | − | ns | − | VN | + |
| Nicotiana tabacum cv. Burley 21 | + | VN | + | VN | + |
| Species | Presence of Marker ACC/CCC 172 | Time of Sampling | Leaves | No. of ELISA-Positive/Tested Plants | Symptoms |
|---|---|---|---|---|---|
| Nicotian mutabilis | + | I | lower | 39/40 | NeS |
| middle | 30/40 | CS/NeS | |||
| upper | 10/40 | CS/NeS | |||
| II | middle | 31/40 | CS/NeS | ||
| upper | 5/40 | CS/NeS | |||
| III | upper | 12/40 | CS/NeS | ||
| Nicotiana alata | + | I | lower | 10/10 | NeS |
| middle | 0/10 | ns | |||
| upper | 0/10 | ns | |||
| II | middle | 0/10 | ns | ||
| upper | 0/10 | ns | |||
| III | upper | 0/10 | ns | ||
| Nicotian forgetiana | + | I | lower | 10/10 | NeS |
| middle | 10/10 | WL | |||
| upper | 10/10 | St | |||
| II | middle | 10/10 | WL | ||
| upper | 10/10 | St | |||
| III | upper | 10/10 | St | ||
| Nicotiana tabacum cv. VAM | − | I | lower | 10/10 | VC/CS |
| middle | 10/10 | VC/CS | |||
| upper | 10/10 | St CA | |||
| II | middle | 10/10 | VC/CS | ||
| upper | 10/10 | St CA | |||
| III | upper | 10/10 | St CA | ||
| Nicotiana tabacum cv. Burley 21 | − | I | lower | 10/10 | VC/CS |
| middle | 10/10 | VC/CS | |||
| upper | 10/10 | St CA | |||
| II | middle | 10/10 | VC/CS | ||
| upper | 10/10 | St CA | |||
| III | upper | 10/10 | St CA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Depta, A.; Doroszewska, T.; Czubacka, A. Resistance Response of the Recently Discovered Species Nicotiana mutabilis to Potato virus Y (PVY) and Tomato spotted wilt virus (TSWV) Compared to Other Sources of Resistance. Agronomy 2021, 11, 1617. https://doi.org/10.3390/agronomy11081617
Depta A, Doroszewska T, Czubacka A. Resistance Response of the Recently Discovered Species Nicotiana mutabilis to Potato virus Y (PVY) and Tomato spotted wilt virus (TSWV) Compared to Other Sources of Resistance. Agronomy. 2021; 11(8):1617. https://doi.org/10.3390/agronomy11081617
Chicago/Turabian StyleDepta, Anna, Teresa Doroszewska, and Anna Czubacka. 2021. "Resistance Response of the Recently Discovered Species Nicotiana mutabilis to Potato virus Y (PVY) and Tomato spotted wilt virus (TSWV) Compared to Other Sources of Resistance" Agronomy 11, no. 8: 1617. https://doi.org/10.3390/agronomy11081617
APA StyleDepta, A., Doroszewska, T., & Czubacka, A. (2021). Resistance Response of the Recently Discovered Species Nicotiana mutabilis to Potato virus Y (PVY) and Tomato spotted wilt virus (TSWV) Compared to Other Sources of Resistance. Agronomy, 11(8), 1617. https://doi.org/10.3390/agronomy11081617






