Next-Generation Proteomics Reveals a Greater Antioxidative Response to Drought in Coffea arabica Than in Coffea canephora
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Water Stress Gradual Exposure and Leaf Water Status
2.3. Protein Extraction and Digestion
2.4. Nano Liquid Chromatography Coupled to High Resolution Mass Spectrometry
2.5. Protein Identification and Label Free Quantification
2.6. Identification of Differentially Abundant Proteins to Drought
2.7. Enriched Gene Ontologies, KEGG Pathways and Protein Networks upon Drought
3. Results
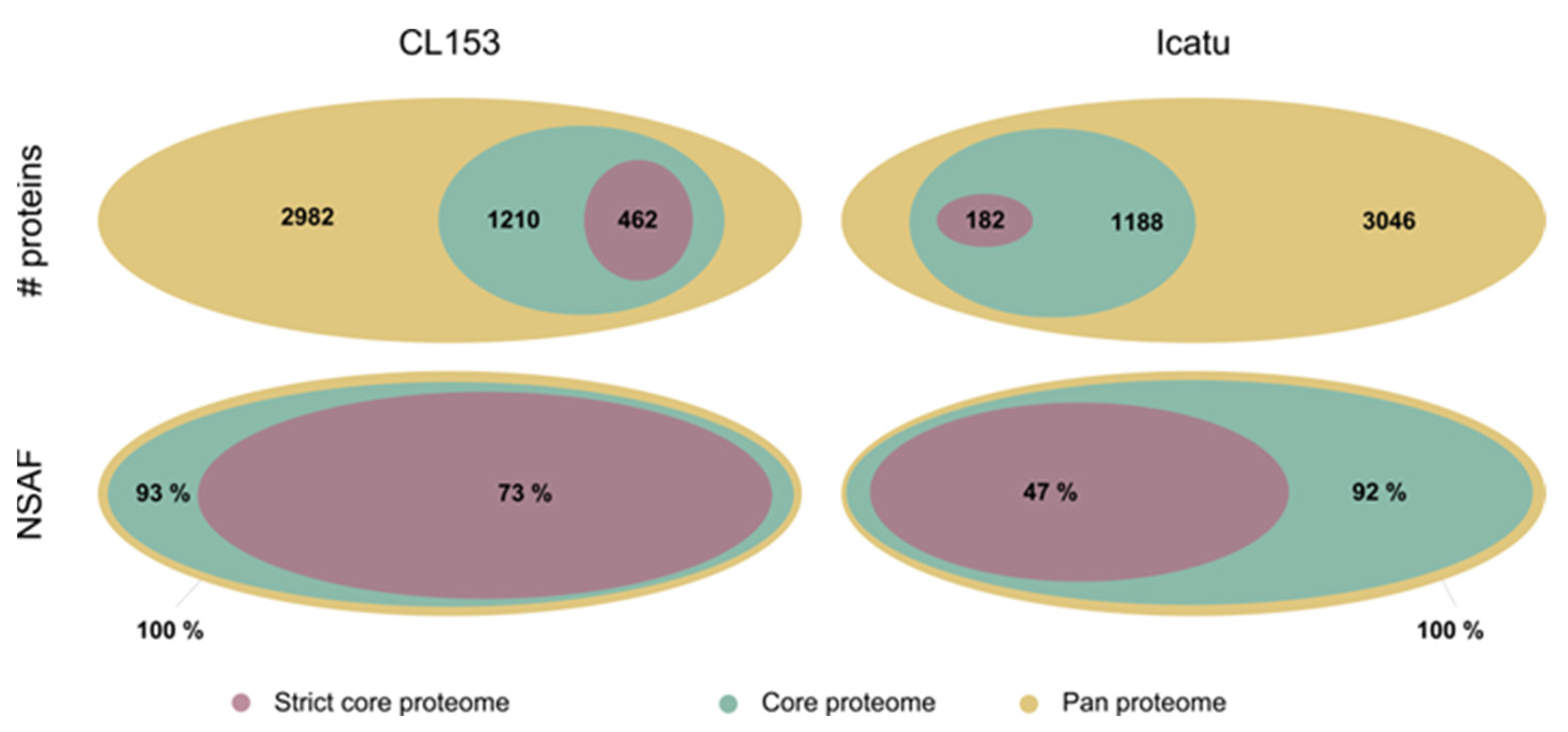
3.1. Main Proteome Features of Coffea canephora and Coffea arabica Regardless of Water Availability
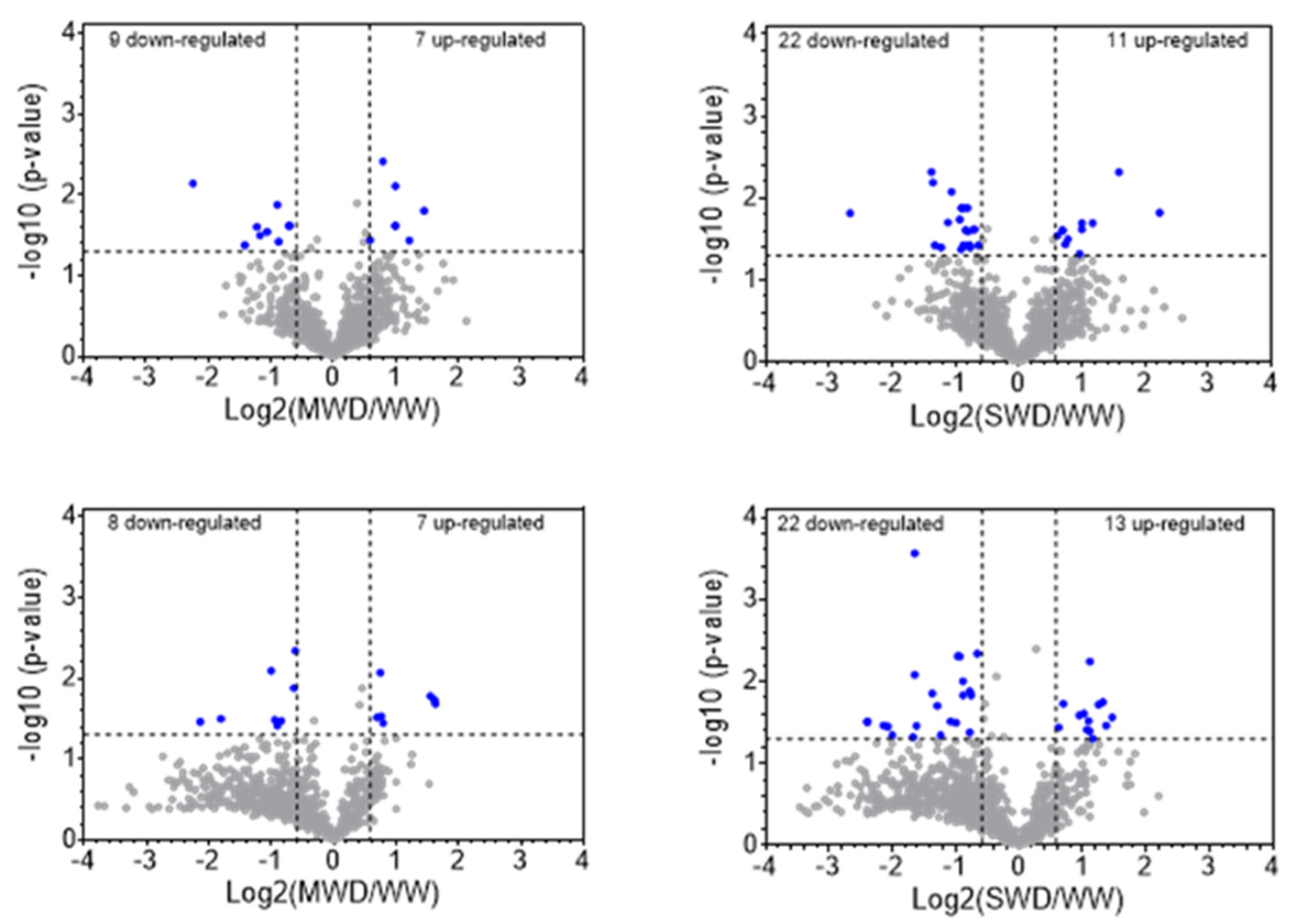
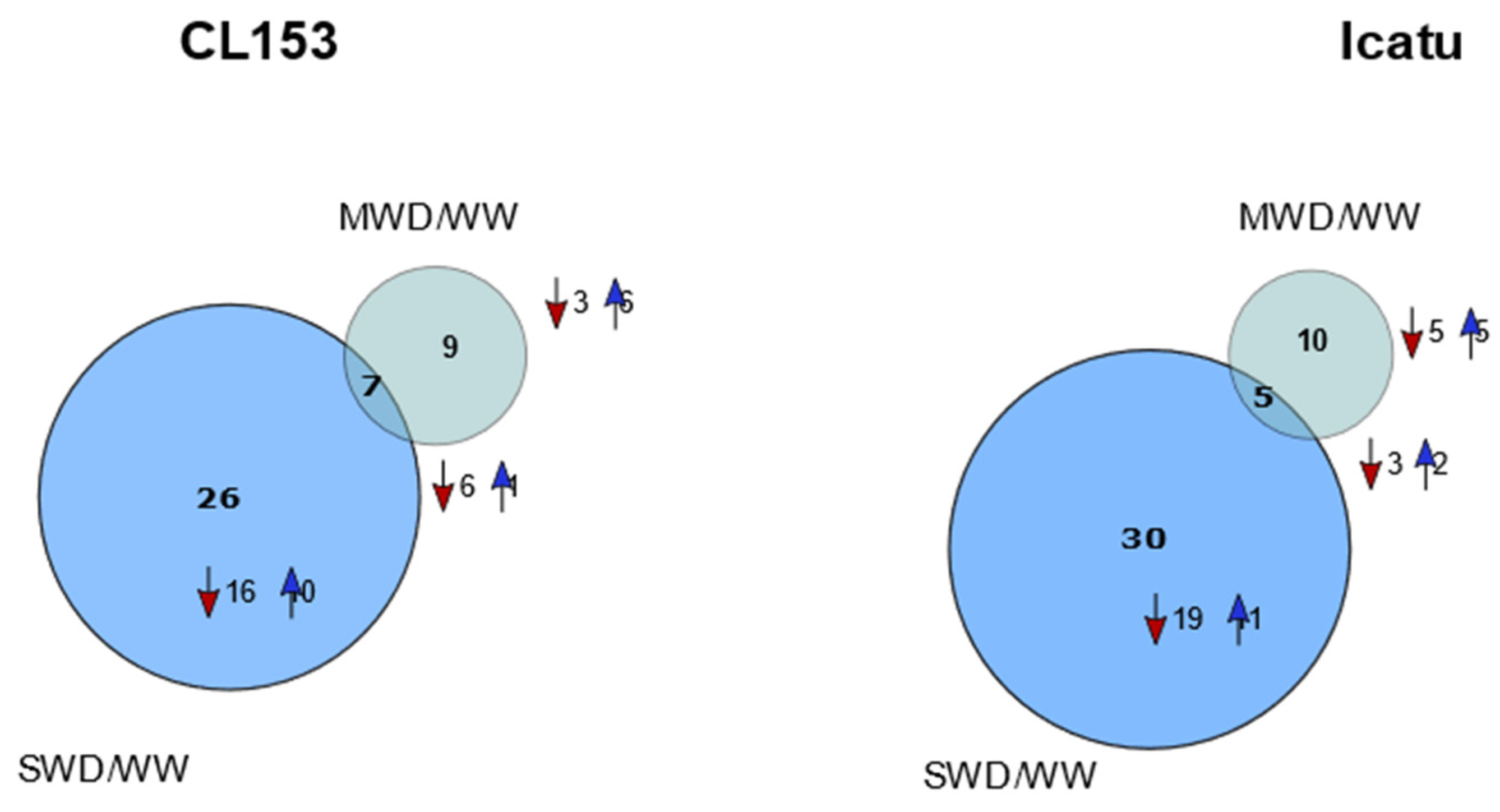
3.2. Altered Protein Abundance in Response to Water Deficit Severity
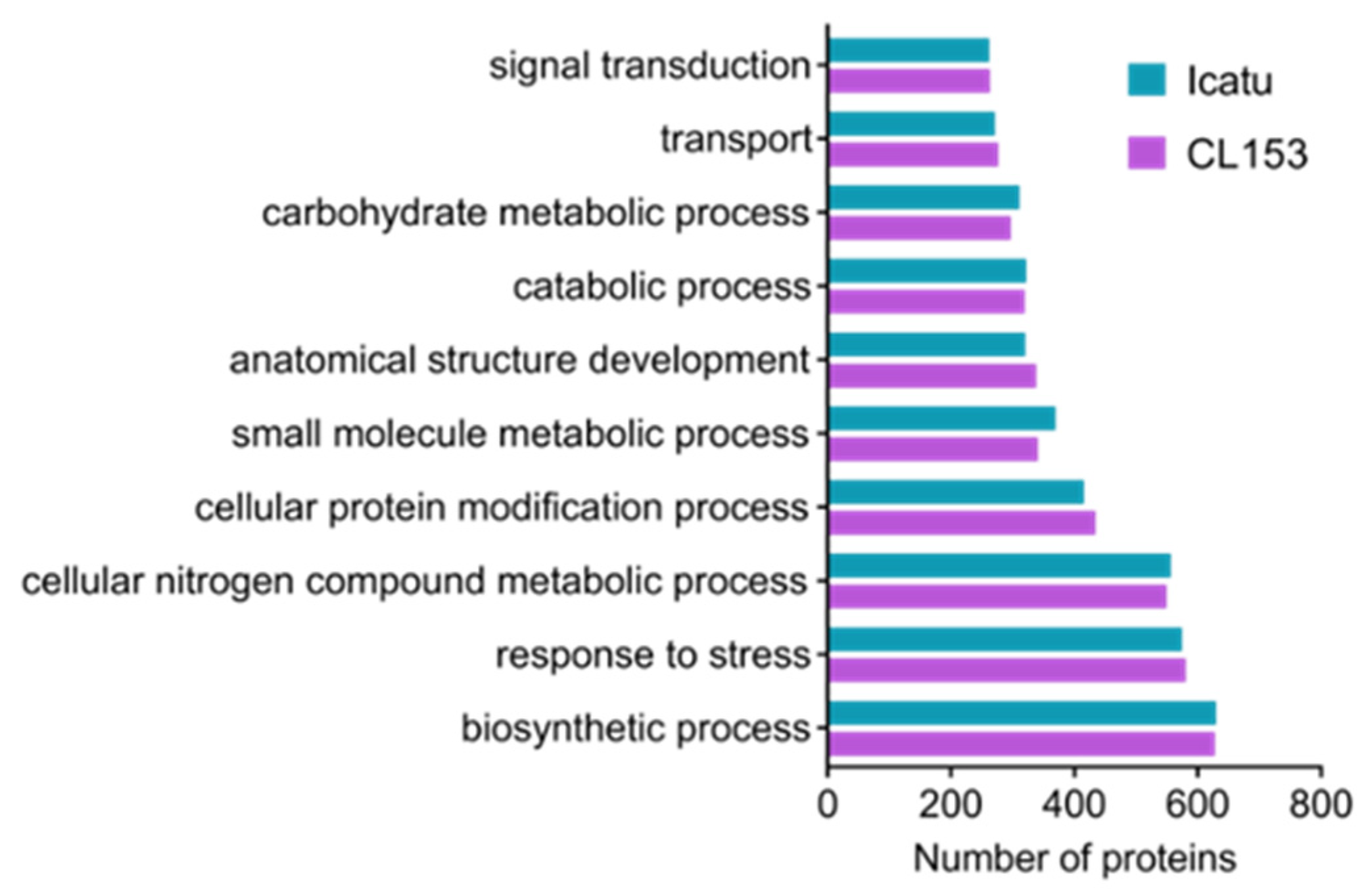
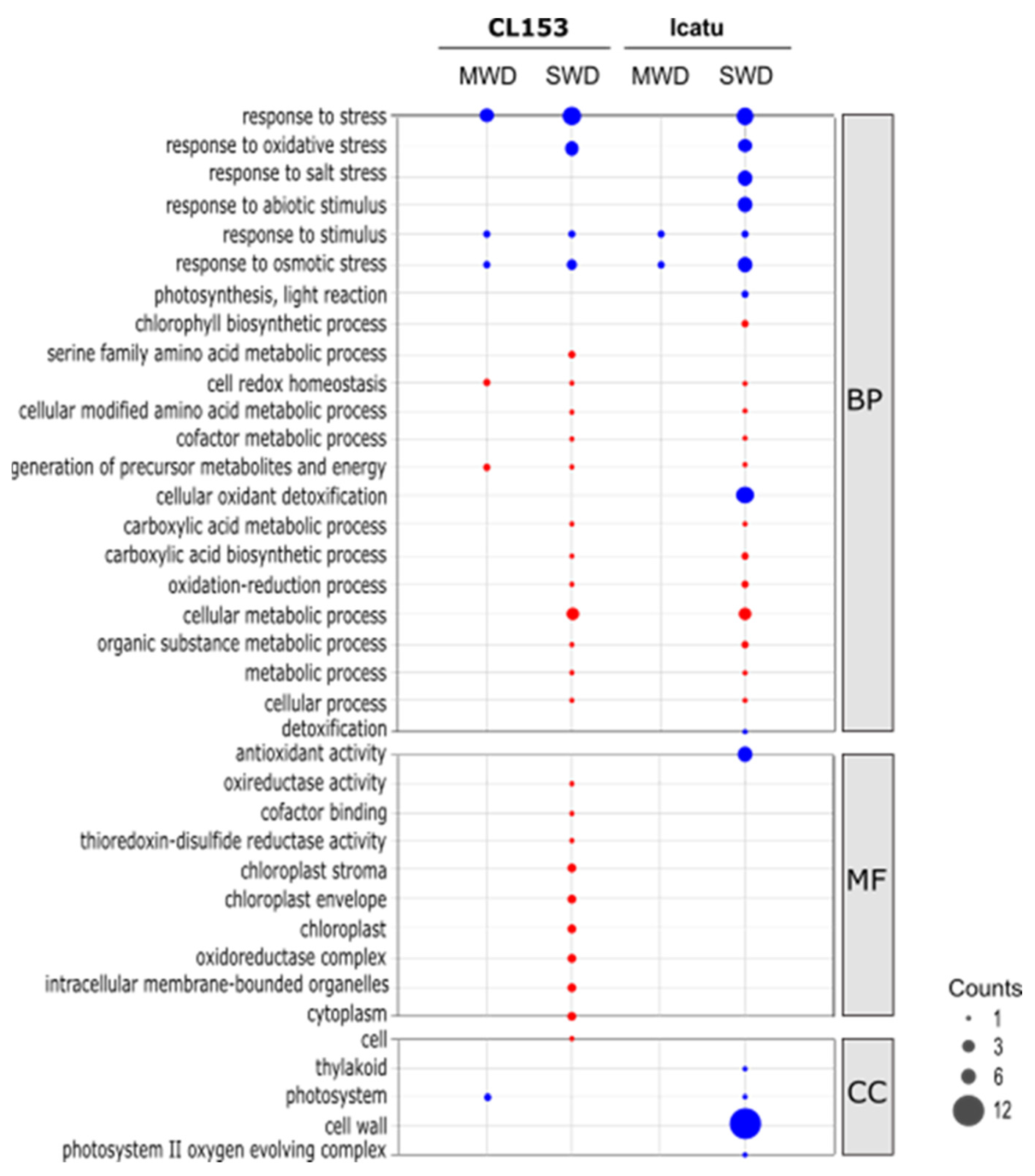
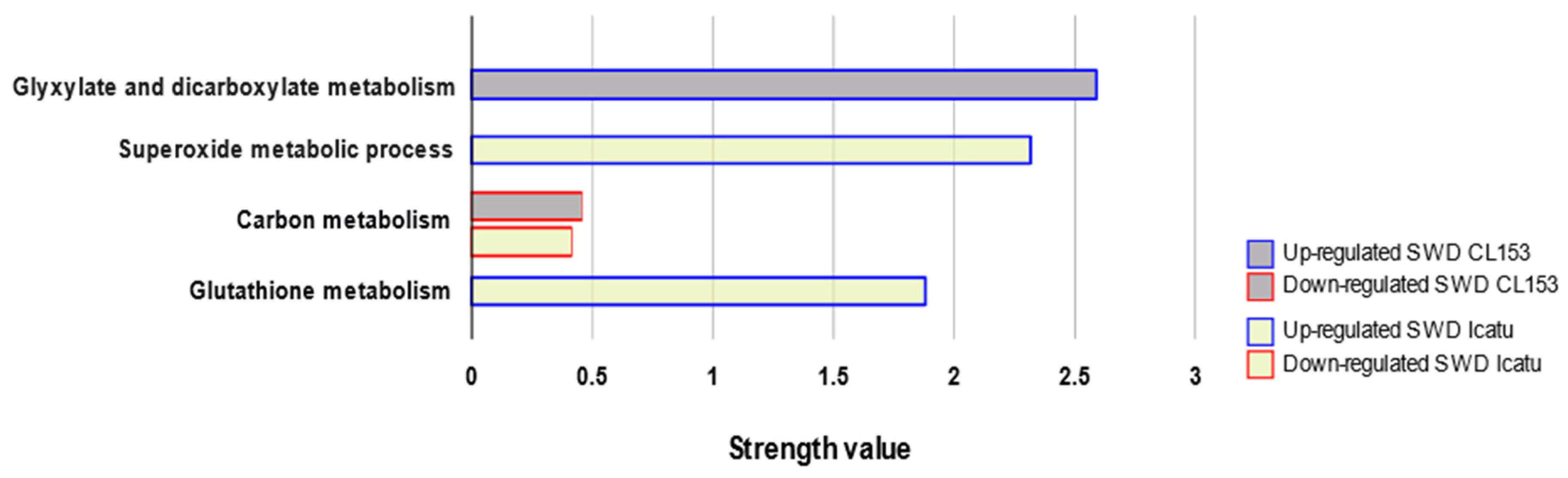
3.3. Functional Characterization of Proteomic Responses to Water Deficit Severity
4. Discussion
4.1. The Global Coffee Proteome
4.2. Differential Proteome Responses to Water Deficit of C. canephora and C. arabica Genotypes
4.3. Functional Characterization of Coffee Proteome Responses to Water Deficit
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ashraf, M.; Akram, N.A.; Al-Qurainy, F.; Foolad, M.R. Drought Tolerance. Roles of Organic Osmolytes, Growth Regulators, and Mineral Nutrients. Adv. Agron. 2011, 111, 249–296. [Google Scholar] [CrossRef]
- Akram, N.A.; Ashraf, M. Regulation in plant stress tolerance by a potential plant growth regulator, 5-aminolevulinic acid. J. Plant Growth Regul. 2013, 32, 663–679. [Google Scholar] [CrossRef]
- Mariotte, P.; Vandenberghe, C.; Kardol, P.; Hagedorn, F.; Buttler, A. Subordinate plant species enhance community resistance against drought in semi-natural grasslands. J. Ecol. 2013, 101, 763–773. [Google Scholar] [CrossRef]
- IPCC. Intergovernmental Panel on Climate Change. In Climate Change 2014: Mitigation of Climate Change. Contribution of Working Group III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Edenhofer, O., Pichs-Madruga, R., Sokona, Y., Farahani, E., Kadner, S., Seyboth, K., Adler, A., Baum, I., Brunner, S., Eickemeier, P., et al., Eds.; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2014. [Google Scholar]
- IPCC—Intergovernmental Panel on Climate Change. Summary for Policymakers; Masson-Delmotte, V., Zhai, P., Pörtner, H.O., Roberts, D., Skea, J., Shukla, P.R., Pirani, A., Moufouma-Okia, W., Péan, C., Pidcock, R., et al., Eds.; World Meteorological Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Lima, A.L.S.; DaMatta, F.M.; Pinheiro, H.A.; Totola, M.R.; Loureiro, M.E. Photochemical responses and oxidative stress in two clones of Coffea canephora under water deficit conditions. Environ. Exp. Bot. 2002, 47, 239–247. [Google Scholar] [CrossRef]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef] [Green Version]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009, 29, 185–212. [Google Scholar] [CrossRef] [Green Version]
- Ramalho, J.C.; DaMatta, F.M.; Rodrigues, A.P.; Scotti-Campos, P.; Pais, I.; Batista-Santos, P.; Partelli, F.L.; Ribeiro, A.; Lidon, F.C.; Leitão, A.E. Cold impact and acclimation response of Coffea spp. plants. Theor. Exp. Plant Physiol. 2014, 26, 5–18. [Google Scholar] [CrossRef]
- Grant, O.M.; Tronina, Ł.; Ramalho, J.C.; Kurz Besson, C.; Lobo-Do-Vale, R.; Santos Pereira, J.; Jones, H.G.; Chaves, M.M. The impact of drought on leaf physiology of Quercus suber L. trees: Comparison of an extreme drought event with chronic rainfall reduction. J. Exp. Bot. 2010, 61, 4361–4371. [Google Scholar] [CrossRef] [Green Version]
- Matos, M.C.; Campos, P.S.; Ramalho, J.C.; Medeira, M.C.; Maia, M.I.; Semedo, J.M.; Marques, N.M.; Matos, A. Photosynthetic activity and cellular integrity of the Andean legume Pachyrhizus ahipa (Wedd.) Parodi under heat and water stress. Photosynthetica 2002, 40, 493–501. [Google Scholar] [CrossRef]
- Chaves, M.M.; Maroco, J.P.; Pereira, J.S. Understanding plant responses to drought - from genes to the whole plant. Funct. Plant Biol. 2003, 30, 239–264. [Google Scholar] [CrossRef]
- Muller, B.; Pantin, F.; Génard, M.; Turc, O.; Freixes, S.; Piques, M.; Gibon, Y. Water deficits uncouple growth from photosynthesis, increase C content, and modify the relationships between C and growth in sink organs. J. Exp. Bot. 2011, 62, 1715–1729. [Google Scholar] [CrossRef] [Green Version]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S.; et al. Crop production under drought and heat stress: Plant responses and management options. Front. Plant Sci. 2017, 8, 1147. [Google Scholar] [CrossRef] [Green Version]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Guo, C.; Hussain, S.; Zhu, B.; Deng, F.; Xue, Y.; Geng, M.; Wu, L. Role of xylo-oligosaccharides in protection against salinity-induced adversities in Chinese cabbage. Environ. Sci. Pollut. Res. 2016, 23, 1254–1264. [Google Scholar] [CrossRef]
- Hussain, S.; Khan, F.; Cao, W.; Wu, L.; Geng, M. Seed priming alters the production and detoxification of reactive oxygen intermediates in rice seedlings grown under sub-optimal temperature and nutrient supply. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Miyake, C.; Okamura, M. Cyclic electron flow within PSII protects PSII from its photoinhibition in thylakoid membranes from spinach chloroplasts. Plant Cell Physiol. 2003, 44, 457–462. [Google Scholar] [CrossRef] [Green Version]
- Dubberstein, D.; Lidon, F.C.; Rodrigues, A.P.; Semedo, J.N.; Marques, I.; Rodrigues, W.P.; Gouveia, D.; Armengaud, J.; Semedo, M.C.; Martins, S.; et al. Resilient and sensitive key points of the photosynthetic machinery of Coffea spp. to the single and superimposed exposure to severe drought and heat stresses. Front. Plant Sci. 2020, 11, 1049. [Google Scholar] [CrossRef]
- Semedo, J.N.; Rodrigues, A.P.; Lidon, F.C.; Pais, I.P.; Marques, I.; Gouveia, D.; Armengaud, J.; Silva, M.J.; Martins, S.; Semedo, M.C.; et al. Intrinsic non-stomatal resilience to drought of the photosynthetic apparatus in Coffea spp. is strengthened by elevated air [CO2]. Tree Physiol. 2021, 41, 708–727. [Google Scholar] [CrossRef]
- Chaves, M.M.; Oliveira, M.M. Mechanisms underlying plant resilience to water deficits: Prospects for water-saving agriculture. J. Exp. Bot. 2004, 55, 2365–2384. [Google Scholar] [CrossRef] [Green Version]
- Hasan, M.M.U.; Ma, F.; Prodhan, Z.H.; Li, F.; Shen, H.; Chen, Y.; Wang, X. Molecular and physio-biochemical characterization of cotton species for assessing drought stress tolerance. Int. J. Mol. Sci. 2018, 19, 2636. [Google Scholar] [CrossRef] [Green Version]
- Gouveia, D.; Grenga, L.; Pible, O.; Armengaud, J. Quick microbial molecular phenotyping by differential shotgun proteomics. Environ. Microbiol. 2020, 22, 2996–3004. [Google Scholar] [CrossRef]
- Ford, K.L.; Cassin, A.; Bacic, A. Quantitative proteomic analysis of wheat cultivars with differing drought stress tolerance. Front. Plant Sci. 2011, 2, 44. [Google Scholar] [CrossRef] [Green Version]
- Hamzelou, S.; Pascovici, D.; Kamath, K.S.; Amirkhani, A.; McKay, M.; Mirzaei, M.; Atwell, B.J.; Haynes, P.A. Proteomic responses to drought vary widely among eight diverse genotypes of rice (Oryza sativa). Int. J. Mol. Sci. 2020, 21, 363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishnan, S.; Ranker, T.A.; Davis, A.P.; Rakotomalala, J.J. An assessment of the genetic integrity of ex situ germplasm collections of three endangered species of Coffea from Madagascar: Implications for the management of field germplasm collections. Genet. Resour. Crop Evol. 2013, 60, 1021–1036. [Google Scholar] [CrossRef]
- DaMatta, F.M.; Ramalho, J.D.C. Impacts of drought and temperature stress on coffee physiology and production: A review. Braz. J. Plant Physiol. 2006, 18, 55–81. [Google Scholar] [CrossRef]
- DaMatta, F.M.; Avila, R.T.; Cardoso, A.A.; Martins, S.C.V.; Ramalho, J.C. Physiological and agronomic performance of the coffee crop in the context of climate change and global warming: A review. J. Agric. Food Chem. 2018, 66, 5264–5274. [Google Scholar] [CrossRef] [PubMed]
- Marques, I.; Fernandes, I.; Paulo, O.S.; Lidon, F.C.; DaMatta, F.M.; Ramalho, J.C.; Ribeiro-Barros, A.I. A transcriptomic approach to understanding the combined impacts of supra-optimal temperatures and CO2 revealed different responses in the polyploid Coffea arabica and its diploid progenitor C. canephora. Int. J. Mol. Sci. 2021, 22, 3125. [Google Scholar] [CrossRef]
- DaMatta, F.M.; Chaves, A.R.M.; Pinheiro, H.A.; Ducatti, C.; Loureiro, M.E. Drought tolerance of two field-grown clones of Coffea canephora. Plant Sci. 2003, 164, 111–117. [Google Scholar] [CrossRef]
- Dias, P.C.; Araujo, W.L.; Moraes, G.A.B.K.; Barros, R.S.; DaMatta, F.M. Morphological and physiological responses of two coffee progenies to soil water availability. J. Plant Physiol. 2007, 164, 1639–1647. [Google Scholar] [CrossRef]
- De Oliveira Santos, M.; Coelho, L.S.; Carvalho, G.R.; Botelho, C.E.; Torres, L.F.; Vilela, D.J.M.; Andrade, A.C.; Silva, V.A. Photochemical efficiency correlated with candidate gene expression promote coffee drought tolerance. Sci. Rep. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, A.M.; Jorge, T.; Osorio, S.; Pott, D.M.; Lidon, F.C.; DaMatta, F.M.; Marques, I.; Ribeiro-Barros, A.I.; Ramalho, J.C.; António, C. Primary metabolite profile changes in Coffea spp. promoted by single and combined exposure to drought and elevated [CO2]. Metabolites 2021, 11, 427. [Google Scholar] [CrossRef] [PubMed]
- Scotti-Campos, P.; Pais, I.P.; Partelli, F.L.; Batista-Santos, P.; Ramalho, J.C. Phospholipids profile in chloroplasts of Coffea spp. genotypes differing in cold acclimation ability. J. Plant Physiol. 2014, 171, 243–249. [Google Scholar] [CrossRef]
- Scotti-Campos, P.; Pais, I.P.; Ribeiro-Barros, A.I.; Martins, L.D.; Tomaz, M.A.; Rodrigues, W.P.; Campostrini, E.; Semedo, J.N.; Fortunato, A.S.; Martins, M.Q.; et al. Lipid profile adjustments may contribute to warming acclimation and to heat impact mitigation by elevated [CO2] in Coffea spp. Environ. Exp. Bot. 2019, 167, 103856. [Google Scholar] [CrossRef]
- Partelli, F.L.; Batista-Santos, P.; Scotti-Campos, P.; Pais, I.P.; Quartin, V.L.; Vieira, H.D.; Ramalho, J.C. Characterization of the main lipid components of chloroplast membranes and cold induced changes in Coffea spp. Environ. Exp. Bot. 2011, 74, 194–204. [Google Scholar] [CrossRef]
- DaMatta, F.M.; Rahn, E.; Läderach, P.; Ghini, R.; Ramalho, J.C. Why could the coffee crop endure climate change and global warming to a greater extent than previously estimated? Clim. Chang. 2019, 152, 167–178. [Google Scholar] [CrossRef] [Green Version]
- Martins, M.Q.; Rodrigues, W.P.; Fortunato, A.S.; Leitão, A.E.; Rodrigues, A.P.; Pais, I.P.; Martins, L.D.; Silva, M.J.; Reboredo, F.H.; Partelli, F.L.; et al. Protective response mechanisms to heat stress in interaction with high [CO2] conditions in Coffea spp. Front. Plant Sci. 2016, 7, 947. [Google Scholar] [CrossRef] [Green Version]
- Crisosto, C.H.; Grantz, D.A.; Meinzer, F.C. Effects of water deficit on flower opening in coffee (Coffea arabica L.). Tree Physiol. 1992, 10, 127–139. [Google Scholar] [CrossRef]
- De Camargo, Â.P.; De Camargo, M.B.P. Definition and outline for the phenological phases of arabic coffee under Brazilian tropical conditions. Bragantia 2001, 60, 65–68. [Google Scholar] [CrossRef] [Green Version]
- CONAB. Série Histórica das Safras. 2017. Available online: https://portaldeinformacoes.conab.gov.br/index.php/safras/cafe-serie-historic (accessed on 10 November 2021).
- Semedo, J.N.; Rodrigues, W.P.; Dubberstein, D.; Martins, M.Q.; Martins, L.D.; Pais, I.P.; Rodrigues, A.P.; Leitão, A.E.; Partelli, F.L.; Campostrini, E.; et al. Coffee responses to drought, warming and high [CO2] in a context of future climate change scenarios. In Climate Change Management; Springer: Berlin/Heidelberg, Germany, 2018; pp. 465–477. [Google Scholar] [CrossRef]
- Ferrão, R.G.; Fonseca, A.F.A.; Ferrão, M.A.G.; Bragança, S.M.; Verdin Filho, A.C.; Volpi, P.S. Cultivares de café Conilon. In Café Conilon; Ferrão, R.G., Fonseca, A.F.A., Bragança, S.M., Ferrão, M.A.G., Muner, L.H., Eds.; Incaper: Linhares, Brazil, 2007; Chapter 7; pp. 203–225. ISBN 978-85-89274-12-8. [Google Scholar]
- Fernandes, I.; Marques, I.; Paulo, O.S.; Batista, D.; Partelli, F.L.; Lidon, F.C.; DaMatta, F.M.; Ramalho, J.C.; Ribeiro-Barros, A.I. Understanding the Impact of Drought in Coffea Genotypes: Transcriptomic Analysis Supports a Common High Resilience to Moderate Water Deficit but a Genotype Dependent Sensitivity to Severe Water Deficit. Agronomy 2021, 11, 2255. [Google Scholar] [CrossRef]
- Ramalho, J.C.; Rodrigues, A.P.; Semedo, J.N.; Pais, I.P.; Martins, L.D.; Simões-Costa, M.C.; Leitão, A.E.; Fortunato, A.S.; Batista-Santos, P.; Palos, I.M.; et al. Sustained photosynthetic performance of Coffea spp. under long-term enhanced [CO2]. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramalho, J.C.; Rodrigues, A.P.; Lidon, F.C.; Marques, L.M.C.; Leitão, A.E.; Fortunato, A.S.; Pais, I.P.; Silva, M.J.; Scotti-Campos, P.; Lopes, A.; et al. Stress cross-response of the antioxidative system promoted by superimposed drought and cold conditions in Coffea spp. PLoS ONE 2018, 13, e0198694. [Google Scholar] [CrossRef]
- Parkhey, S.; Chandrakar, V.; Naithani, S.C.; Keshavkant, S. Efficient extraction of proteins from recalcitrant plant tissue for subsequent analysis by two-dimensional gel electrophoresis. J. Sep. Sci. 2015, 38, 3622–3628. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M. A Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Hartmann, E.M.; Allain, F.; Gaillard, J.C.; Pible, O.; Armengaud, J. Taking the shortcut for high-throughput shotgun proteomic analysis of bacteria. Methods Mol. Biol. 2014, 1197, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Klein, G.; Mathé, C.; Biola-Clier, M.; Devineau, S.; Drouineau, E.; Hatem, E.; Marichal, L.; Alonso, B.; Gaillard, J.C.; Lagniel, G.; et al. RNA-binding proteins are a major target of silica nanoparticles in cell extracts. Nanotoxicology 2016, 10, 1555–1564. [Google Scholar] [CrossRef] [Green Version]
- Denoeud, F.; Carretero-Paulet, L.; Dereeper, A.; Droc, G.; Guyot, R.; Pietrella, M.; Zheng, C.; Alberti, A.; Anthony, F.; Aprea, G.; et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 2014, 345, 1181–1184. [Google Scholar] [CrossRef] [Green Version]
- Christie-Oleza, J.A.; Fernandez, B.; Nogales, B.; Bosch, R.; Armengaud, J. Proteomic insights into the lifestyle of an environmentally relevant marine bacterium. ISME J. 2012, 6, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Hulsen, T.; de Vlieg, J.; Alkema, W. BioVenn—A web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genom. 2008, 9, 488. [Google Scholar] [CrossRef] [Green Version]
- Otasek, D.; Morris, J.H.; Bouças, J.; Pico, A.R.; Demchak, B. Cytoscape Automation: Empowering workflow-based network analysis. Genome Biol. 2019, 20, 185. [Google Scholar] [CrossRef] [Green Version]
- Armengaud, J.; Trapp, J.; Pible, O.; Geffard, O.; Chaumot, A.; Hartmann, E.M. Non-model organisms, a species endangered by proteogenomics. J. Proteom. 2014, 105, 5–18. [Google Scholar] [CrossRef]
- Cao, Y.; Luo, Q.; Tian, Y.; Meng, F. Physiological and proteomic analyses of the drought stress response in Amygdalus mira (Koehne) Yü et Lu roots. BMC Plant Biol. 2017, 17, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boguszewska-Mańkowska, D.; Gietler, M.; Nykiel, M. Comparative proteomic analysis of drought and high temperature response in roots of two potato cultivars. Plant Growth Regul. 2020, 92, 345–363. [Google Scholar] [CrossRef]
- Mann, K.; Mann, M. In-depth analysis of the chicken egg white proteome using an LTQ Orbitrap Velos. Proteome Sci. 2011, 9, 7. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, G.K.; Sarkar, A.; Righetti, P.G.; Pedreschi, R.; Carpentier, S.; Wang, T.; Barkla, B.J.; Kohli, A.; Ndimba, B.K.; Bykova, N.V.; et al. A decade of plant proteomics and mass spectrometry: Translation of technical advancements to food security and safety issues. Mass Spectrom. Rev. 2013, 32, 335–365. [Google Scholar] [CrossRef] [Green Version]
- Casas-Vila, N.; Bluhm, A.; Sayols, S.; Dinges, N.; Dejung, M.; Altenhein, T.; Kappei, D.; Altenhein, B.; Roignant, J.Y.; Butter, F. The developmental proteome of Drosophila melanogaster. Genome Res. 2017, 27, 1273–1285. [Google Scholar] [CrossRef] [Green Version]
- Scalabrin, S.; Toniutti, L.; Di Gaspero, G.; Scaglione, D.; Magris, G.; Vidotto, M.; Pinosio, S.; Cattonaro, F.; Magni, F.; Jurman, I.; et al. A single polyploidization event at the origin of the tetraploid genome of Coffea arabica is responsible for the extremely low genetic variation in wild and cultivated germplasm. Sci. Rep. 2020, 10, 65. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, W.P.; Martins, M.Q.; Fortunato, A.S.; Rodrigues, A.P.; Semedo, J.N.; Simões-Costa, M.C.; Pais, I.P.; Leitão, A.E.; Colwell, F.; Goulao, L.; et al. Long-term elevated air [CO2] strengthens photosynthetic functioning and mitigates the impact of supra-optimal temperatures in tropical Coffea arabica and C. canephora species. Glob. Chang. Biol. 2016, 22, 415–431. [Google Scholar] [CrossRef]
- Marques, I.; Fernandes, I.; David, P.H.C.; Paulo, O.S.; Goulao, L.F.; Fortunato, A.S.; Lidon, F.C.; DaMatta, F.M.; Ramalho, J.C.; Ribeiro-Barros, A.I. Transcriptomic leaf profiling reveals differential responses of the two most traded coffee species to elevated [CO2]. Int. J. Mol. Sci. 2020, 21, 9211. [Google Scholar] [CrossRef]
- Salehin, M.; Li, B.; Tang, M.; Katz, E.; Song, L.; Ecker, J.R.; Kliebenstein, D.J.; Estelle, M. Auxin-sensitive Aux/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels. Nat. Commun. 2019, 10, 4021. [Google Scholar] [CrossRef] [Green Version]
- Zhu, B.; Coleman, G.D. Phytochrome-mediated photoperiod perception, shoot growth, glutamine, calcium, and protein phosphorylation influence the activity of the poplar bark storage protein gene promoter (bspA). Plant Physiol. 2001, 126, 342–351. [Google Scholar] [CrossRef] [Green Version]
- Pettengill, E.A.; Pettengill, J.B.; Coleman, G.D. Elucidating the evolutionary history and expression patterns of nucleoside phosphorylase paralogs (vegetative storage proteins) in Populus and the plant kingdom. BMC Plant Biol. 2013, 13, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skorupa, M.; Gołȩbiewski, M.; Kurnik, K.; Niedojadło, J.; Kȩsy, J.; Klamkowski, K.; Wójcik, K.; Treder, W.; Tretyn, A.; Tyburski, J. Salt stress vs. salt shock—The case of sugar beet and its halophytic ancestor. BMC Plant Biol. 2019, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sultana, N.; Islam, S.; Juhasz, A.; Yang, R.; She, M.; Alhabbar, Z.; Zhang, J.; Ma, W. Transcriptomic study for identification of major nitrogen stress responsive genes in australian bread wheat cultivars. Front. Genet. 2020, 11. [Google Scholar] [CrossRef]
- Xiong, D.; Yu, T.; Liu, X.; Li, Y.; Peng, S.; Huang, J. Heterogeneity of photosynthesis within leaves is associated with alteration of leaf structural features and leaf N content per leaf area in rice. Funct. Plant Biol. 2015, 42, 687–696. [Google Scholar] [CrossRef]
- Gojon, A.; Nacry, P.; Davidian, J.C. Root uptake regulation: A central process for NPS homeostasis in plants. Curr. Opin. Plant Biol. 2009, 12, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.Z.; Tian, W.H.; Liu, X.X.; Lin, X.Y.; Jin, C.W.; Zheng, S.J. Alleviation of proton toxicity by nitrate uptake specifically depends on nitrate transporter 1.1 in Arabidopsis. New Phytol. 2016, 211, 149–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Kim, S.G.; Kang, K.Y.; Kim, J.G.; Park, S.R.; Gupta, R.; Kim, Y.H.; Wang, Y.; Kim, S.T. Overexpression of a pathogenesis-related protein 10 enhances biotic and abiotic stress tolerance in rice. Plant Pathol. J. 2016, 32, 552–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akbudak, M.A.; Yildiz, S.; Filiz, E. Pathogenesis related protein-1 (PR-1) genes in tomato (Solanum lycopersicum L.): Bioinformatics analyses and expression profiles in response to drought stress. Genomics 2020, 112, 4089–4099. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Mao, X.; Wang, R.; Li, A.; Zhao, G.; Zhao, J.; Jing, R. Identification of wheat stress-responding genes and TaPR-1-1 function by screening a cDNA yeast library prepared following abiotic stress. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Chakhchar, A.; Wahbi, S.; Lamaoui, M.; Ferradous, A.; El Mousadik, A.; Ibnsouda-Koraichi, S.; Filali-Maltouf, A.; El Modafar, C. Physiological and biochemical traits of drought tolerance in Argania spinosa. J. Plant Interact. 2015, 10, 252–261. [Google Scholar] [CrossRef]
- Horn, R.; Paulsen, H. Folding in vitro of light-harvesting chlorophyll a/b protein is coupled with pigment binding. J. Mol. Biol. 2002, 318, 547–556. [Google Scholar] [CrossRef]
- Jia, Y.; Wong, D.C.J.; Sweetman, C.; Bruning, J.B.; Ford, C.M. New insights into the evolutionary history of plant sorbitol dehydrogenase. BMC Plant Biol. 2015, 15, 101. [Google Scholar] [CrossRef] [Green Version]
- Yancey, P.H.; Clark, M.E.; Hand, S.C.; Bowlus, R.D.; Somero, G.N. Living with water stress: Evolution of osmolyte systems. Science 1982, 217, 1214–1222. [Google Scholar] [CrossRef]
- Conde, A.; Regalado, A.; Rodrigues, D.; Costa, J.M.; Blumwald, E.; Chaves, M.M.; Gerós, H. Polyols in grape berry: Transport and metabolic adjustments as a physiological strategy for water-deficit stress tolerance in grapevine. J. Exp. Bot. 2015, 66, 889–906. [Google Scholar] [CrossRef] [Green Version]
- Sage, R.F.; Way, D.A.; Kubien, D.S. Rubisco, Rubisco activase, and global climate change. J. Exp. Bot. 2008, 59, 1581–1595. [Google Scholar] [CrossRef] [Green Version]
- Perdomo, J.A.; Capó-Bauçà, S.; Carmo-Silva, E.; Galmés, J. Rubisco and rubisco activase play an important role in the biochemical limitations of photosynthesis in rice, wheat, and maize under high temperature and water deficit. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, T.; Kumar, K.R.R.; Kirti, P.B. Constitutive expression of a trypsin protease inhibitor confers multiple stress tolerance in transgenic tobacco. Plant Cell Physiol. 2009, 50, 541–553. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Liu, Q.; Han, J.Y.; Gao, J.Y. Different pollinator assemblages ensure reproductive success of Cleisostoma linearilobatum (Orchidaceae) in fragmented holy hill forest and traditional tea garden. Sci. Rep. 2016, 6, 21435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Ramalho, J.C.; Campos, P.S.; Teixeira, M.; Nunes, M.A. Nitrogen dependent changes in antioxidant system and in fatty acid composition of chloroplast membranes from Coffea arabica L. plants submitted to high irradiance. Plant Sci. 1998, 135, 115–124. [Google Scholar] [CrossRef]
- Fortunato, A.S.; Lidon, F.C.; Batista-Santos, P.; Eduardo Leitão, A.; Pais, I.P.; Ribeiro, A.I.; Ramalho, J.C. Biochemical and molecular characterization of the antioxidative system of Coffea sp. under cold conditions in genotypes with contrasting tolerance. J. Plant Physiol. 2010, 167, 333–342. [Google Scholar] [CrossRef]
- Chen, T.H.H.; Murata, N. Enhancement of tolerance of abiotic stress by metabolic engineering of betaines and other compatible solutes. Curr. Opin. Plant Biol. 2002, 5, 250–257. [Google Scholar] [CrossRef]
- Yasar, F.; Uzal, O.; Tufenkci, S.; Yildiz, K. Ion accumulation in different organs of green bean genotypes grown under salt stress. Plant Soil Environ. 2006, 52, 476–480. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Zhang, M.; Guo, R.; Shi, D.; Liu, B.; Lin, X.; Yang, C. Effects of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice (Oryza sativa L.). BMC Plant Biol. 2012, 12. [Google Scholar] [CrossRef] [Green Version]
- Tomé, F.; Nägele, T.; Adamo, M.; Garg, A.; Marco-Llorca, C.; Nukarinen, E.; Pedrotti, L.; Peviani, A.; Simeunovic, A.; Tatkiewicz, A.; et al. The low energy signaling network. Front. Plant Sci. 2014, 5, 353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.L.; Ye, W.W.; Wang, J.J.; Song, L.Y.; Fan, W.L.; Cui, Y.P. Constructing SSH library of cotton under drought stress and analysis of drought associated genes. Acta Agron. Sin. 2010, 36, 2035–2044. [Google Scholar] [CrossRef]
- Voss, I.; Sunil, B.; Scheibe, R.; Raghavendra, A.S. Emerging concept for the role of photorespiration as an important part of abiotic stress response. Plant Biol. 2013, 15, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Lou, L.; Li, X.; Chen, J.; Li, Y.; Tang, Y.; Lv, J. Photosynthetic and ascorbate-glutathione metabolism in the flag leaves as compared to spikes under drought stress of winter wheat (Triticum aestivum L.). PLoS ONE 2018, 13, e0194625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruz De Carvalho, M.H. Drought stress and reactive oxygen species: Production, scavenging and signaling. Plant Signal. Behav. 2008, 3, 156–165. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, N.; Malagoli, M.; Wirtz, M.; Hell, R. Drought stress in maize causes differential acclimation responses of glutathione and sulfur metabolism in leaves and roots. BMC Plant Biol. 2016, 16, 247. [Google Scholar] [CrossRef]

| Protein Group | CL153 NSAF % | Icatu NSAF % | Biological Process |
|---|---|---|---|
| Auxin-binding protein ABP20 | 2.940 | 2.869 | auxin-activated signaling pathway |
| Putative bark storage protein A | 3.095 | 0.545 # | nucleoside metabolic process |
| Oxygen-evolving enhancer protein 1 | 1.978 | 1.868 | Photosynthesis; photosystem II stabilization |
| Photosystem I reaction center subunit II | 1.434 | 1.705 | photosynthesis |
| Oxygen-evolving enhancer protein 2 | 1.793 | 2.015 | photosynthesis |
| Histone H4 | 1.051 | 1.075 | nucleosome assembly |
| Photosystem II 10 kDa polypeptide | 1.009 | 0.678 | photosynthesis |
| Plastocyanin | 0.948 | 0.450 # | electron transport |
| Glyceraldehyde-3-phosphate dehydrogenase | 0.934 | 0.898 | glucose metabolic process |
| Ribulose bisphosphate carboxylase small chain SSU11A | 0.928 | 1.095 | carbon fixation |
| Phosphoglycerate kinase | 0.881 | 0.759 | glycolysis |
| Peroxisomal (S)-2-hydroxy-acid oxidase | 0.729 | 0.679 | metabolic process |
| Photosystem I reaction center subunit III | 0.659 | 0.748 | photosynthesis |
| Chlorophyll a–b binding protein CP29.2 | 0.654 | 0.748 | photosynthesis |
| Polyphenol oxidase I | 0.590 | 0.232 # | metabolic process |
| Carbonic anhydrase | 0.565 | 0.386 # | carbon utilization |
| Ribulose bisphosphate carboxylase/oxygenase activase 1 | 0.510 # | 0.713 | photosynthesis |
| Photosystem II 22 kDa protein | 0.472 # | 0.693 | photosynthesis |
| Chlorophyll a–b binding protein CP26 | 0.395 # | 0.626 | photosynthesis |
| Photosystem I reaction center subunit VI | 0.463 # | 0.619 | photosynthesis |
| Putative Acidic endochitinase | 0.147 # | 0.577 | carbohydrate metabolic process |
| Basic endochitinase | 0.251 # | 0.552 | carbohydrate metabolic process |
| Uncharacterized protein At2g37660 | 0.315 # | 0.495 | metabolic process |
| Quinone oxidoreductase-like protein At1g23740 | 0.478 # | 0.480 | oxidation–reduction process |
| Peptidyl-prolyl cis–trans isomerase | 0.336 # | 0.470 | protein folding |
| CL153 | log2 FC | Biological Process |
| Glycerate dehydrogenase | 0.594 | cellular response to water deprivation; oxidative photosynthetic carbon pathway |
| Glutamate–glyoxylate aminotransferase 2 | 0.798 | biosynthetic process; photorespiration |
| Carbonic anhydrase | 1.000 | carbon utilization |
| Putative Ubiquilin-1 | 1.000 | ubiquitin-dependent protein catabolic process |
| Protein-L-isoaspartate O-methyltransferase | 1.000 | protein methylation; protein repair |
| Putative UDP-rhamnose | 1.222 | carbohydrate transport; plant-type primary cell wall biogenesis |
| Pathogenesis-related protein 1B | 1.459 | Plant defense |
| Icatu | log2 FC | Biological process |
| Putative Protein tas | 0.696 | cellular response to amino acid starvation |
| Fructose-1,6-bisphosphatase | 0.746 | fructose 1,6-bisphosphate metabolic process; starch catabolic process; sucrose biosynthetic process; photosynthesis |
| ADP, ATP carrier protein 3 | 0.763 | mitochondrial transmembrane transport |
| L-ascorbate peroxidase 3 | 0.793 | response to oxidative stress |
| Ferritin-1 | 1.547 | leaf development; photosynthesis; iron ion homeostasis |
| Carotenoid 9,10(9′,10′)-cleavage dioxygenase | 1.615 | carotene, carotenoid catabolic process |
| Polyphenol oxidase I | 1.627 | ion binding |
| CL153 | log2 FC | Biological Process |
| PsbP domain-containing protein 4 | 0.613 | photosynthesis |
| Histone H2B | 0.692 | nucleosome assembly |
| NAD-dependent malic enzyme 62 kDa isoform | 0.692 | malate metabolic process; pyruvate metabolic process |
| 40S ribosomal protein SA | 0.737 | cytoplasmic translation; response to osmotic stress |
| Xylose isomerase | 0.778 | carbohydrate metabolic process |
| Acidic endochitinase | 0.963 | cellular response to water deprivation; response to stress |
| Gamma carbonic anhydrase-like 2 | 0.999 | photorespiration; response to stress |
| Thioredoxin H-type 1 | 1.000 | cell redox homeostasis |
| UDP-glucose 4-epimerase GEPI48 | 1.170 | metabolic process |
| Protein-L-isoaspartate O-methyltransferase | 1.585 | protein repair |
| NADP-dependent D-sorbitol-6-phosphate dehydrogenase | 2.228 | oxidoreductase activity |
| Icatu | log2 FC | Biological process |
| Chlorophyll a–b binding protein 4 | 0.625 | photosynthesis |
| Putative Protein tas | 0.697 | cellular response to amino acid starvation |
| Probable lactoylglutathione lyase | 0.951 | catabolic process; response to cold |
| L-ascorbate peroxidase T | 1.023 | cellular response to oxidative stress |
| Short-chain dehydrogenase TIC 32 | 1.070 | protein transport |
| Elongation factor 1-gamma 2 | 1.097 | protein biosynthesis |
| Serpin-ZX | 1.111 | protease inhibitor; response to stress |
| L-ascorbate peroxidase 3 | 1.116 | response to oxidative stress |
| Malate dehydrogenase [NADP] | 1.164 | carbohydrate metabolic process; malate metabolic process; tricarboxylic acid cycle |
| Ribulose bisphosphate carboxylase/oxygenase activase 1 | 1.256 | photosynthesis |
| Probable glutathione S-transferase | 1.322 | auxin signaling pathway |
| V-type proton ATPase catalytic subunit A | 1.376 | Golgi organization; ion transport |
| NADP-dependent D-sorbitol-6-phosphate dehydrogenase | 1.469 | oxidoreductase activity |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marques, I.; Gouveia, D.; Gaillard, J.-C.; Martins, S.; Semedo, M.C.; Lidon, F.C.; DaMatta, F.M.; Ribeiro-Barros, A.I.; Armengaud, J.; Ramalho, J.C. Next-Generation Proteomics Reveals a Greater Antioxidative Response to Drought in Coffea arabica Than in Coffea canephora. Agronomy 2022, 12, 148. https://doi.org/10.3390/agronomy12010148
Marques I, Gouveia D, Gaillard J-C, Martins S, Semedo MC, Lidon FC, DaMatta FM, Ribeiro-Barros AI, Armengaud J, Ramalho JC. Next-Generation Proteomics Reveals a Greater Antioxidative Response to Drought in Coffea arabica Than in Coffea canephora. Agronomy. 2022; 12(1):148. https://doi.org/10.3390/agronomy12010148
Chicago/Turabian StyleMarques, Isabel, Duarte Gouveia, Jean-Charles Gaillard, Sónia Martins, Magda C. Semedo, Fernando C. Lidon, Fábio M. DaMatta, Ana I. Ribeiro-Barros, Jean Armengaud, and José C. Ramalho. 2022. "Next-Generation Proteomics Reveals a Greater Antioxidative Response to Drought in Coffea arabica Than in Coffea canephora" Agronomy 12, no. 1: 148. https://doi.org/10.3390/agronomy12010148
APA StyleMarques, I., Gouveia, D., Gaillard, J.-C., Martins, S., Semedo, M. C., Lidon, F. C., DaMatta, F. M., Ribeiro-Barros, A. I., Armengaud, J., & Ramalho, J. C. (2022). Next-Generation Proteomics Reveals a Greater Antioxidative Response to Drought in Coffea arabica Than in Coffea canephora. Agronomy, 12(1), 148. https://doi.org/10.3390/agronomy12010148














