Abstract
Italian ryegrass (Lolium multiflorum Lam.) was introduced into China as a kind of turfgrass and has invaded wheat fields of the Huang-Huai-Hai Plain, causing great losses to grain yield. The acetyl–CoA carboxylase (ACCase) inhibitor clodinafop-propargyl and the acetolactate synthase (ALS) inhibitor mesosulfuron-methyl are highly efficient herbicides that have been widely used for control of this species, which has also resulted in its resistance evolution. However, the resistance status of L. multiflorum in the Huang-Huai-Hai Plain of China remains unclear, which hinders the integrated management of this weed in winter wheat production systems. In the current study, a total of 37 L. multiflorum populations were collected from the wheat fields across the region, and their susceptibility to clodinafop-propargyl and mesosulfuron-methyl was assessed. Of these, 13 populations were resistant or evolving resistance to clodinafop-propargyl (R?, RR, and RRR) with resistance index (RI) ranging from 2.62 to 830.05, and 8 populations were resistant or evolving resistance to mesosulfuron-methyl (R? and RR) with RI ranging from 3.89 to 16.68. Seven populations showed multiple-resistance to both clodinafop-propargyl and mesosulfuron-methyl. Three ACCase (I1781L, I2041N, and D2078G) and four ALS (P197T, P197S, P197A, and W574L) resistance mutations were identified in the herbicide-resistant populations, and I1781L and P197T were predominant ACCase and ALS mutations, respectively. Real-time quantitative PCR assays showed that compared with the susceptible population, the ACCase expression was slightly upregulated in some of the clodinafop-propargyl-resistant populations (AH-01 and AH-05) following clodinafop-propargyl treatment, while the ALS expression in the mesosulfuron-methyl-resistant populations showed no significant change following mesosulfuron-methyl treatment. Whole-plant dose-response testing showed that the AH-01 population carrying an ACCase gene I2041N mutation exhibited cross-resistance to all the ACCase inhibitors tested and multiple-resistance to the ALS inhibitor bispyribac-sodium, the AH-05 population carrying an ACCase gene I1781L mutation and an ALS gene P197T mutation showed cross-resistance to all the ACCase and ALS inhibitors tested, and the HN-07 population carrying an ACCase gene D2078G mutation showed cross-resistance to all the ACCase inhibitors tested and multiple-resistance to some ALS inhibitors. All the resistant populations remained susceptible to the 5-enolpyruvylshikimate-3-phosphate inhibitor glyphosate and the photosystem II inhibitor isoproturon. This study has clarified the distributions of clodinafop-propargyl- and/or mesosulfuron-methyl-resistant L. multiflorum in the Huang-Huai-Hai Plain of China, and target gene mutation was one of the most common mechanisms responsible for the resistance.
1. Introduction
Italian ryegrass (Lolium multiflorum Lam.) originated along the Mediterranean coast and was introduced into China as a lawn grass in the 1930s [1]. It is a diploid (2n = 2x = 14) or tetraploid (2n = 4x = 28), obligate outcrossing species that is widely planted as turf, pasture, and cover crop [2,3]. Currently, L. multiflorum is becoming a rampant grass severely threatening agricultural production due to the dispersal of its seeds in cultivated land and crop fields [2]. In 1987, the first case of diclofop-methyl-resistant L. multiflorum population was reported in Oregon, USA [4], and resistance evolved rapidly thereafter. In recent years, herbicide-resistant L. multiflorum has been found in wheat fields in Provinces of Hubei, Henan, and Jiangsu, China [5,6]. Resistant weeds have always been one of the major threats to agricultural production. Monitoring the distribution, spread, and evolution of resistant weeds in real time will help to develop effective weed management strategies.
Acetyl–CoA carboxylase (ACCase; EC 6.4.1.2) catalyzes the formation of malonyl–CoA and regulates de novo fatty acid biosynthesis and oxidation [7]. Two types of ACCase have been recognized: the heteromeric prokaryotic ACCase is composed of multiple subunits, whereas the homomeric eukaryotic ACCase is a large multidomain protein [8,9]. Plants have both the cytosolic and the plastidic ACCase [9]. In grasses, the plastidic ACCase is homomeric and is the target site for three classes of ACCase inhibitors [10,11,12]. ACCase inhibitors were first introduced into the market in 1978 with the appearance of diclofop-methyl, including aryloxyphenoxypropionate (APP), cyclohexanedione (CHD), and phenylpyrazoline (PPZ) chemical groups [7,13]. The multidomain type found in the cytosol of all plants and the multiple-subunit type found in plastids of dicots are insensitive to APPs and CHDs [7]. In contrast, the plastidic ACCase in grasses is herbicide-sensitive [7]. However, their repeated and extensive use for many years has led to the evolution of herbicide resistant populations in many important grass species, such as wild oat (Avena fatua L.) [14,15], annual ryegrass (Lolium rigidum Gaud.) [16,17], black-grass (Alopecurus myosuroides Huds.) [18,19], Asia minor bluegrass (Polypogon fugax Nees ex Steud.) [20], and Chinese sprangletop (Leptochloa chinensis (L.) Nees) [21]. Up to now, there have been reported 50 species with resistance to these herbicides [22].
Acetolactate synthase (ALS, EC 2.2.1.6), also referred to as acetohydroxyacid synthase (AHAS), is the first enzyme in the biosynthetic pathway for the branched-chain amino acids valine, leucine, and isoleucine [23]. ALS is the target site for a large number of herbicides across the dissimilar sulfonylureas (SU), imidazolinones (IMI), triazolopyrimidines (TP), pyrimidinyl-thiobenzoates (PTB), and sulfonylaminocarbonyl-triazolinones (SCT) herbicide chemistries [24]. ALS-inhibiting herbicides can control diverse weed species, have low mammalian toxicity, and are selective in major crops [25], which ensure their global and intensive use in different crops over huge areas. However, the evolution of herbicide-resistant populations rapidly followed, and a total of 170 ALS-resistant biotypes have now been reported [22].
The knowledge of resistance mechanisms is important for the design of effective weed control strategies to delay the onset of resistance to herbicides. Herbicide resistance includes target-site-based resistance (TSR) and non-TSR (NTSR) [26]. TSR is essentially caused by the changes of single amino acid, target enzyme activity, or the overexpression of a specific gene encoding the target protein [27]. In most cases, target-site mutations often lead to high-level resistance to herbicides with a same mode of action (MOA). Up to now, a total of seven and eight codon positions of ACCase and ALS can show a variety of mutations to confer ACCase and ALS resistance, respectively [13,28]. By contrast, NTSR can be due to any one or a combination of mechanisms that limit to a nonlethal dose the amount of herbicide reaching a target site [26], such as decreased herbicide penetration into the plant, decreased rates of herbicide translocation, and increased rates of herbicide sequestration/metabolism [26].
Today, resistance to herbicides with eight MOAs has been reported in L. multiflorum globally, with the most severe resistance occurring to ACCase and ALS inhibitors [22]. In China, Zhang et al. [29] first reported the ACCase-resistant L. multiflorum populations collected from the Provinces of Henan and Jiangsu, with varying degrees of cross-resistance to haloxyfop-R-methyl, quizalofop-P-ethyl, and sethoxydim. In the past few years, farmers in the Huang-Huai-Hai Plain have found that an increasing number of L. multiflorum populations are becoming resistant to the commonly used herbicides clodinafop-propargyl and mesosulfuron-methyl. To determine the distribution of herbicide-resistant L. multiflorum and the extent of resistance and thus aid in developing effective weed management strategies, a total of 37 populations of L. multiflorum were collected from different sites in this region where the wheat fields were severely infested with this species. The objectives of this study were to (1) test the susceptibility of different populations to clodinafop-propargyl and mesosulfuron-methyl, (2) identify potential ACCase and ALS mutations from the resistant populations, (3) assess the effects of clodinafop-propargyl and mesosulfuron-methyl on the expression of ACCase and ALS genes, respectively, in different resistant populations, and (4) clarify their cross- and multiple-resistance patterns to different herbicides.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
During May 2013 to May 2021, mature seeds of 37 L. multiflorum populations were collected from the winter wheat fields with repeated clodinafop-propargyl and/or mesosulfuron-methyl use in Provinces of Shaanxi (SHX), Hubei (HB), Henan (HN), Anhui (AH), and Jiangsu (JS) across the Huang-Huai-Hai Plain of China (Table 1, Figure 1). The seeds of each population were randomly collected from more than 100 mature plants, after which they were dried and stored in paper bags at 4 °C until further use.

Table 1.
Information on the L. multiflorum seeds collected in this study and the susceptibility of different L. multiflorum populations to clodinafop-propargyl and mesosulfuron-methyl.
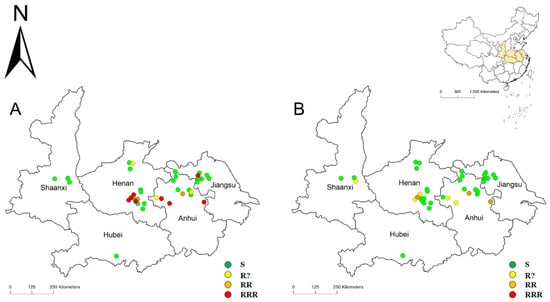
Figure 1.
The distribution of 37 L. multiflorum populations collected from the Huang-Huai-Hai Plain, China. The susceptibility of different populations to clodinafop-propargyl (A) and mesosulfuron-methyl (B) was classified according the “R” rating system. RRR means resistance confirmed, highly likely to reduce herbicide performance; RR means resistance confirmed, probably reducing herbicide performance; R? means early indications that resistance may be developing, possibly reducing herbicide performance; and S means susceptible.
Plump seeds were randomly selected from each population and placed into a 9 cm diameter Petri dish containing two pieces of filter paper moistened with 5 mL of distilled water. Then, the Petri dishes were transferred into an artificial growth chamber for incubation (200 μmol m−2 s−1 photosynthetic photonflux density, 12 h photoperiod, 25 °C constant temperature, with ∼75% relative humidity). After germination, 10 seeds were sown into each plastic pot (10 cm × 10 cm × 8.5 cm) filled with loam soils and grown in a naturally lit glasshouse (25/15 °C day/night) with regular watering and fertilization. When the seedlings had reached the two- to three-leaf stage, they were thinned to eight uniformly sized seedlings per pot.
2.2. Initial Resistance Screening
Weed seedlings at the three- to four-leaf stage were treated with clodinafop-propargyl (56 g a.i. ha−1) or mesosulfuron-methyl (11.25 g a.i. ha−1) at the recommended field rate (RFR). An untreated control for each population was treated with water. Both herbicide and water were applied using a laboratory cabinet sprayer (3WP-2000, Nanjing Mechanization Research Institute of the Ministry of Agriculture, Nanjing, China), which delivered 450 L ha−1 of liquid at 0.275 MPa. At 21 d after treatment (DAT), the aboveground fresh weight per pot was recorded. Each treatment had three replicates, and the experiment was conducted twice.
According to the results of initial screening, the L. multiflorum populations were assigned into four resistance categories (RRR, RR, R?, and S) using the “R” rating system proposed by Moss et al. [30]. Of the four resistance categories, “S” means that the population is susceptible, and its fresh weight reduction ranged from 9*SS/10 to SS; “R?” means that the resistance may be developing, possibly reducing herbicide performance, and its fresh weight reduction ranged from 4*SS/5 to 9*SS/10; “RR” means that the resistance is confirmed, possibly reducing herbicide performance, and its fresh weight reduction ranged from 2*SS/5 to 4*SS/5; and “RRR” means that the resistance is confirmed, high likely reducing herbicide performance, and its fresh weight reduction ranged from 0 to 2*SS/5. In this current study, the standard susceptible (SS) population AH-07 was collected from a roadside without any herbicide use history, and a discriminating dose of clodinafop-propargyl at 56 g a.i. ha−1 or mesosulfuron-methyl at 11.25 g a.i. ha−1 caused a fresh weight reduction of 96.44% or 98.28%, respectively.
2.3. Whole-Plant Dose-Response Experiment
Based on the results of initial screening, the L. multiflorum population AH-07 susceptible to both clodinafop-propargyl and mesosulfuron-methyl was used as the susceptible (S) population. All the resistant populations (i.e., the R?, RR, and RRR populations) and the S population were then tested for herbicide susceptibility level by whole-plant dose-response experiments. Information on the herbicides used in the experiments is shown in Table 2. At 21 DAT, the aerial parts of the plants in each pot were cut off, and their fresh weight was recorded. Each treatment had three replicates, and the experiment was conducted twice.

Table 2.
Herbicides and their doses used in the whole-plant dose-response experiments for different L. multiflorum populations.
2.4. ACCase and ALS Sequencing
Fresh leaf tissues were collected from the individuals of the S plant (AH-07) and all the resistant plants (i.e., the R?, RR, and RRR populations) at their three- to four-leaf stage and stored at −20 °C. Genomic DNA was then extracted from the leaf tissues using the classical cetyltrimethylammonium bromide method [31]. To determine the frequency of plants with specific mutations in different resistant populations, at least 10 plants from each population were randomly selected. Primers used to amplify ACCase and ALS genes have been reported elsewhere (Table 3), and the amplified regions contained all the known resistance mutation sites [29,32]. Polymerase chain reaction (PCR) was conducted in a 25 µL volume that consisted of 1 µL of genomic DNA, 1 µL of each primer (10 μM), 9.5 µL of double-distilled H2O, and 12.5 µL of 2 × Es Taq MasterMix (Dye). PCR cycling settings were as follows: 94 °C for 2 min, 35 cycles of 94 °C for 30 s, x °C for 30 s, and 72 °C for 30 s, followed by a final extension step of 5 min at 72 °C. The x refers to the respective annealing temperatures used for each primer pair (Table 3). The targeted PCR products were purified from agarose gels using a FastPure® Gel DNA Extraction Mini Kit (Vazyme, Nanjing, China) and were directly sequenced by Tsingke Biotech (Beijing, China) from both directions. The resulting sequences were aligned and compared using DNAMAN v 6.0 software (Lynnon, Quebec, Canada).

Table 3.
Primers used for ACCase and ALS sequencing in different L. multiflorum populations.
2.5. ACCase and ALS Expression Analysis
Three clodinafop-propargyl-resistant populations (AH-01, AH-05, and HN-07) each carrying a specific ACCase resistance mutation and two mesosulfuron-methyl-resistant populations (AH-05 and HN-08) each carrying a specific ALS resistance mutation were chosen to detect the relative expression levels of ACCase and ALS, respectively. The population AH-07 susceptible to both herbicides was used as a control. Weed plants at the three- to four-leaf stage were treated with clodinafop-propargyl or mesosulfuron-methyl at their RFRs, respectively. Fresh leaf tissues were then sampled from their individual plant at 0 (CK), 2, 6, 12, 24, 48, 72, and 120 h after treatment (HAT) and immediately frozen in liquid nitrogen. RNA was then extracted from the leaf samples using the Trizol reagent (Ambition, life technologies, USA), and complementary DNA (cDNA) was obtained using a HiFiScript gDNA Removal and cDNA Synthesis Kit (CWBIO, Beijing, China).
In this current study, elongation factor 1-alpha (EF1α) was used as the reference gene, and the primers for the reference gene and target genes were reported elsewhere [33,34]. The real-time quantitative PCR (RT-qPCR) was performed using the CFX96 Real-time PCR system (Bio-Rad Laboratories, Hercules, CA, USA) with a ChamQ SYBR qPCR Master Mix (Vazyme, Nanjing, China). The reaction system and cycling profile were prepared according to the manufacturer’s instructions, and target gene expression relative to that of EF1α was analyzed by the 2−ΔΔCt method [35]. Each experiment had six biological replicates (six individual plants), and each biological replicate had three technical replicates. Two threshold values including the fold-change (2-fold) and the Student’s t-test (p < 0.05) were used to determine the up- or downregulation of target genes in the R relative to the S plants.
2.6. Cross- and Multiple-Resistance Patterns of Specific Populations to Other Herbicides
To determine the potential alternatives for control of the resistant L. multiflorum plants, three populations AH-01, AH-05, and HN-07 each carrying specific target-site mutations were selected to determine their cross- and multiple-resistance to different herbicides. Weed plants of the S (AH-07) and the three resistant biotypes at the three- to four-leaf stage were treated with a series of doses of different herbicides (Table 4). An untreated control was established for each population and treated with water. At 21 DAT, the aboveground fresh weight of L. multiflorum was recorded. Each treatment had three replicates, and the experiment was conducted twice.

Table 4.
Herbicides and their doses used in cross- and multiple- resistance experiments for different L. multiflorum populations.
2.7. Data Analysis
All the dose-response data from the two repeated experiments were compared and analyzed by variance (ANOVA) using the SPSS software (Version 22.0; IBM Corporation, Armonk, NY, USA). Since the two repeated experiments did not show significant difference (p > 0.05), the datasets were combined and fitted to a nonlinear logistic model using SigmaPlot software (Version 14.0; Systat Software Inc., CA, USA). The herbicide dose that reduced the aboveground fresh weight by 50% (GR50) was calculated according to the following equation.
where y is the response at the herbicide dose x, C is the lower limit, D is the upper limit, and b is the relative slope at GR50. Resistance index (RI) was calculated as the ratio of GR50 values between the resistant and susceptible populations.
3. Results
3.1. Susceptibility of L. multiflorum to Clodinafop-Propargyl and Mesosulfuron-Methyl
Single-dose testing was first conducted to determine the susceptibility of different L. multiflorum populations to clodinafop-propargyl and mesosulfuron-methyl. Of the populations tested, 13 populations had evolved resistance to clodinafop-propargyl, and eight populations had evolved resistance to mesosulfuron-methyl (Table 1). Meanwhile, seven of the resistant populations showed multiple resistance to both clodinafop-propargyl and mesosulfuron-methyl (Table 1). For the SS population AH-07, all plants died at the RFRs of clodinafop-propargyl and mesosulfuron-methyl, with the fresh weight inhibitions confirmed to be 96.44% and 98.28%, respectively (Table 1). Based on the “R” rating system, of the 13 clodinafop-propargyl-resistant populations, 10 were classified as “RRR” and “RR”, and three were classified as “R?”; of the eight mesosulfuron-methyl-resistant populations, three were classified as “RR”, and five were classified as “R?” (Table 1).
The resistance distributions of L. multiflorum to clodinafop-propargyl and mesosulfuron-methyl were then plotted by combining the geographical information and the results of single-dose testing (Figure 1). Based on the data, the herbicide-resistant L. multiflorum were mainly distributed in Provinces of Henan, Anhui, and Jiangsu.
3.2. Whole-Plant Dose-Response Experiment
The susceptibility of the S population AH-07 was confirmed by the whole-plant dose-response experiment. Clodinafop-propargyl applied at 1.61 g a.i. ha−1 or mesosulfuron-methyl applied at 0.53 g a.i. ha−1 provided a 50% growth reduction in the AH-07 population, which was much lower than their respective RFRs (Table 5). Compared with AH-07, the clodinafop-propargyl-resistant populations that classed as “R?, RR, and RRR” had GR50 values ranging from 4.22 to 1336.38 g a.i. ha−1, which were 2.62- and 830.05-fold more resistant (Table 5). Similarly, the mesosulfuron-methyl-resistant populations that classed as “R? and RR” had GR50 values ranging from 2.06 to 8.84 g a.i. ha−1, which were 3.89- to 16.68-fold more resistant than the AH-07 population (Table 5).

Table 5.
The GR50 valuesa and RIsb to clodinafop-propargyl and mesosulfuron-methyl in different resistant populations of L. multiflorum.
3.3. ACCase and ALS Resistance Mutations in L. multiflorum
Following PCR amplifications, the ACCase and ALS genes spanning all the known resistance codon positions were obtained from the ACCase- and ALS-resistant populations, respectively. Of the 13 clodinafop-propargyl-resistant populations (Table 1), 10 populations were detected for ACCase mutations, with I2041N detected in five populations, I1781L detected in four populations, and D2078G detected in one population (Table 6, Figure 2). No known resistance mutation was detected in the ACCase of three clodinafop-propargyl-“R?” populations (AH-08, AH-12, and HN-14). Similarly, of the eight mesosulfuron-methyl-resistant populations (Table 1), ALS mutations were detected in only three populations, and a total of four mutations at codon positions 197 and 574 of ALS were identified (P197T, P197S, P197A, and W574L) (Table 6, Figure 2). In the HN-08 population, three mutations (P197S/A/T) were simultaneously identified in ALS of its individuals (Table 6). No known resistance mutation was detected in the ALS of five mesosulfuron-methyl-“R?” populations. Notably, mutations in ACCase and ALS were simultaneously detected in individuals from the AH-05, AH-11, or HN-08 population (Table 6).

Table 6.
Mutations and their frequencies in different L. multiflorum populations.
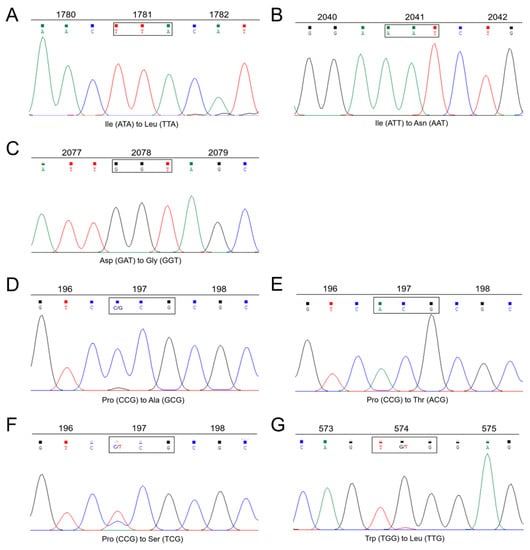
Figure 2.
Partial ACCase and ALS gene sequences from different resistant populations of L. multiflorum. ACCase sequences showing (A) Ile-1781-Leu, (B) Ile-2041-Asn, and (C) Asp-2078-Gly mutations. ALS sequences showing (D) Pro-197-Ala, (E) Pro-197-Thr, (F) Pro-197-Ser, and (G) Trp-574-Leu mutations.
3.4. ACCase or ALS Expression Analysis
In this current study, three ACCase-resistant populations (AH-01, AH-05, and HN-07) and two ALS-resistant populations (AH-05 and HN-08) were selected for gene expression assays. The total expression of ACCase or ALS was normalized relative to that of EF1α and compared between the R and S biotypes of L. multiflorum at 0 (control), 2, 6, 12, 24, 48, 72, and 120 HAT (Figure 3). According to the results, no significant difference was observed in the ACCase expression between the ACCase-resistant population HN-07 and the S population AH-07 at each time point (Figure 3A). However, the ACCase expression in the ACCase-resistant populations AH-01 and AH-05 was upregulated more than three-fold at 12 and 2 HAT when compared with the S population, respectively (Figure 3A). In addition, no significant difference in the ALS expression was revealed in the ALS-resistant populations AH-05 and HN-08 when compared with the S population before and after herbicide treatment (Figure 3B).
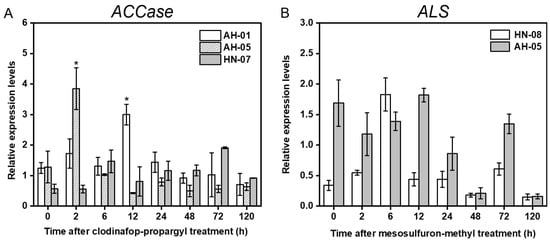
Figure 3.
Relative expression levels of (A) ACCase and (B) ALS in ACCase- and ALS-resistant L. multiflorum plants at 0 (control), 2, 6, 12, 24, 48, 72, and 120 h after clodinafop-propargyl or mesosulfuron-methyl treatment, respectively. Asterisk indicates significant upregulation (fold change ≥2 and p < 0.05) of target gene in resistant plants compared with the susceptible plants (AH-07).
3.5. Cross- and Multiple-Resistance Patterns of L. multiflorum to Other Herbicides
In this current study, all the herbicides tested showed great control efficacies against the plants from the S population AH-07 (Table 7, Figure 4). In comparison, the AH-01 population carrying an ACCase I2041N mutation showed high resistance to haloxyfop-R-methyl, quizalofop-P-ethyl, and cyhalofop-butyl, moderate resistance to fenoxaprop-P-ethyl and metamifop, low resistance to clethodim, sethoxydim, pinoxaden, and bispyribac-sodium, and was susceptible to imazethapyr, imazamox, flucarbazone-Na, glyphosate, and isoproturon. The AH-05 population carrying both an ACCase gene I1781L mutation and an ALS gene P197T mutation showed high resistance to fenoxaprop-P-ethyl, haloxyfop-R-methyl, quizalofop-P-ethyl, cyhalofop-butyl, metamifop, sethoxydim, pinoxaden, and bispyribac-sodium, moderate resistance to imazethapyr, low resistance to clethodim, pyroxsulam, imazamox, and flucarbazone-Na, and was susceptible to glyphosate and isoproturon. The HN-07 population carrying an ACCase D2078G mutation showed high resistance to fenoxaprop-P-ethyl, quizalofop-P-ethyl, sethoxydim, moderate resistance to haloxyfop-R-methyl, metamifop, pyroxsulam, and bispyribac-sodium, low resistance level to clethodim, imazamox, and imazethapyr, and was susceptible to flucarbazone-Na, glyphosate, and isoproturon (Table 7, Figure 4).

Table 7.
The GR50 values a and Rls b of susceptible (AH-07) and resistant (AH-01, AH-05, and HN-07) populations of L. multiflorum to different herbicides.
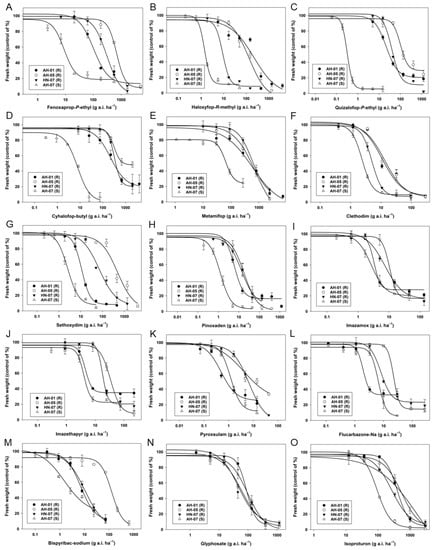
Figure 4.
The doses-response curves of the aboveground fresh weights of the susceptible (AH-07) and different resistant (AH-01, AH-05, and HN-07) populations of L. multiflorum to (A) fenoxaprop-P-ethyl, (B) haloxyfop-R-methyl, (C) quizalofop-P-ethyl, (D) cyhalofop-butyl, (E) metamifop, (F) clethodim, (G) sethoxydim, (H) pinoxaden, (I) imazamox, (J) imazethapyr, (K) pyroxsulam, (L) flucarbazone-Na, (M) bispyribac-sodium, (N) glyphosate, and (O) isoproturon. Vertical bars represent the SE of the means.
4. Discussion
Lolium multiflorum, an invasive grass weed in wheat cropping systems in China, can occur even after application of the ACCase inhibitor clodinafop-propargyl and/or the ALS inhibitor mesosulfuron-methyl. Here, a total of 37 L. multiflorum populations were collected from the wheat fields across the Huang-Huai-Hai Plain, and 13 and 8 of the populations were confirmed to be resistant to clodinafop-propargyl and mesosulfuron-methyl, respectively (Figure 1). Most of the clodinafop-propargyl-resistant populations showed high- to moderate-level resistance, but the mesosulfuron-methyl-resistant populations mostly displayed low-level resistance. These results indicated that the herbicide resistance of L. multiflorum was prevalent in the Huang-Huai-Hai Plain, and the ACCase resistance seemed to be more severe than the ALS resistance.
In recent years, the infestation of L. multiflorum in wheat fields has been observed in some Provinces of China such as Henan and Shaanxi, and its effective control heavily relies on the chemical herbicides clodinafop-propargyl and mesosulfuron-methyl [5,6]. Zhang et al. [36] first reported that the L. multiflorum populations from Henan and Jiangsu had evolved resistance to fenoxaprop-P-ethyl and showed cross-resistance to different ACCase inhibitors. In addition, Wu et al. [37] recently found that a L. multiflorum population collected from Henan Province had evolved multiple herbicide resistance due to both a target-site mutation and the P450s-mediated enhanced metabolism. In this present study, different L. multiflorum populations were collected from the wheat fields across Provinces of Shaanxi, Henan, Hubei, Jiangsu, and Anhui, and their susceptibility to clodinafop-propargyl and mesosulfuron-methyl was determined by a single-dose herbicide-resistance testing and whole-plant dose-response experiments. The results showed that the herbicide-resistant populations mainly distributed in Henan, Jiangsu, and Anhui, and almost no resistant population was identified in Shaanxi and Hubei (Figure 1). This result may partially be due to the fact that only several populations of L. multiflorum were collected in the latter two Provinces, which may not be able to reflect the practical situation in most parts of the region. Moreover, the agricultural departments in Shaanxi Province have issued relevant warnings of the infestation of L. multiflorum in its wheat fields previously [5,6], suggesting that continuous attention should be paid to the possible resistance evolution in these regions. It is noteworthy that most populations exhibiting high-level clodinafop-propargyl resistance had simultaneously evolved mesosulfuron-methyl resistance (Table 5), which may be owing to the relevant herbicide application history. According to the local farmers, mesosulfuron-methyl has been commonly used as an alternative for control of the ACCase-resistant grass weeds in wheat fields, which may thus accelerate the evolution of multiple-herbicide-resistant weed biotypes.
In this study, three ACCase mutations including I1781L, I2041N, and D2078G were identified in 10 of the 13 clodinafop-propargyl-resistant populations (Table 6). Mutations at codon positions 1781, 2041, and 2078 of ACCase conferring ACCase resistance have been documented in diverse weed species [26]. Notably, among the 10 mutant populations, 5 were identified to have an I2041N mutation, 4 had an I1781L mutation, and only 1 had a D2078G mutation (Table 6). The rarity of the D2078G mutation may be partially due to its possible adverse pleiotropic effects on plant growth by changing enzyme functionality [38]. ACCase resistance conferred by the above three mutations has been identified in different weed species, such as A. fatua [14], shortawn foxtail (Alopecurus aequalis Sobol.) [39], and A. myosuroides [17]. In addition, four ALS mutations including P197A, P197S, P197T, and W574L were also identified in three of the eight mesosulfuron-methyl-resistant populations (Table 6). The codon positions 197 and 574 of ALS seem the most prone to mutation [22], and mutations at these two positions have been frequently identified in a variety of ALS-resistant weeds, such as wild radish (Raphanus raphanistrum L.) [24], L. rigidum [40], and A. aequalis [41]. Previous studies have found that P197S and W574L had no significant effect on ALS dynamics and plant vegetative growth, which may partially explain why these two mutations are often evolved under the selection of ALS herbicides [42]. In this study, no resistance mutation was identified in some ACCase- and ALS-resistant populations that were classed as “R?” (Table 6), which indicates the potential presence of NTSR in these populations.
Target gene overexpression is also a TSR mechanism responsible for herbicide resistance, which has been frequently identified in the glyphosate-resistant weeds and rarely reported in the other herbicide-resistant weeds. Notably, Laforest et al. [43] found that transcription of ACCase was 3.4- to 9.3-times higher in the ACCase-resistant crabgrass (Digitaria sanguinalis (L.) Scop.) biotype than in the susceptible biotype, which was the first report of ACCase overexpression conferring ACCase resistance. Zhao et al. [44] also found that the ALS expression was significantly higher in the mesosulfuron-methyl-resistant than in the susceptible plants before and after mesosulfuron-methyl treatment. In the current study, the relative expression of ACCase or ALS in some herbicide-resistant populations of L. multiflorum was also determined (Figure 3). Compared with the susceptible population, the ACCase expression in the ACCase-resistant populations AH-01 and AH-05 was slightly but significantly upregulated at 12 and 2 HAT, respectively. However, the ALS expression in the ALS-resistant populations AH-05 and HN-08 showed no significant difference when compared with the susceptible population. These results indicated that the ACCase resistance in these populations may simultaneously involve the target-site mutations and the target-gene overexpression, while the ALS resistance was mainly due to the target-site mutations.
Cross- and multiple-resistance are defined as the expressions of genetically endowed mechanisms, including TSR and NTSR mechanisms, that can confer resistance to different herbicides of the same or different chemical groups [45]. Determining the resistance patterns of herbicide-resistant weed populations to the commonly used herbicides can help inform proactive or reactive resistant weed management by identifying less resistance-prone alternatives for growers. In this study, three target-site mutant populations, AH-01 with an ACCase I2041N mutation, AH-05 with both an ACCase I1781L mutation and an ALS P197T mutation, and HN-07 with an ACCase D2078G mutation, were selected for characterizing their cross- and multiple-resistance. Previously, the I1781L and D2078G mutations have been reported to confer resistance to all the three groups of ACCase inhibitors in canarygrass (Phalaris minor Retz.), A. aequalis, southern crabgrass (Digitaria ciliaris (Retz.) Koeler), and perennial ryegrass (Lolium perenne L.) [46,47,48], while the I2041N-mutant A. myosuroides, A. aequalis, and A. fatua showed high-level APP resistance [17,49,50]. In this current study, all the three ACCase mutations (I1781L, I2041N, and D2078G) conferred resistance to all the ACCase inhibitors tested, including APPs, CHDs, and PPZ. It is noteworthy that the AH-05 population with an ACCase I1781L mutation also carrying an ALS P197T mutation, which may be responsible for its resistance to the ALS inhibitors tested. Interestingly, two populations (AH-01 and HN-07) exhibiting moderate to low resistance to some ALS inhibitors had no ALS resistance mutations, indicating the possible participation of NTSR mechanisms.
In this study, all the mutant populations were confirmed susceptible to glyphosate and isoproturon. Isoproturon is a dominant herbicide widely used in the wheat field, which can be used as an alternative for control of the ACCase- and/or ALS-resistant L. multiflorum. However, we do not recommend its intensive and extensive use since the repeated use of the same-MOA herbicides is the greatest risk factor for fueling herbicide-resistance evolution [51]. Combining chemical control with other weed management methods may be wiser strategies to slow down the spread of resistance in these herbicide-resistant populations [21].
5. Conclusions
In summary, this study has clarified the resistance distributions and resistance levels of L. multiflorum to clodinafop-propargyl and mesosulfuron-methyl in wheat fields across the Huang-Huai-Hai Plain. Mutations at 1781, 2041, and 2078 codons of ACCase and at 197 and 574 codons of ALS were identified as the important TSR mechanisms to clodinafop-propargyl and mesosulfuron-methyl in L. multiflorum, respectively, and the ACCase overexpression may also be involved in the ACCase resistance. The comprehensive cross- and multiple-resistance patterns were characterized for the resistance populations carrying different resistance mutations, and alternative herbicides were also identified for control of the resistant populations. Further studies are still required to figure out the exact mechanisms responsible for the resistance in the resistant populations without target-site mutations.
Author Contributions
Conceptualization, N.Z. and H.C.; methodology, W.L., C.W., M.W., M.J., J.Z., M.L., H.C. and N.Z.; software, W.L. and M.J.; validation, N.Z., H.C., J.Z. and M.L.; formal analysis, W.L., C.W., M.W., M.J., J.Z., M.L., H.C. and N.Z.; investigation, W.L., C.W., M.W., M.J., J.Z., M.L., H.C. and N.Z.; resources, M.L., H.C. and N.Z.; data curation, W.L., C.W. and M.W.; writing—original draft preparation, W.L.; writing—review and editing, N.Z. and M.J.; visualization, N.Z.; supervision, N.Z.; project administration, N.Z.; funding acquisition, N.Z., H.C. and M.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (No. 32102237), the Anhui Provincial Natural Science Foundation (No. 2108085QC115), and the Talent Research Project of Anhui Agricultural University (No. rc342004).
Data Availability Statement
The data presented in this study are available on request from the corresponding authors.
Acknowledgments
The authors would like to thank Tao Jin (KingAgroot Co., Ltd., Qingdao, China) for providing a portion of seeds.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhang, X.; Li, Y.; Gou, W.; Zhang, R. Research development of Italian ryegrass. Pratacultural Sci. 2009, 26, 55–60. [Google Scholar]
- Brunharo, C.A.; Hanson, B.D. Multiple herbicide-Resistant Italian ryegrass [Lolium perenne L. spp. multiflorum (Lam.) Husnot] in California perennial crops: Characterization, mechanism of resistance, and chemical management. Weed Sci. 2018, 66, 696–701. [Google Scholar] [CrossRef]
- Bobadilla, L.K.; Hulting, A.G.; Berry, P.A.; Moretti, M.L.; Mallory-Smith, C. Frequency, distribution, and ploidy diversity of herbicide-resistant Italian ryegrass (Lolium perenne spp. multiflorum) populations of western Oregon. Weed Sci. 2021, 69, 177–185. [Google Scholar] [CrossRef]
- Stanger, C.E.; Appleby, A.P. Italian ryegrass (Lolium multiflorum) accessions tolerant to diclofop. Weed Sci. 1989, 37, 350–352. [Google Scholar] [CrossRef]
- Wang, H.; Song, Y.; Li, J.; Yang, G.; He, W.; Lou, Z. Precaution against malaria weed ryegrass in wheat field. Plant Prot. 2008, 34, 149–151. [Google Scholar]
- Hao, P.; Zhang, L. Highly alert to the spread of harmful ryegrass in weeds. Shanxi J. Agric. Sci. 2015, 61, 50–51. [Google Scholar]
- Liu, W.; Harrison, D.K.; Chalupska, D.; Gornicki, P.; O’donnell, C.C.; Adkins, S.W.; Williams, R.R. Single-site mutations in the carboxyltransferase domain of plastid acetyl-CoA carboxylase confer resistance to grass-specific herbicides. Proc. Natl. Acad. Sci. USA 2007, 104, 3627–3632. [Google Scholar] [CrossRef]
- Tate, T.M. Characterization of Acetyl Coenzyme an Inhibitor Resistance in Turfgrass and Grassy Weeds. Ph.D. Thesis, University of Georgia, Athens, GA, USA, 2012. [Google Scholar]
- Harwood, J.L. Fatty-acid metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1988, 39, 101–138. [Google Scholar] [CrossRef]
- Takano, H.K.; Ovejero, R.F.L.; Belchior, G.G.; Maymone, G.P.L.; Dayan, F.E. ACCase-inhibiting herbicides: Mechanism of action, resistance evolution and stewardship. Sci. Agric. 2020, 78, e20190102. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, Z.; Shen, Y.; Tong, L. Crystal structure of the carboxyltransferase domain of acetyl-coenzyme A carboxylase. Science 2003, 299, 2064–2067. [Google Scholar] [CrossRef]
- Egli, M.A.; Gengenbach, B.G.; Gronwald, J.W.; Somers, D.A.; Wyse, D.L. Characterization of maize acetyl-coenzyme A carboxylase. Plant Physiol. 1993, 101, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Kaundun, S.S. Resistance to acetyl-CoA carboxylase-inhibiting herbicides. Pest Manag. Science 2014, 70, 1405–1417. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Ahmad-Hamdani, M.S.; Han, H.; Christoffers, M.J.; Powles, S.B. Herbicide resistance-endowing ACCase gene mutations in hexaploid wild oat (Avena fatua): Insights into resistance evolution in a hexaploid species. Heredity 2013, 110, 220–231. [Google Scholar] [CrossRef]
- Alwarnaidu Vijayarajan, V.B.; Forristal, P.D.; Cook, S.K.; Staples, J.; Schilder, D.; Hennessy, M.; Barth, S. First report on assessing the severity of herbicide resistance to ACCase inhibitors pinoxaden, propaquizafop and cycloxydim in six Avena fatua populations in Ireland. Agronomy 2020, 10, 1362. [Google Scholar] [CrossRef]
- Yu, Q.; Cairns, A.; Powles, S. Glyphosate, paraquat and ACCase multiple herbicide resistance evolved in a Lolium rigidum biotype. Planta 2007, 225, 499–513. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Abdallah, I.; Han, H.; Owen, M.; Powles, S. Distinct non-target site mechanisms endow resistance to glyphosate, ACCase and ALS-inhibiting herbicides in multiple herbicide-resistant Lolium rigidum. Planta 2009, 230, 713–723. [Google Scholar] [CrossRef]
- Délye, C.; Matéjicek, A.; Michel, S. Cross-resistance patterns to ACCase-inhibiting herbicides conferred by mutant ACCase isoforms in Alopecurus myosuroides Huds. (black-grass), re-examined at the recommended herbicide field rate. Pest Manag. Science 2008, 64, 1179–1186. [Google Scholar] [CrossRef]
- Bailly, G.C.; Dale, R.P.; Archer, S.A.; Wright, D.J.; Kaundun, S.S. Role of residual herbicides for the management of multiple herbicide resistance to ACCase and ALS inhibitors in a black-grass population. Crop Prot. 2012, 34, 96–103. [Google Scholar] [CrossRef]
- Zhao, N.; Yang, J.; Jiang, M.; Liao, M.; Cao, H. Identification of essential genes involved in metabolism-based resistance mechanism to fenoxaprop-P-ethyl in Polypogon fugax. Pest Manag. Sci. 2022, 78, 1164–1175. [Google Scholar] [CrossRef]
- Zhao, N.; Jiang, M.; Li, Q.; Gao, Q.; Zhang, J.; Liao, M.; Cao, H. Cyhalofop-butyl resistance conferred by a novel Trp-2027-Leu mutation of acetyl-CoA carboxylase and enhanced metabolism in Leptochloa chinensis. Pest Manag. Sci. 2022, 78, 1176–1186. [Google Scholar] [CrossRef]
- Heap, I. International Herbicide Resistance Weed Database. Available online: www.weedscience.org (accessed on 24 June 2022).
- Garcia, M.D.; Nouwens, A.; Lonhienne, T.G.; Guddat, L.W. Comprehensive understanding of acetohydroxyacid synthase inhibition by different herbicide families. Proc. Natl. Acad. Sci. USA 2017, 114, E1091–E1100. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Han, H.; Li, M.; Purba, E.; Walsh, M.J.; Powles, S.B. Resistance evaluation for herbicide resistance–Endowing acetolactate synthase (ALS) gene mutations using Raphanus raphanistrum populations homozygous for specific ALS mutations. Weed Res. 2012, 52, 178–186. [Google Scholar] [CrossRef]
- Shaner, D.L. Herbicide safety relative to common targets in plants and mammals. Pest Manag. Sci. 2004, 60, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Gaines, T.A.; Duke, S.O.; Morran, S.; Rigon, C.A.; Tranel, P.J.; Küpper, A.; Dayan, F.E. Mechanisms of evolved herbicide resistance. J. Biol. Chem. 2020, 295, 10307–10330. [Google Scholar] [CrossRef]
- Powles, S.B.; Yu, Q. Evolution in action: Plants resistant to herbicides. Annu. Rev. Plant Biol. 2010, 61, 317–347. [Google Scholar] [CrossRef]
- Yu, Q.; Powles, S.B. Resistance to AHAS inhibitor herbicides: Current understanding. Pest Manag. Sci. 2014, 70, 1340–1350. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, H.; Xu, H.; Gao, Y.; Zhang, W.; Dong, L. Mechanism of fenoxaprop-P-ethyl resistance in Italian ryegrass (Lolium perenne ssp. multiflorum) from China. Weed Sci. 2017, 65, 710–717. [Google Scholar] [CrossRef]
- Moss, S.R.; Perryman, S.A.; Tatnell, L.V. Managing herbicide-resistant blackgrass (Alopecurus myosuroides): Theory and practice. Weed Technol. 2007, 21, 300–309. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Report. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Yu, Q.; Han, H.; Powles, S.B. Mutations of the ALS gene endowing resistance to ALS-inhibiting herbicides in Lolium rigidum populations. Pest Manag. Sci. 2008, 64, 1229–1236. [Google Scholar] [CrossRef]
- Zhang, P. Fenoxaprop-P-ethyl Resistance and the Control of Lolium multiflorum in Wheat Fields. Ph.D. Thesis, Nanjing Agricultural University, Nanjing, China, 2018. [Google Scholar]
- Duhoux, A.; Delye, C. Reference genes to study herbicide stress response in Lolium sp.: Up-regulation of P450 genes in plants resistant to acetolactate-synthase inhibitors. PLoS ONE 2013, 8, e63576. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhang, Y.; Chen, X.; Dong, L. Cross resistance patterns to acetyl-CoA carboxylase inhibiting herbicides associated with different mutations in Italian ryegrass from China. Crop Prot. 2021, 143, 105479. [Google Scholar] [CrossRef]
- Wu, C.; Song, M.; Zhang, T.; Zhou, C.; Liu, W.; Jin, T.; Zhao, N. Target-site mutation and cytochrome P450s confer resistance to multiple herbicides in Italian ryegrass (Lolium multiflorum Lam.) from China. Crop Prot. 2022, 161, 106068. [Google Scholar] [CrossRef]
- Zhao, N.; Yan, Y.; Du, L.; Zhang, X.; Liu, W.; Wang, J. Unravelling the effect of two herbicide resistance mutations on acetolactate synthase kinetics and growth traits. J. Exp. Bot. 2020, 71, 3535–3542. [Google Scholar] [CrossRef]
- Guo, W.; Chi, Y.; Feng, L.; Tian, X.; Liu, W.; Wang, J. Fenoxaprop-P-ethyl and mesosulfuron-methyl resistance status of shortawn foxtail (Alopecurus aequalis Sobol.) in eastern China. Pestic. Biochem. Physiol. 2018, 148, 126–132. [Google Scholar] [CrossRef]
- Tan, M.K.; Preston, C.; Wang, G.X. Molecular basis of multiple resistance to ACCase-inhibiting and ALS-inhibiting herbicides in Lolium rigidum. Weed Res. 2007, 47, 534–541. [Google Scholar] [CrossRef]
- Marshall, R.; Moss, S.R. Characterisation and molecular basis of ALS inhibitor resistance in the grass weed Alopecurus myosuroides. Weed Res. 2008, 48, 439–447. [Google Scholar] [CrossRef]
- Yu, Q.; Han, H.; Vila-Aiub, M.M.; Powles, S.B. AHAS herbicide resistance endowing mutations: Effect on AHAS functionality and plant growth. J. Exp. Bot. 2010, 61, 3925–3934. [Google Scholar] [CrossRef]
- Laforest, M.; Soufiane, B.; Simard, M.J.; Obeid, K.; Page, E.; Nurse, R.E. Acetyl-CoA carboxylase overexpression in herbicide-resistant large crabgrass (Digitaria sanguinalis). Pest Manag. Sci. 2017, 73, 2227–2235. [Google Scholar] [CrossRef]
- Zhao, N.; Yan, Y.; Wang, H.; Bai, S.; Wang, Q.; Liu, W.; Wang, J. Acetolactate synthase overexpression in mesosulfuron-methyl-resistant shortawn foxtail (Alopecurus aequalis Sobol.): Reference gene selection and herbicide target gene expression analysis. J. Agric. Food Chem. 2018, 66, 9624–9634. [Google Scholar] [CrossRef] [PubMed]
- Powles, S.B.; Preston, C. Herbicide cross resistance and multiple resistance in plants. Herbic. Resist. Action Comm. Monogr. 1995, 2, 1–19. [Google Scholar]
- Basak, S.; McElroy, J.S.; Brown, A.M.; Gonçalves, C.G.; Patel, J.D.; McCullough, P.E. Plastidic ACCase Ile-1781-Leu is present in pinoxaden-resistant southern crabgrass (Digitaria ciliaris). Weed Sci. 2020, 68, 41–50. [Google Scholar] [CrossRef]
- Cruz-Hipolito, H.; Fernandez, P.; Alcantara, R.; Gherekhloo, J.; Osuna, M.D.; De Prado, R. Ile-1781-Leu and Asp-2078-Gly mutations in ACCase gene, endow cross-resistance to APP, CHD, and PPZ in Phalaris minor from Mexico. Int. J. Mol. Sci. 2015, 16, 21363–21377. [Google Scholar] [CrossRef]
- Yanniccari, M.; Gigón, R. Cross-resistance to acetyl-CoA carboxylase–inhibiting herbicides conferred by a target-site mutation in perennial ryegrass (Lolium perenne) from Argentina. Weed Sci. 2020, 68, 116–124. [Google Scholar] [CrossRef]
- Zhao, N.; Yan, Y.; Ge, L.A.; Zhu, B.; Liu, W.; Wang, J. Target site mutations and cytochrome P450s confer resistance to fenoxaprop-P-ethyl and mesosulfuron-methyl in Alopecurus aequalis. Pest Manag. Sci. 2019, 75, 204–214. [Google Scholar] [CrossRef]
- Cruz-Hipolito, H.; Osuna, M.D.; Dominguez-Valenzuela, J.A.; Espinoza, N.; De Prado, R. Mechanism of resistance to ACCase-inhibiting herbicides in wild oat (Avena fatua) from Latin America. J. Agric. Food Chem. 2011, 59, 7261–7267. [Google Scholar] [CrossRef]
- Norsworthy, J.K.; Ward, S.M.; Shaw, D.R.; Llewellyn, R.S.; Nichols, R.L.; Webster, T.M.; Barrett, M. Reducing the risks of herbicide resistance: Best management practices and recommendations. Weed Sci. 2012, 60 (Suppl. 1), 31–62. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).