Identifying Nematode Damage on Soybean through Remote Sensing and Machine Learning Techniques
Abstract
1. Introduction
2. Materials and Methods
2.1. Sites Description
2.2. Data Collection
2.2.1. Pathogen and Plant Sampling
2.2.2. Nematode Extraction, Identification, and Qualification
2.2.3. Multispectral Data and VI Calculation
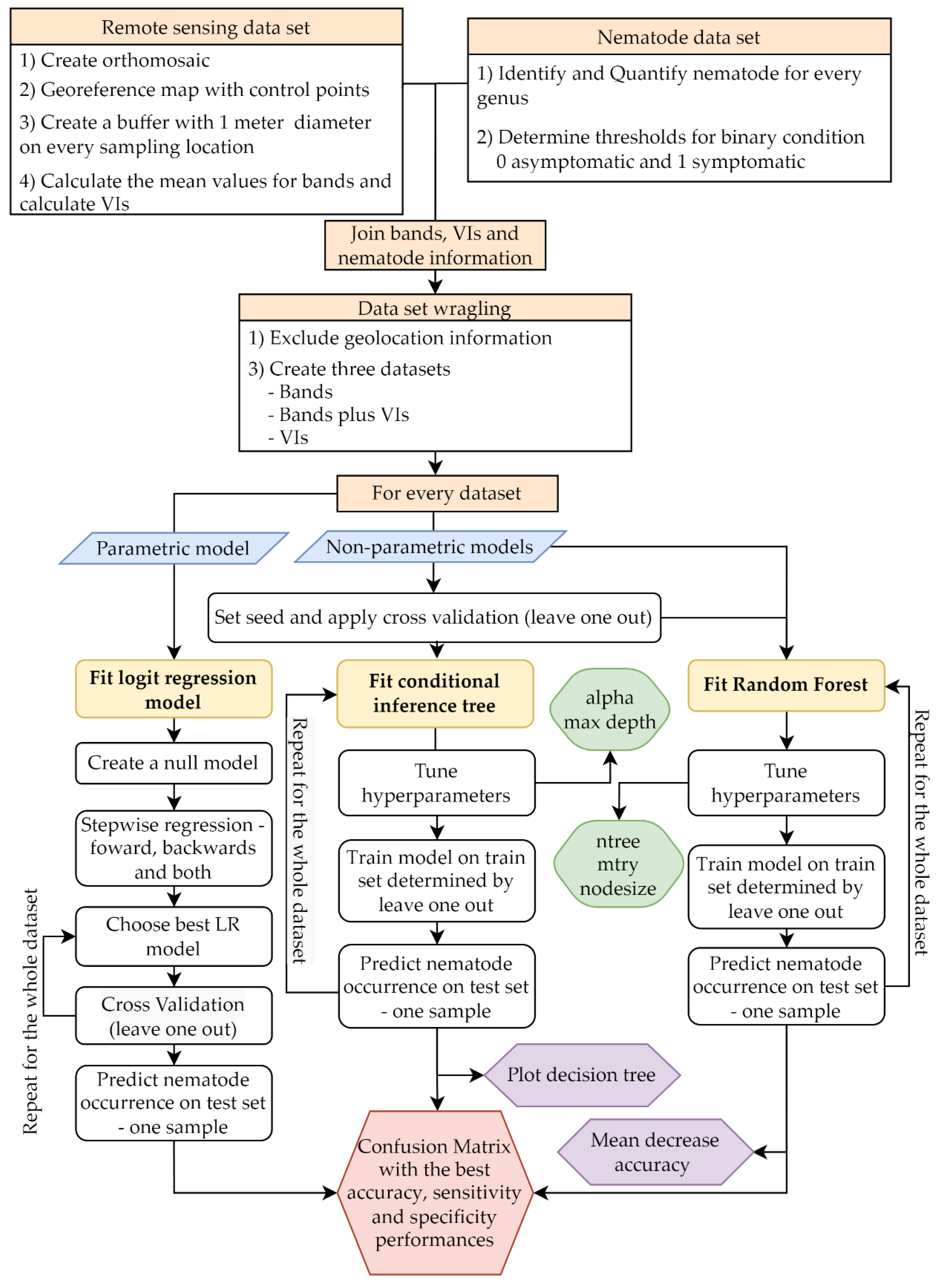
2.3. Data Analysis
2.3.1. Nematode Infection Classification
2.3.2. Bivariate Analysis of Individual Bands and VIs
2.3.3. Nematode Infection Prediction
2.3.4. Algorithm Performance Evaluation
3. Results
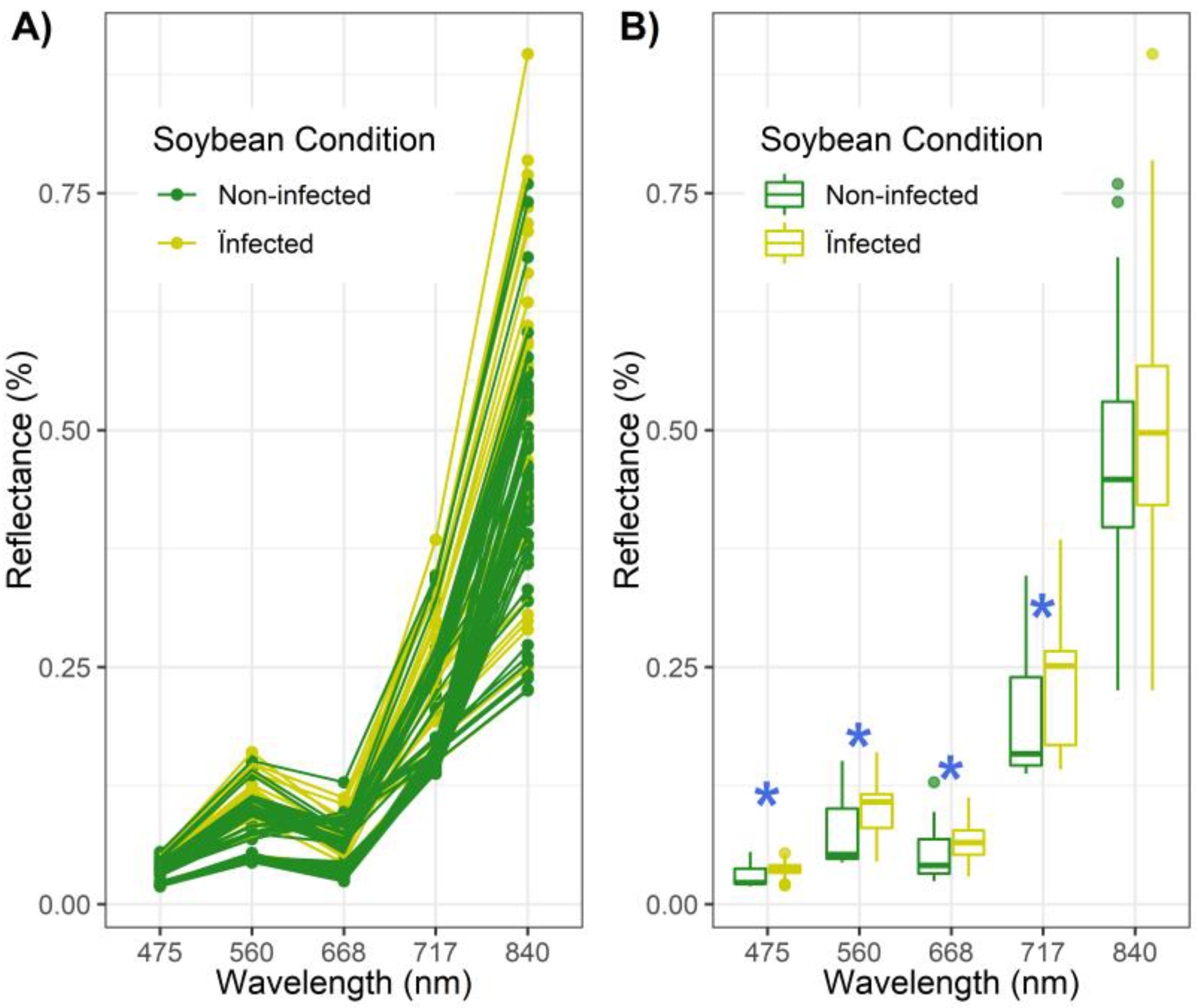
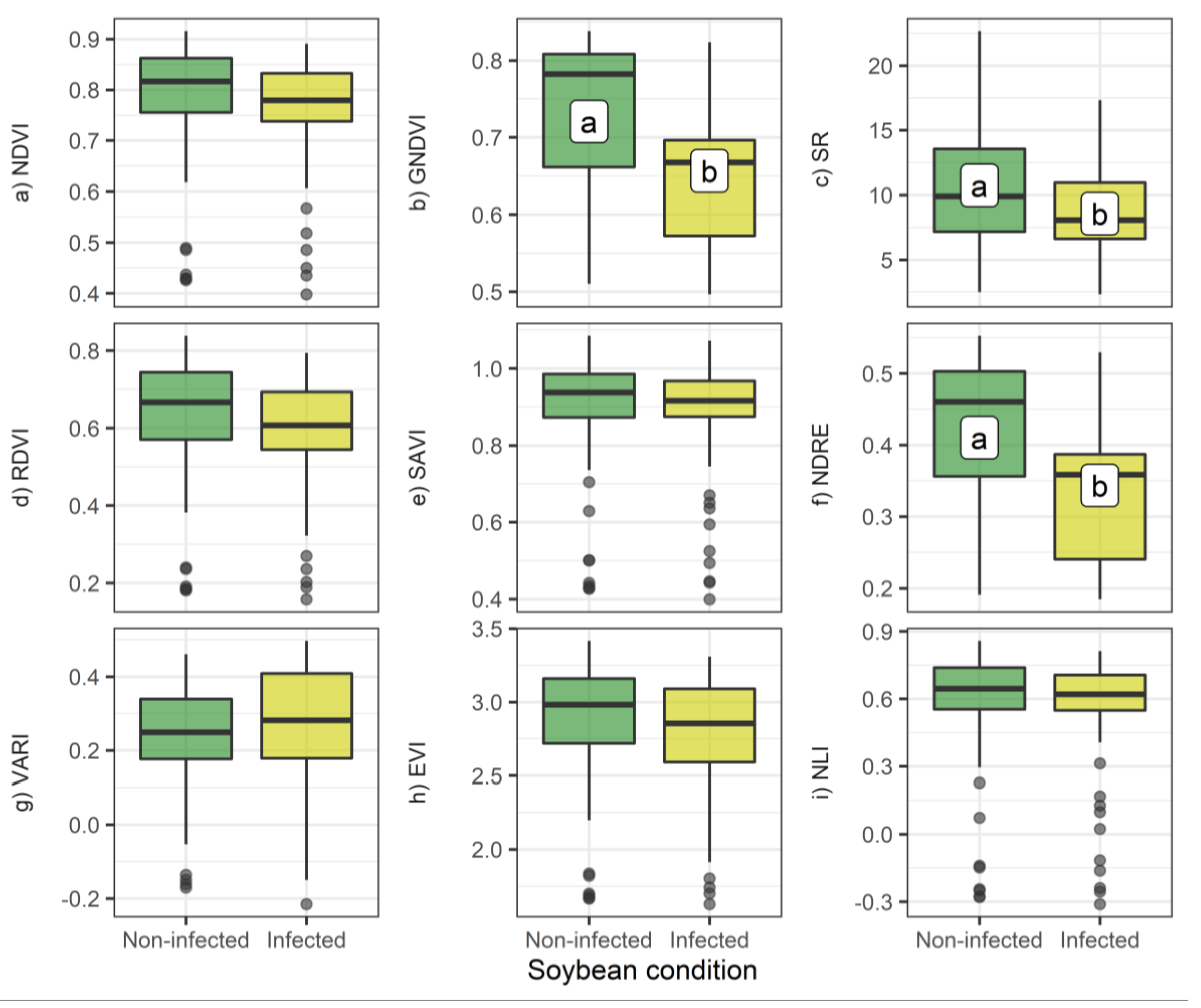
3.1. Bivariate Analysis of Individual Bands and VIs
3.2. Algorithms Performance
3.2.1. Logistic Regression
3.2.2. Random Forest
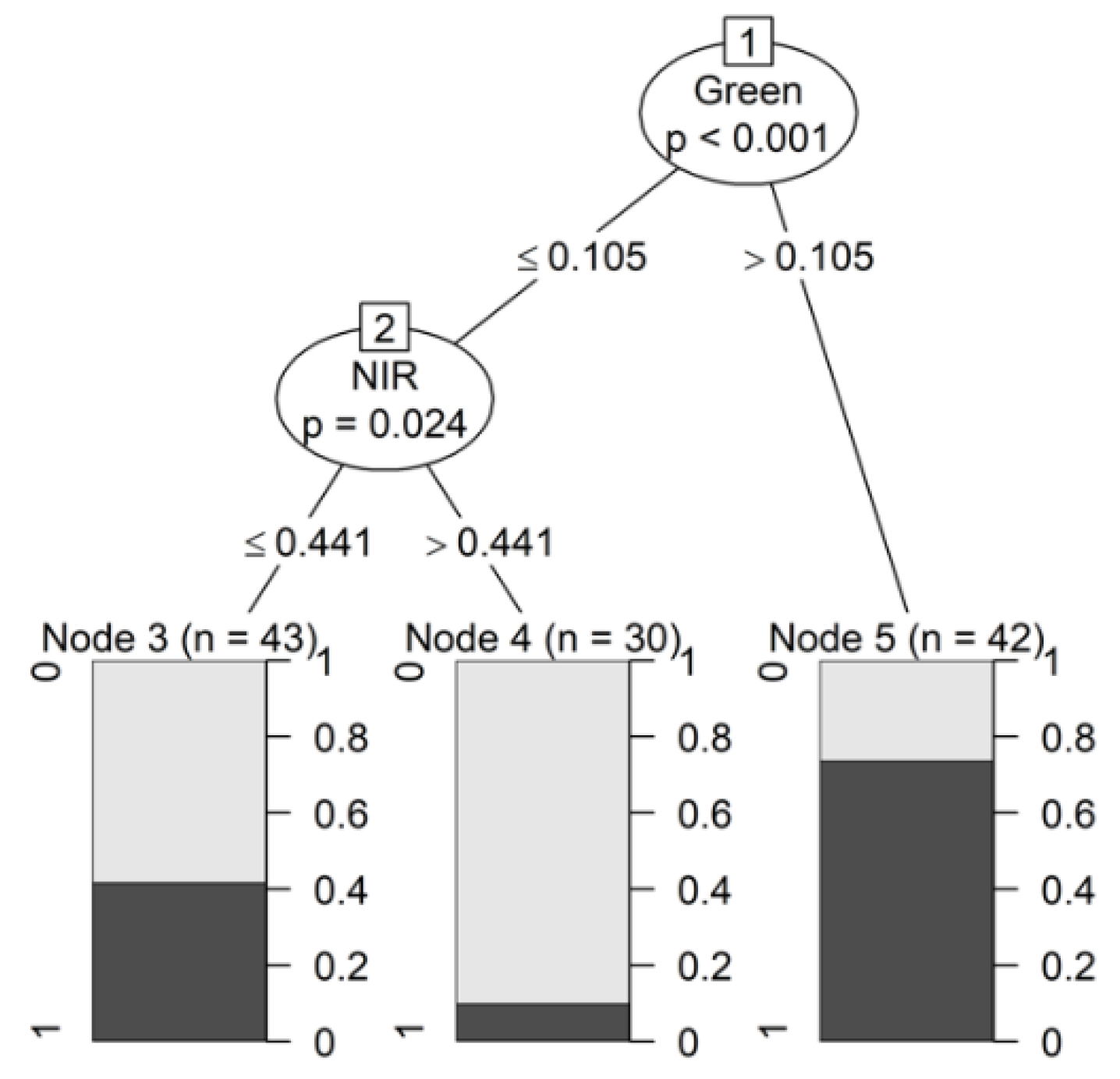
3.2.3. Conditional Inference Tree
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Askary, T.H.; Martinelli, P.P. Biocontrol Agents of Phytonematodes; CABI: Wallingford, UK, 2015. [Google Scholar]
- Allen, T.W.; Bradley, C.A.; Sisson, A.J.; Byamukama, E.; Chilvers, M.I.; Coker, C.M.; Collins, A.A.; Damicone, J.P.; Dorrance, A.E.; Dufault, N.S.; et al. Soybean Yield Loss Estimates Due to Diseases in the United States and Ontario, Canada, from 2010 to 2014. Plant Health Prog. 2017, 18, 19–27. [Google Scholar] [CrossRef]
- Zhao, L.; Jiang, P.; Humble, L.M.; Sun, J. Within-Tree Distribution and Attractant Sampling of Propagative Pinewood Nematode, Bursaphelenchus xylophilus: An Early Diagnosis Approach. For. Ecol. Manag. 2009, 258, 1932–1937. [Google Scholar] [CrossRef]
- Carneiro, R.G.; Mazzafera, P.; Ferraz, L.C.C.B.; Muraoka, T.; Trivelin, P.C.O. Uptake and Translocation of Nitrogen, Phosphorus and Calcium in Soybean Infected with Meloidogyne incognita and M. javanica. Fitopatol. Bras. 2002, 27, 141–150. [Google Scholar] [CrossRef]
- Blevins, D.G.; Dropkin, V.H.; Luedders, V.D. Macronutrient Uptake, Translocation, and Tissue Concentration of Soybeans Infested with the Soybean Cyst Nematode and Elemental Composition of Cysts Isolated from Roots1. J. Plant Nutr. 1995, 18, 579–591. [Google Scholar] [CrossRef]
- Hillnhütter, C.; Mahlein, A.K.; Sikora, R.A.; Oerke, E.C. Remote Sensing to Detect Plant Stress Induced by Heterodera Schachtii and Rhizoctonia Solani in Sugar Beet Fields. Field Crops Res. 2011, 122, 70–77. [Google Scholar] [CrossRef]
- Martins, G.D.; Galo, M.d.L.B.T.; Vieira, B.S. Detecting and Mapping Root-Knot Nematode Infection in Coffee Crop Using Remote Sensing Measurements. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 10, 5395–5403. [Google Scholar] [CrossRef]
- Wiesel, L.; Daniell, T.J.; King, D.; Neilson, R. Determination of the Optimal Soil Sample Size to Accurately Characterise Nematode Communities in Soil. Soil Biol. Biochem. 2015, 80, 89–91. [Google Scholar] [CrossRef]
- Garcia-Ruiz, F.; Sankaran, S.; Maja, J.M.; Lee, W.S.; Rasmussen, J.; Ehsani, R. Comparison of Two Aerial Imaging Platforms for Identification of Huanglongbing-Infected Citrus Trees. Comput. Electron. Agric. 2013, 91, 106–115. [Google Scholar] [CrossRef]
- Bajwa, S.G.; Rupe, J.C.; Mason, J. Soybean Disease Monitoring with Leaf Reflectance. Remote Sens. 2017, 9, 127. [Google Scholar] [CrossRef]
- Delalieux, S.; van Aardt, J.; Keulemans, W.; Schrevens, E.; Coppin, P. Detection of Biotic Stress (Venturia inaequalis) in Apple Trees Using Hyperspectral Data: Non-Parametric Statistical Approaches and Physiological Implications. Eur. J. Agron. 2007, 27, 130–143. [Google Scholar] [CrossRef]
- Steddom, K.; Heidel, G.; Jones, D.; Rush, C.M. Remote Detection of Rhizomania in Sugar Beets. Phytopathology 2003, 93, 720–726. [Google Scholar] [CrossRef] [PubMed]
- Adelabu, S.; Mutanga, O.; Adam, E. Evaluating the Impact of Red-Edge Band from Rapideye Image for Classifying Insect Defoliation Levels. ISPRS J. Photogramm. Remote Sens. 2014, 95, 34–41. [Google Scholar] [CrossRef]
- Rodriguez-Galiano, V.F.; Ghimire, B.; Rogan, J.; Chica-Olmo, M.; Rigol-Sanchez, J.P. An Assessment of the Effectiveness of a Random Forest Classifier for Land-Cover Classification. ISPRS J. Photogramm. Remote Sens. 2012, 67, 93–104. [Google Scholar] [CrossRef]
- Hothorn, T.; Hornik, K.; Zeileis, A. Unbiased Recursive Partitioning: A Conditional Inference Framework. J. Comput. Graph. Stat. 2006, 15, 651–674. [Google Scholar] [CrossRef]
- Yuan, Q.; Shen, H.; Li, T.; Li, Z.; Li, S.; Jiang, Y.; Xu, H.; Tan, W.; Yang, Q.; Wang, J.; et al. Deep Learning in Environmental Remote Sensing: Achievements and Challenges. Remote Sens. Environ. 2020, 241, 111716. [Google Scholar] [CrossRef]
- Susič, N.; Žibrat, U.; Širca, S.; Strajnar, P.; Razinger, J.; Knapič, M.; Vončina, A.; Urek, G.; Stare, B.G. Discrimination between Abiotic and Biotic Drought Stress in Tomatoes Using Hyperspectral Imaging. Sens. Actuators B Chem. 2018, 273, 842–852. [Google Scholar] [CrossRef]
- Jenkins, W.R. A Rapid Centrifugal-Flotation Technique for Separating Nematodes from Soil. Plant Dis. Report. 1964, 48, 692. [Google Scholar]
- Coolen, W.A.; D’herde, C.J.A.; D’herde, C.J. A Method for the Quantitative Extraction of Nematodes from Plant Tissue; State Agricultural Research Centre: Ghent, Belgium, 1972. [Google Scholar]
- Tihohod, D. Nematologia Agrícola, 2nd ed.; Funep: Jaboticabal, Brazil, 1993. [Google Scholar]
- Southey, J.F. Laboratory Methods for Work with Plant and Soil Nematodes; Technical bulletin 2; Her Majesty’s Stationery Office: London, UK, 1986; ISBN 0112427545. [Google Scholar]
- Shepherd, A.M. Extraction and Estimation of Cyst Nematodes. In Laboratory Methods for Work with Plant and Soil Nematodes; Her Majesty’s Stationery Office: London, UK, 1986; pp. 31–49. [Google Scholar]
- Rouse, R.W.H.; Haas, J.A.W.; Deering, D.W. Monitoring Vegetation Systems in the Great Plains with ERTS; NASA: Washington, DC, USA, 1974. [Google Scholar]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a Green Channel in Remote Sensing of Global Vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Jordan, C.F. Derivation of Leaf-Area Index from Quality of Light on the Forest Floor. Ecology 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Roujean, J.; Breon, F. Estimating PAR Absorbed by Vegetation from Bidirectional Reflectance Measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Huete, A.R. A Soil-Adjusted Vegetation Index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Barnes, E.M.; Clarke, T.R.; Richards, S.E.; Colaizzi, P.D.; Haberland, J.; Kostrzewski, M.; Waller, P.M.; Choi, C.Y.; Riley, E.; Thompson, T.E.; et al. Coincident Detection of Crop Water Stress, Nitrogen Status and Canopy Density Using Ground-Based Multispectral Data. In Proceedings of the Fifth International Conference on Precision Agriculture, Madison, WI, USA, 16–19 July 2000. [Google Scholar]
- Liu, H.Q.; Huete, A. A Feedback Based Modification of the NDVI to Minimize Canopy Background and Atmospheric Noise. IEEE Trans. Geosci. Remote Sens. 1995, 33, 457–465. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Stark, R.; Rundquist, D. Novel Algorithms for Remote Estimation of Vegetation Fraction. Remote Sens. Environ. 2002, 80, 76–87. [Google Scholar] [CrossRef]
- Goel, N.S.; Qin, W. Influences of Canopy Architecture on Relationships between Various Vegetation Indices and LAI and FPAR: A Computer Simulation. Remote Sens. Rev. 1994, 10, 309–347. [Google Scholar] [CrossRef]
- Kinloch, R.A. Review: The Control of Nematodes Injurious to Soybean. Nematropica 1980, 141–153. [Google Scholar]
- Koenning, S.R.; Wrather, J.A.; Kirkpatrick, T.L.; Walker, N.R.; Starr, J.L.; Mueller, J.D. Plant-parasitic nematodes attacking cotton in the United States: Old and emerging production challenges. Plant Dis. 2004, 88, 100–113. [Google Scholar] [CrossRef]
- Dickerson, O.; Blake, J.; Lewis, S. Nematode Guidelines for South Carolina. In Clemson Extension Bulletin; Clemson University: Clemson, SC, USA, 2000. [Google Scholar]
- Liaw, A.; Wiener, M. Classification and Regression by RandomForest. R News 2002, 2, 18–22. [Google Scholar]
- Kuhn, M. Building predictive models in R using the caret package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Nutter, F.W.; Tylka, G.L.; Guan, J.; Moreira, A.J.D.; Marett, C.C.; Rosburg, T.R.; Basart, J.P.; Chong, C.S. Use of Remote Sensing to Detect Soybean Cyst Nematode-Induced Plant Stress. J. Nematol. 2002, 34, 222–231. [Google Scholar]
- Haseeb, A.; Srivastava, N.K.; Pandey, R. The Influence of Meloidogyne incognita on Growth, Physiology, Nutrient Concentration and Alkaloid Yield of Hyoscy Amus Niger. Nematol. Mediterr. 1990, 18, 127–129. [Google Scholar]
- Behmann, J.; Steinrücken, J.; Plümer, L. Detection of Early Plant Stress Responses in Hyperspectral Images. ISPRS J. Photogramm. Remote Sens. 2014, 93, 98–111. [Google Scholar] [CrossRef]
- Sims, D.A.; Gamon, J.A. Relationships between Leaf Pigment Content and Spectral Reflectance across a Wide Range of Species, Leaf Structures and Developmental Stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Chappelle, E.W.; McMurtrey, J.E.; Wood, F.M.; Newcomb, W.W. Laser-Induced Fluorescence of Green Plants 2: LIF Caused by Nutrient Deficiencies in Corn. Appl. Opt. 1984, 23, 139. [Google Scholar] [CrossRef]
- Curran, P.J.; Dungan, J.L.; Gholz, H.L. Exploring the Relationship between Reflectance Red Edge and Chlorophyll Content in Slash Pine. Tree Physiol. 1990, 7, 33–48. [Google Scholar] [CrossRef] [PubMed]
- Gitelson, A.A.; Merzlyak, M.N.; Chivkunova, O.B. Optical Properties and Nondestructive Estimation of Anthocyanin Content in Plant Leaves. Photochem. Photobiol. 2001, 74, 38–45. [Google Scholar] [CrossRef]
- Baret, F.; Guyot, G. Potentials and Limits of Vegetation Indices for LAI and APAR Assessment. Remote Sens. Environ. 1991, 35, 161–173. [Google Scholar] [CrossRef]
- Martins, G.D.; de Lourdes Bueno Trindade Galo, M. Caracterização Espectral Da Cana-de-Açúcar Infectada Por Nematoides e Migdolus Fryanus Por Espectrorradiometria de Campo. Boletim Ciências Geodésicas 2015, 21, 783–796. [Google Scholar] [CrossRef][Green Version]
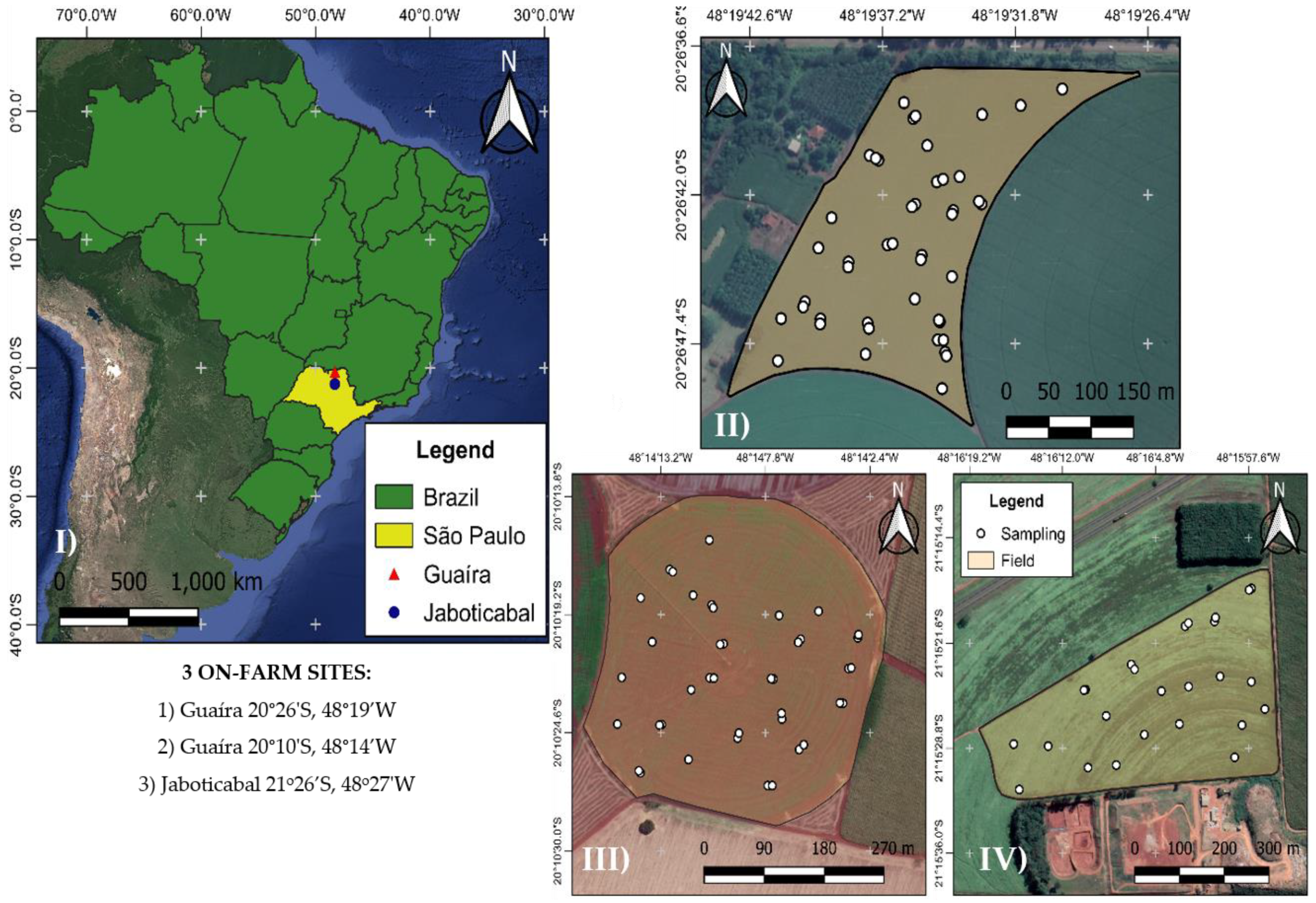
| Site | Elevation (m) * | Nematode Species Identified | Area (ha) |
|---|---|---|---|
| I | 502 | Heterodera glycines, Meloidogyne incognita and Pratylenchus brachyurus | 16.1 |
| II | 540 | M. incognita, P. brachyurus | 8.2 |
| III | 630 | M. incognita, Rotylenchulus reniformis and P. brachyurus | 19.7 |
| VI | Description | Equation | Reference |
|---|---|---|---|
| NDVI | Normalized difference vegetation index | (NIR – R)/(NIR + R) | [23] |
| GNDVI | Green normalized difference vegetation index | (NIR – Green)/(NIR + Green) | [24] |
| SR | Simple ratio | NIR/R | [25] |
| RDVI | Renormalized difference vegetation index | (NIR – R)/√ (NIR + R) | [26] |
| SAVI | Soil adjusted vegetation index | (NIR-R)/(NIR + R + L) × (1 + L) | [27] |
| NDRE | Normalized difference red edge | (NIR – RE) / (NIR + RE) | [28] |
| EVI | Enhanced vegetation index | 2.5 × (NIR – R)/(NIR + 6 × R – 7.5× B) + 1 | [29] |
| VARI | Visible atmospherically resistant index | (G – R)/(G + R – B) | [30] |
| NLI | Non-linear index | (NIR2 – R)/(NIR2 + R) | [31] |
| Nematode Genus | Soil | Roots |
|---|---|---|
| H. glycines (Cysts) | 5 | - |
| H. glycines | 400 | - |
| P. brachyurus | 100 | 800 |
| M. incognita | 300 | 120 |
| R. reniformis | 300 | - |
| Metric | Bands | VIs | Bands Plus VIs | ||||||
|---|---|---|---|---|---|---|---|---|---|
| LR | CIT | RF | LR | CIT | RF | LR | CIT | RF | |
| Accuracy | 0.66 | 0.71 | 0.70 | 0.65 | 0.67 | 0.65 | 0.64 | 0.71 | 0.68 |
| Sensitivity | 0.71 | 0.72 | 0.67 | 0.67 | 0.60 | 0.62 | 0.66 | 0.72 | 0.66 |
| Specificity | 0.62 | 0.71 | 0.71 | 0.62 | 0.78 | 0.67 | 0.62 | 0.71 | 0.69 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, L.B.; Bastos, L.M.; de Oliveira, M.F.; Soares, P.L.M.; Ciampitti, I.A.; da Silva, R.P. Identifying Nematode Damage on Soybean through Remote Sensing and Machine Learning Techniques. Agronomy 2022, 12, 2404. https://doi.org/10.3390/agronomy12102404
Santos LB, Bastos LM, de Oliveira MF, Soares PLM, Ciampitti IA, da Silva RP. Identifying Nematode Damage on Soybean through Remote Sensing and Machine Learning Techniques. Agronomy. 2022; 12(10):2404. https://doi.org/10.3390/agronomy12102404
Chicago/Turabian StyleSantos, Letícia Bernabé, Leonardo Mendes Bastos, Mailson Freire de Oliveira, Pedro Luiz Martins Soares, Ignacio Antonio Ciampitti, and Rouverson Pereira da Silva. 2022. "Identifying Nematode Damage on Soybean through Remote Sensing and Machine Learning Techniques" Agronomy 12, no. 10: 2404. https://doi.org/10.3390/agronomy12102404
APA StyleSantos, L. B., Bastos, L. M., de Oliveira, M. F., Soares, P. L. M., Ciampitti, I. A., & da Silva, R. P. (2022). Identifying Nematode Damage on Soybean through Remote Sensing and Machine Learning Techniques. Agronomy, 12(10), 2404. https://doi.org/10.3390/agronomy12102404









