Simulation of Multi-Species Plant Communities in Perturbed and Nutrient-Limited Grasslands: Development of the Growth Model ModVege
Abstract
:1. Introduction
1.1. Plant Diversity in Grasslands
1.2. Functional Typology
1.3. Grassland Modelling
2. Materials and Methods
2.1. The Modelling Framework
2.1.1. Generic Grassland Model (ModVege)
2.1.2. Community Simulation Model (CoSMo)
2.2. Study-Site and Experimental Design
2.3. Experimental Data
2.4. Simulation Design and Model Evaluation
3. Results
3.1. Evaluation of Modelling Solutions for Grassland Biomass Production
3.2. Evaluation of Modelling Solutions for Relative Abundances
3.2.1. Relative Abundance of Plant (Grass) Functional Types
3.2.2. Relative Abundance of Plant Species
4. Discussion
4.1. Plant Biomass Simulation
4.2. Relative Abundance Simulation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Theau, J.-P.; Carrié, R.; Sirami, C.; Prud’homme, F. Grassland plant diversity: Definition. Dict. D’agroecologie. 2022. Available online: https://dicoagroecologie.fr/en/encyclopedia/grassland-plant-diversity (accessed on 7 October 2022).
- Bengtsson, J.; Bullock, J.M.; Egoh, B.; Everson, C.; Everson, T.; O’Connor, T.; O’Farrell, P.J.; Smith, H.G.; Lindborg, R. Grasslands—more important for ecosystem services than you might think. Ecosphere 2019, 10, e02582. [Google Scholar] [CrossRef]
- Perronne, R.; Mauchamp, L.; Mouly, A.; Gillet, F. Contrasted taxonomic, phylogenetic and functional diversity patterns in semi-natural permanent grasslands along an altitudinal gradient. Plant Ecol. Evol. 2014, 147, 165–175. [Google Scholar] [CrossRef] [Green Version]
- Tilman, D.; Knops, J.; Wedin, D.; Reich, P.; Ritchie, M.; Siemann, E. The influence of functional diversity and composition on ecosystem processes. Science 1997, 277, 1300–1302. [Google Scholar] [CrossRef] [Green Version]
- Tilman, D.; Reich, P.B.; Knops, J.M.H. Biodiversity and ecosystem stability in a decade-long grassland experiment. Nature 2006, 441, 629–632. [Google Scholar] [CrossRef]
- Marquard, E.; Weigelt, A.; Roscher, C.; Gubsch, M.; Lipowsky, A.; Schmid, B. Positive biodiversity-productivity relationship due to increased plant density. J. Ecol. 2009, 97, 696–704. [Google Scholar] [CrossRef] [Green Version]
- Kraft, N.J.B.; Godoy, O.; Levine, J.M. Plant functional traits and the multidimensional nature of species coexistence. Proc. Natl. Acad. Sci. USA 2015, 112, 797–802. [Google Scholar] [CrossRef] [Green Version]
- Roscher, C.; Gubsch, M.; Lipowsky, A.; Schumacher, J.; Weigelt, A.; Buchmann, N.; Schulze, E.-D.; Schmid, B. Trait means, trait plasticity and trait differences to other species jointly explain species performances in grasslands of varying diversity. Oikos 2018, 127, 865. [Google Scholar] [CrossRef] [Green Version]
- Chapin, F.S. Effects of plant traits on ecosystem and regional processes: A conceptual framework for predicting the consequences of global change. Ann. Bot. 2003, 91, 455–463. [Google Scholar] [CrossRef] [Green Version]
- Ansquer, P.; Duru, M.; Theau, J.P.; Cruz, P. Functional traits as indicators of fodder provision over a short time scale in species-rich grasslands. Ann. Bot. 2009, 103, 117–126. [Google Scholar] [CrossRef] [Green Version]
- Zheng, S.; Li, W.; Lan, Z.; Ren, H.; Wang, K. Functional trait responses to grazing are mediated by soil moisture and plant functional group identity. Sci. Rep. 2015, 5, 18163. [Google Scholar] [CrossRef]
- Isbell, F.; Craven, D.; Connolly, J.; Loreau, M.; Schmid, B.; Beierkuhnlein, C.; Bezemer, T.M.; Bonin, C.; Bruelheide, H.; de Luca, E.; et al. Biodiversity increases the resistance of ecosystem productivity to climate extremes. Nature 2015, 526, 574. [Google Scholar] [CrossRef]
- Díaz, S.; Lavorel, S.; de Bello, F.; Quétier, F.; Grigulis, K.; Robson, M. Incorporating plant functional diversity effects in ecosystem service assessments. Proc. Natl. Acad. Sci. USA 2007, 104, 20684–20689. [Google Scholar] [CrossRef] [Green Version]
- Klumpp, K.; Soussana, J.-F. Using functional traits to predict grassland ecosystem change: A mathematical test of the response-and-effect trait approach. Glob. Change Biol. 2009, 15, 2921–2934. [Google Scholar] [CrossRef]
- Lavorel, S. Plant functional effects on ecosystem services. J. Ecol. 2013, 101, 4–8. [Google Scholar] [CrossRef]
- Quétier, F.; Lavorel, S.; Thuiller, W.; Davies, I. Plant-trait-based modeling assessment of ecosystem-service sensitivity to land-use change. Ecol. Appl. 2007, 17, 2377–2386. [Google Scholar] [CrossRef]
- Lamarque, P.; Lavorel, S.; Mouchet, M.; Quétier, F. Plant trait-based models identify direct and indirect effects of climate change on bundles of grassland ecosystem services. Proc. Natl. Acad. Sci. USA 2014, 111, 13751–13756. [Google Scholar] [CrossRef] [Green Version]
- Carrère, P.; Chabalier, C.; Landrieaux, J.; Orth, D.; Piquet, M.; Rivière, J.; Seytre, L. Une typologie multifonctionnelle des prairies des systèmes laitiers AOP du Massif Central combinant des approches agronomiques et écologiques. Fourrages 2012, 209, 9–22. (In French) [Google Scholar]
- Cruz, P.; Duru, M.; Therond, O.; Theau, J.P.; Ducourtieux, C.; Jouany, C.; Al Haj Khaled, R.; Ansquer, P. Une nouvelle approche pour caractériser les prairies naturelles et leur valeur d’usage. Fourrages 2002, 172, 335–354. (In French) [Google Scholar]
- Theau, J.-P.; Pauthenet, Y.; Cruz, P. Une typologie des espèces non graminéennes pour mieux caractériser la diversité et la valeur d’usage des prairies permanentes. Fourrages 2017, 232, 321–329. (In French) [Google Scholar]
- Violle, C.; Navas, M.-L.; Vile, D.; Kazakou, E.; Fortunel, C.; Hummel, I.; Garnier, E. Let the concept of trait be functional. Oikos 2007, 116, 882–892. [Google Scholar] [CrossRef]
- Michaud, A.; Andueza, D.; Picard, F.; Plantureux, S.; Baumont, R. Seasonal dynamics of biomass production and herbage quality of three grasslands with contrasting functional compositions. Grass Forage Sci. 2012, 67, 64–76. [Google Scholar] [CrossRef]
- Duru, M.; Cruz, P.; Al Haj Khaled, R.; Ducourtieux, C.; Theau, J.-P. Relevance of plant functional types based on leaf dry matter content for assessing digestibility of native grass species and species-rich grassland communities in spring. Agron. J. 2008, 100, 1622–1630. [Google Scholar] [CrossRef]
- Cruz, P.; Theau, J.P.; Lecloux, E.; Jouany, C.; Duru, M. Typologie fonctionnelle de graminées fourragères pérennes: Une classification multitraits. Fourrages 2010, 201, 11–17. (In French) [Google Scholar]
- Jouven, M.; Carrère, P.; Baumont, R. Model predicting dynamics of biomass, structure and digestibility of herbage in managed permanent pastures. 1. Model description. Grass Forage Sci. 2006, 61, 112–124. [Google Scholar] [CrossRef]
- Jouven, M.; Carrère, P.; Baumont, R. Model predicting dynamics of biomass, structure and digestibility of herbage in managed permanent pastures. 2. Model evaluation. Grass Forage Sci. 2006, 61, 125–133. [Google Scholar] [CrossRef]
- Ehrhardt, F.; Soussana, J.-F.; Bellocchi, G.; Grace, P.; McAuliffe, R.; Recous, S.; Sándor, R.; Smith, P.; Snow, V.; de Antoni Migliorati, M.; et al. Assessing uncertainties in crop and pasture ensemble model simulations of productivity and N2O emissions. Glob. Change Biol. 2018, 24, e603–e616. [Google Scholar] [CrossRef] [Green Version]
- Sándor, R.; Ehrhardt, F.; Grace, P.; Recous, S.; Smith, P.; Snow, V.; Soussana, J.-F.; Basso, B.; Bhatia, A.; Brilli, L.; et al. Ensemble modelling of carbon fluxes in grasslands and croplands. Field Crops Res. 2020, 252, 107791. [Google Scholar] [CrossRef]
- Li, F.Y.; Snow, V.; Holzworth, D.R. Modelling the seasonal and geographical pattern of pasture production in New Zealand. N. Z. J. Agric. Res. 2011, 54, 331–352. [Google Scholar] [CrossRef] [Green Version]
- Corre-Hellou, G.; Faure, M.; Launay, M.; Brisson, N.; Crozat, Y. Adaptation of the STICS intercrop model to simulate crop growth and N accumulation in pea-barley intercrops. Field Crops Res. 2009, 113, 72–81. [Google Scholar] [CrossRef]
- Jégo, G.; Rotz, C.A.; Bélanger, G.; Tremblay, G.F.; Charbonneau, É.; Pellerin, D. Simulating forage crop production in a northern climate with the integrated farm system model. Can. J. Plant Sci. 2015, 95, 745–757. [Google Scholar] [CrossRef] [Green Version]
- Thivierge, M.-N.; Jégo, G.; Bélanger, G.; Bertrand, A.; Tremblay, G.F.; Rotz, C.A.; Qian, B. Predicted yield and nutritive value of an alfalfa-timothy mixture under climate change and elevated atmospheric carbon dioxide. Agron. J. 2016, 108, 585–603. [Google Scholar] [CrossRef] [Green Version]
- Wirth, S.B.; Taubert, F.; Tietjen, B.; Müller, C.; Rolisnki, S. Do details matter? Disentangling the processes related to plant species interactions in two grassland models of different complexity. Ecol. Model. 2021, 460, 109737. [Google Scholar] [CrossRef]
- Moulin, T.; Perasso, A.; Gillet, F. Modelling vegetation dynamics in managed grasslands: Responses to drivers depend on species richness. Ecol. Model. 2018, 374, 22–36. [Google Scholar] [CrossRef]
- Tilman, D. Resources: A graphical-mechanistic approach to competition and predation. Am. Nat. 1980, 116, 362–393. [Google Scholar] [CrossRef]
- Moulin, T.; Perasso, A.; Calanca, P.; Gillet, F. DynaGraM: A process-based model to simulate multi-species plant community dynamics in managed grasslands. Ecol. Model. 2021, 439, 109345. [Google Scholar] [CrossRef]
- Confalonieri, R. CoSMo: A simple approach for reproducing plant community dynamics using a single instance of generic crop simulators. Ecol. Model. 2014, 286, 1–10. [Google Scholar] [CrossRef]
- Confalonieri, R.; Francone, C.; Cappelli, G.; Stella, T.; Frasso, N.; Carpani, M.; Bregaglio, S.; Acutis, M.; Tubiello, F.N.; Fernandes, E. A multi-approach software library for estimating crop suitability to environment. Comput. Electron. Agric. 2013, 90, 170–175. [Google Scholar] [CrossRef] [Green Version]
- van Oijen, M.; Barcza, Z.; Confalonieri, R.; Korhonen, P.; Kröel-Dulay, G.; Lellei-Kovács, E.; Louarn, G.; Louault, F.; Martin, R.; Moulin, T.; et al. Incorporating biodiversity into biogeochemistry models to improve prediction of ecosystem services in temperate grasslands: Review and roadmap. Agronomy 2020, 10, 259. [Google Scholar] [CrossRef] [Green Version]
- Stöckle, C.O.; Donatelli, M.; Nelson, R. CropSyst, a cropping systems simulation model. Eur. J. Agron. 2003, 18, 289–307. [Google Scholar] [CrossRef]
- van Keulen, H.; Wolf, J. Modelling of Agricultural Production: Weather Soils and Crops; Pudoc: Wageningen, The Netherlands, 1986. [Google Scholar]
- Movedi, E.; Bellocchi, G.; Argenti, G.; Paleari, L.; Vesely, F.; Staglianò, N.; Dibari, C.; Confalonieri, R. Development of generic crop models for simulation of multi-species plant communities in mown grasslands. Ecol. Model. 2019, 401, 111–128. [Google Scholar] [CrossRef]
- Carrère, P.; Force, C.; Soussana, J.-F.; Louault, F.; Dumont, B.; Baumont, R. Design of a spatial model of a perennial grassland grazed by a herd of ruminants: The vegetation sub-model. In Grassland Science in Europe; Durand, J.-L., Emile, J.-C., Huyghes, C., Lemaire, G., Eds.; European Grassland Federation: Zürich, Switzerland, 2002; Volume 7, pp. 282–283. [Google Scholar]
- Carrère, P.; Sosinski, E.E., Jr.; Louault, F.; Soussana, J.-F. Validation of a model simulating grassland vegetation dynamics using plant traits measured along a gradient of disturbance. In Grassland Science in Europe; Lüscher, A., Jeangros, B., Kessler, W., Huguenin, O., Lobsiger, M., Millar, N., Suter, D., Eds.; European Grassland Federation: Zürich, Switzerland, 2004; Volume 9, pp. 784–786. [Google Scholar]
- Graux, A.-I.; Klumpp, K.; Ma, S.; Martin, R.; Bellocchi, G. Plant trait-based assessment of the Pasture Simulation model. In Proceedings of the 8th International Congress on Environmental Modelling and Software, Toulouse, France, 10–14 July 2016; Sauvage, S., Sánchez-Pérez, J.M., Rizzoli, A.E., Eds.; International Environmental Modelling and Software Society: Toulouse, France, 2016; Volume 2, pp. 518–525. [Google Scholar]
- Schapendonk, A.H.C.M.; Stol, W.; van Kraalingen, D.W.G.; Bouman, B.A.M. LINGRA, a sink/source model to simulate grassland productivity in Europe. Eur. J. Agron. 1998, 9, 87–100. [Google Scholar] [CrossRef]
- Bélanger, G.; Gastal, F.; Warembourg, F.R. Carbon balance of tall fescue (Festuca arundinacea Schreb.): Effects of nitrogen fertilization and the growing season. Ann. Bot. 1994, 74, 653–659. [Google Scholar] [CrossRef]
- Hurtado-Uria, C.; Hennessy, D.; Shalloo, L.; Schulte, R.P.O.; Delaby, L.; O’Connor, D. Evaluation of three grass growth models to predict grass growth in Ireland. J. Agric. Sci. 2013, 151, 91–104. [Google Scholar] [CrossRef] [Green Version]
- Calanca, P.; Deléglise, C.; Martin, R.; Carrère, P.; Mosimann, E. Testing the ability of a simple grassland model to simulate the seasonal effects of drought on herbage growth. Field Crops Res. 2016, 187, 12–23. [Google Scholar] [CrossRef]
- Ruelle, E.; Delaby, L.; O’Donovan, M. La prévision de la croissance d’herbe en Irlande: Une information attendue, de l’éleveur au gouvernement. Fourrages 2021, 247, 33–39. (In French) [Google Scholar]
- Ruelle, E.; Hennessy, D.; Delaby, L. Development of the Moorepark St Gilles grass growth model (MoSt GG model): A predictive model for grass growth for pasture based systems. Eur. J. Agron. 2018, 99, 80–91. [Google Scholar] [CrossRef]
- Louault, F.; Pottier, J.; Note, P.; Vile, D.; Soussana, J.-F.; Carrère, P. Complex plant community responses to modifications of disturbance and nutrient availability in productive permanent grasslands. J. Veg. Sci. 2017, 28, 538–549. [Google Scholar] [CrossRef]
- De Martonne, E. Nouvelle carte mondiale de l’indice d’aridité. Ann. De Géographie 1942, 51, 242–250. (In French) [Google Scholar] [CrossRef]
- Diodato, N.; Ceccarelli, M. Multivariate indicator Kriging approach using a GIS to classify soil degradation for Mediterranean agricultural lands. Ecol. Indic. 2004, 4, 177–187. [Google Scholar] [CrossRef]
- Grant, S.A. Resource description: Vegetation and sward components. In Sward Measurement Handbook, 2nd ed.; Davies, A., Baker, R.D., Grant, S.A., Laidlaw, A.S., Eds.; British Grassland Society: Reading, UK, 1993; pp. 69–97. [Google Scholar]
- Piseddu, F.; Bellocchi, G.; Picon-Cochard, C. Mowing and warming effects on grassland species richness and harvested biomass: Meta-analyses. Agron. Sustain. Dev. 2021, 41, 74. [Google Scholar] [CrossRef]
- Jackson, R.B.; Mooney, H.A.; Schulze, E.D. A global budget for fine root biomass, surface area, and nutrient contents. Proc. Natl. Acad. Sci. USA 1997, 94, 7362–7366. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Frelich, L.E. Flowering phenology and height growth pattern are associated with maximum plant height, relative growth rate and stem tissue mass density in herbaceous grassland species. J. Ecol. 2011, 99, 991–1000. [Google Scholar] [CrossRef]
- Zhang, H.; Sun, Y.; Chang, L.; Qin, Y.; Chen, J.; Qin, Y.; Du, J.; Yi, S.; Wang, Y. Estimation of grassland canopy height and aboveground biomass at the quadrat scale using unmanned aerial vehicle. Remote Sens. 2018, 10, 851. [Google Scholar] [CrossRef] [Green Version]
- Bourdôt, G.W. A Study of the Growth and Development of Yarrow (Achillea millefolium L.); Lincoln College, University of Canterbury: Canterbury, UK, 1980. [Google Scholar]
- Ianovici, N.; Veres, M.; Catrina, R.G.; Pîrvulescu, A.M.; Tanase, R.M.; Datcu, D.A. Methods of biomonitoring in urban environment: Leaf area and fractal dimension. Ann. West Univ. Timisoara. Ser. Biol. 2015, 18, 169–178. [Google Scholar]
- Gulías, J.; Flexas, J.; Mus, M.; Cifre, J.; Lefi, E.; Medrano, H. Relationship between maximum leaf photosynthesis, nitrogen content and specific leaf area in Balearic endemic and non-endemic Mediterranean species. Ann. Bot. 2003, 92, 215–222. [Google Scholar] [CrossRef]
- Poorter, H.; de Jong, R. A comparison of specific leaf area, chemical composition and leaf construction costs of field plants from 15 habitats differing in productivity. New Phytol. 1999, 143, 163–176. [Google Scholar] [CrossRef]
- Nölke, I.; Tonn, B.; Isselstein, J. Seasonal plasticity is more important than population variability in effects on white clover architecture and productivity. Ann. Bot. 2021, 128, 73–82. [Google Scholar] [CrossRef]
- Bellocchi, G.; Rivington, M.; Matthews, K.; Donatelli, M. Validation of biophysical models: Issues and methodologies. A review. Agron. Sustain. Dev. 2010, 30, 109–130. [Google Scholar] [CrossRef] [Green Version]
- Snow, V.; Rotz, C.A.; Moore, A.D.; Martin-Clouaire, R.; Johnson, I.R.; Hutchings, N.J.; Eckard, R.J. The challenges—and some solutions—to process-based modelling of grazed agricultural systems. Environ. Model. Softw. 2014, 62, 420–436. [Google Scholar] [CrossRef]
- Vuichard, N.; Soussana, J.-F.; Ciais, P.; Viovy, N.; Ammann, C.; Calanca, P.; Clifton-Brown, J.; Fuhrer, J.; Jones, M.; Martin, C. Estimating the greenhouse gas fluxes of European grasslands with a process-based model: 1. Model evaluation from in situ measurements. Global Biogeochem. Cycles 2007, 21, GB1004. [Google Scholar] [CrossRef] [Green Version]
- Gomára, I.; Bellocchi, G.; Martin, R.; Rodriguez-Fonseca, B.; Ruíz-Ramos, M. Influence of climate variability on the potential forage production of a mown permanent grassland in the French Massif Central. Agric. For. Meteorol. 2020, 280, 107768. [Google Scholar] [CrossRef]
- Sándor, R.; Picon-Cochard, C.; Martin, R.; Louault, F.; Klumpp, K.; Borras, D.; Bellocchi, G. Plant acclimation to temperature: Developments in the Pasture Simulation model. Field Crops Res. 2018, 222, 238–255. [Google Scholar] [CrossRef]
- Zwicke, M.; Alessio, G.A.; Thiery, L.; Falcimagne, R.; Baumont, R.; Rossignol, N.; Soussana, J.-F.; Picon-Cochard, C. Lasting effects of climate disturbance on perennial grassland above-ground biomass production under two cutting frequencies. Glob. Change Biol. 2013, 19, 3435–3448. [Google Scholar] [CrossRef] [PubMed]
- Kollas, C.; Kersebaum, K.C.; Nendel, C.; Manevski, K.; Müller, C.; Palosuo, T.; Armas-Herrera, C.M.; Beaudoin, N.; Bindi, M.; Charfeddine, M.; et al. Crop rotation modelling—a European model intercomparison. Eur. J. Agron. 2015, 70, 98–111. [Google Scholar] [CrossRef]
- Sándor, R.; Barcza, Z.; Acutis, M.; Doro, L.; Hidy, D.; Köchy, M.; Minet, J.; Lellei-Kovács, E.; Ma, S.; Perego, A.; et al. Multi-model simulation of soil temperature, soil water content and biomass in Euro-Mediterranean grasslands: Uncertainties and ensemble performance. Eur. J. Agron. 2017, 88, 22–40. [Google Scholar] [CrossRef] [Green Version]
- Durand, J.-L.; Gonzales-Dugo, V.; Gastal, F. How much do water deficits alter the nitrogen nutrition status of forage crops? Nutr. Cycl. Agroecosyst. 2010, 88, 231–243. [Google Scholar] [CrossRef]
- Fuchs, K.; Merbold, L.; Buchmann, N.; Bellocchi, G.; Bindi, M.; Brilli, L.; Conant, R.T.; Dorich, C.D.; Ehrhardt, F.; Fitton, N.; et al. Evaluating the potential of legumes to mitigate N2O emissions from permanent grassland using process-based models. Global Biogeochem. Cy. 2020, 34, e2020GB006561. [Google Scholar] [CrossRef]
- Chaves, M.M.; Pereira, J.S.; Maroco, J.; Rodrigues, M.L.; Ricardo, C.P.P.; Osorio, M.L.; Carvalho, I.; Faria, T.; Pinheiro, C. How plants cope with water stress in the field: Photosynthesis and growth. Ann. Bot. 2002, 89, 907–916. [Google Scholar] [CrossRef] [Green Version]
- Volaire, F. Plant traits and functional types to characterise drought survival of pluri-specific perennial herbaceous swards in Mediterranean areas. Eur. J. Agron. 2008, 29, 116–124. [Google Scholar] [CrossRef]
- Volaire, F.; Lelièvre, F. Drought survival in Dactylis glomerata and Festuca arundinacea under similar rooting conditions in tubes. Plant Soil 2001, 229, 225–234. [Google Scholar] [CrossRef]
- Gilgen, A.K.; Signarbieux, C.; Feller, U.; Buchmann, N. Competitive advantage of Rumex obtusifolius L. might increase in intensively managed temperate grasslands under drier climate. Agric. Ecosyst. Environ. 2010, 135, 15–23. [Google Scholar] [CrossRef] [Green Version]
- Mariotte, P.; Vandenberghe, C.; Kardol, P.; Hagedorn, F.; Buttler, A. Subordinate plant species enhance community resistance against drought in semi-natural grasslands. J. Ecol. 2013, 101, 763–773. [Google Scholar] [CrossRef]
- Weisser, W.W.; Roscher, C.; Meyer, S.T.; Ebeling, A.; Luo, G.; Allan, E.; Beßler, H.; Barnard, R.L.; Buchmann, N.; Buscot, F.; et al. Biodiversity effects on ecosystem functioning in a 15-year grassland experiment: Patterns, mechanisms, and open questions. Basic Appl. Ecol. 2017, 23, 1–73. [Google Scholar] [CrossRef]
- Soussana, J.F.; Maire, V.; Gross, N.; Bachelet, V.; Pagès, L.; Martin, R.; Hill, D.; Wirth, C. GEMINI: A grassland model simulating the role of plant traits for community dynamics and ecosystem functioning. Parametrization and evaluation. Ecol. Model. 2012, 231, 134–145. [Google Scholar] [CrossRef]
- Taubert, F.; Hetzer, J.; Schmid, J.S.; Huth, A. Confronting an individual-based simulation model with empirical community patterns of grasslands. PLoS ONE 2020, 15, e0236546. [Google Scholar] [CrossRef]
- Amiaud, B.; Touzard, B.; Bonis, A.; Bouzillé, J.-B. After grazing exclusion, is there any modification of strategy for two guerrilla species: Elymus repens (L.) Gould and Agrostis stolonifera (L.)? Plant Ecol. 2000, 197, 107–117. [Google Scholar] [CrossRef]
- Ringselle, B.; Bergkvist, G.; Aronsson, H.; Andersson, L. Under-sown cover crops and post-harvest mowing as measures to control Elymus repens. Weed Res. 2015, 55, 309–319. [Google Scholar] [CrossRef]
- Snow, D.W.; Perrins, C.M. The Birds of the Western Palearctic; Oxford University Press: Oxford, UK, 1998. [Google Scholar]
- Ružičková, H.; Banásová, V.; Kalivoda, H. Morava River alluvial meadows on the Slovak–Austrian border (Slovak part): Plant community dynamics, floristic and butterfly diversity—threats and management. J. Nat. Conserv. 2004, 12, 157–169. [Google Scholar] [CrossRef]
- Franzluebbers, A.J.; Nazih, N.; Stuedemann, J.A.; Fuhrmann, J.J.; Schomberg, H.H.; Hartel, P.G. Soil carbon and nitrogen pools under low- and high-endophyte-infected tall fescue. Soil Sci. Soc. Am. J. 1999, 63, 1687–1694. [Google Scholar] [CrossRef] [Green Version]
- McDonagh, J.; O’Donovan, M.; McEvoy, M.; Gilliland, T.J. Genetic gain in perennial ryegrass (Lolium perenne) varieties 1973 to 2013. Euphytica 2016, 212, 187–199. [Google Scholar] [CrossRef] [Green Version]
- Martin, G.; Durand, J.-L.; Duru, M.; Gastal, F.; Julier, B.; Litrico, I.; Louarn, G.; Médiène, S.; Moreau, D.; Valentin-Morison, M.; et al. Role of ley pastures in tomorrow’s cropping systems. A review. Agron. Sustain. Dev. 2020, 40, 17. [Google Scholar] [CrossRef]
- Defelice, M.S. Catchweed bedstraw or cleavers, Galium aparine L.—A very “sticky” subject. Weed Technol. 2002, 16, 467–472. [Google Scholar] [CrossRef]
- Pavlů, L.; Pavlů, V.; Gaisler, J.; Hejcman, M.; Mikulka, J. Effect of long-term cutting versus abandonment on the vegetation of a mountain hay meadow (Polygono-Trisetion) in Central Europe. Flora: Morphol. Distrib. Funct. Ecol. Plants 2011, 206, 1020–1029. [Google Scholar] [CrossRef]
- Jernej, I.; Bohner, A.; Walcher, R.; Hussain, R.I.; Arnberger, A.; Zaller, J.G.; Frank, T. Impact of land-use change in mountain semi-dry meadows on plants, litter decomposition and earthworms. Web Ecol. 2019, 19, 53–63. [Google Scholar] [CrossRef]
- Garnier, E.; Cortez, J.; Billès, G.; Navas, M.-L.; Roumet, C.; Debussche, M.; Laurent, G.; Blanchard, A.; Aubry, D.; Bellmann, A.; et al. Plant functional markers capture ecosystem properties during secondary succession. Ecology 2004, 85, 2630–2637. [Google Scholar] [CrossRef]
- Dubuis, A.; Rossier, L.; Pottier, J.; Pellissier, L.; Vittoz, P.; Guisan, A. Predicting current and future spatial community patterns of plant functional traits. Ecography 2013, 36, 1158–1168. [Google Scholar] [CrossRef]




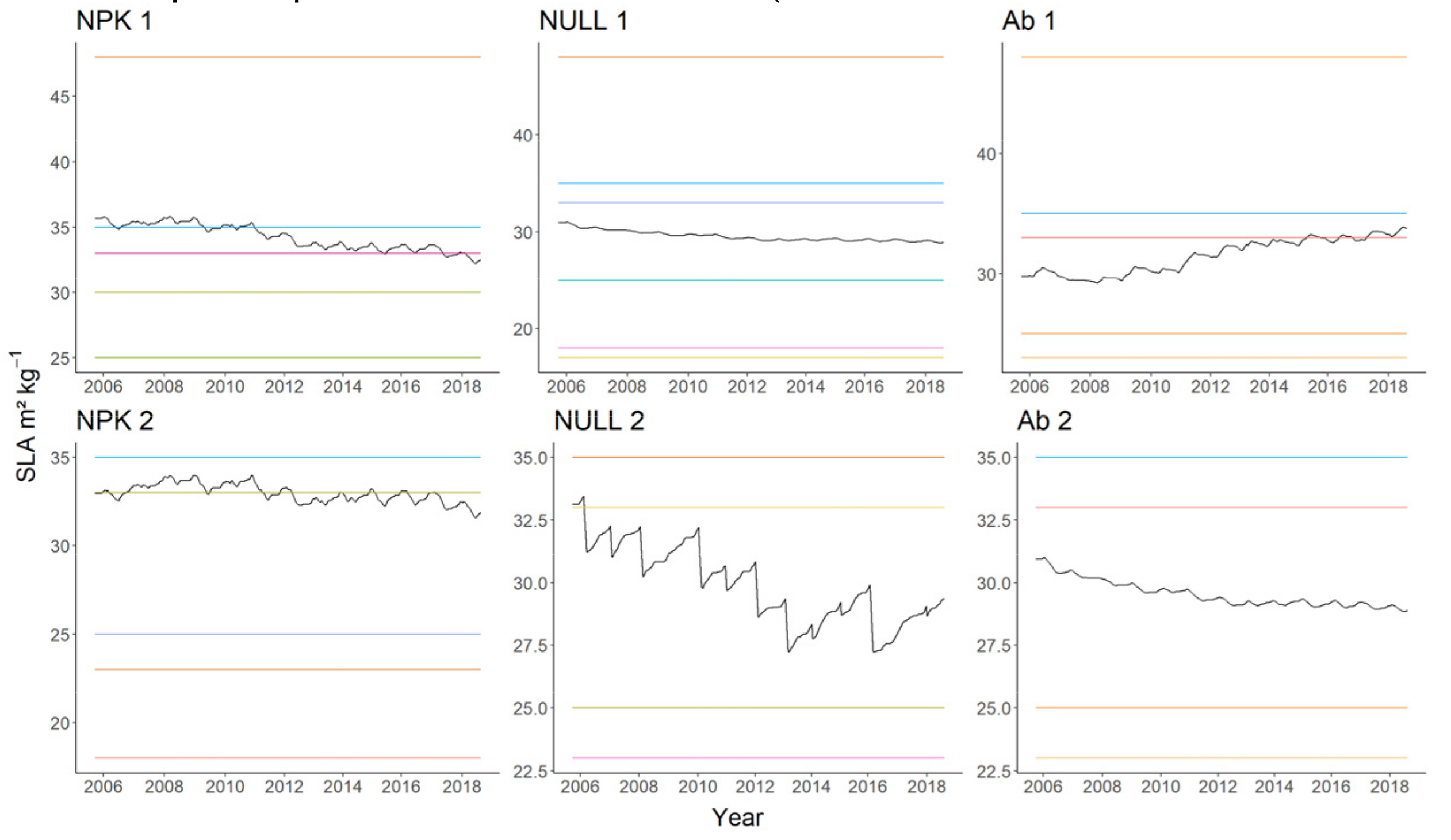
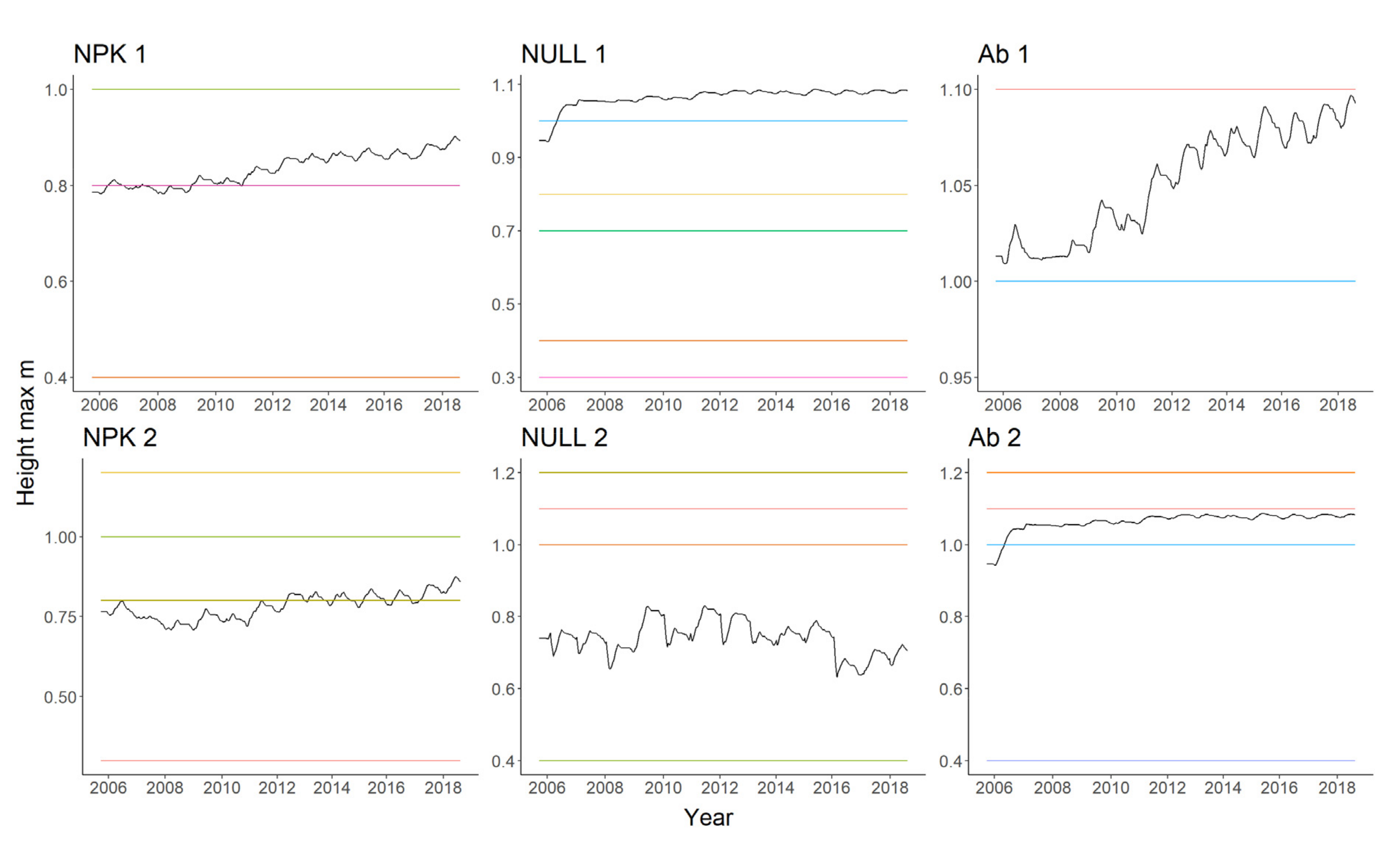
| Soil properties | Unit | Block 1 | Block 2 | ||
|---|---|---|---|---|---|
| Layer thickness | m | 0.00–0.20 | 0.20–0.40 | 0.00–0.20 | 0.20–0.40 |
| Clay | % | 19.7 | 17.0 | 23.0 | 25.0 |
| Silt | % | 26.9 | 27.4 | 26.1 | 24.2 |
| Sand | % | 53.4 | 55.6 | 51.0 | 50.8 |
| Carbon content | g kg−1 | 40.3 | 18.5 | 43.1 | 15.1 |
| pH | - | 5.9 | 6.2 | 6.0 | 6.5 |
| Bulk density | g cm−3 | 0.94 | 1.23 | 0.89 | 1.18 |
| NPK | Null | Ab | ||||||
|---|---|---|---|---|---|---|---|---|
| Species | Relative Abundances | Species | Relative Abundances | Species | Relative Abundances | |||
| Obs | Rec | Obs | Rec | Obs | Rec | |||
| Block 1 | ||||||||
| Elymus repens | 0.26 | 0.31 | Achillea millefolium | 0.04 | 0.06 | Alopecurus pratensis | 0.13 | 0.17 |
| Festuca arundinacea | 0.09 | 0.11 | Elymus repens | 0.05 | 0.07 | Arrhenatherum elatius | 0.04 | 0.05 |
| Lolium multiflorum | 0.07 | 0.09 | Festuca arundinacea | 0.09 | 0.13 | Dactylis glomerata | 0.04 | 0.05 |
| Poa pratensis | 0.15 | 0.17 | Holcus lanatus | 0.05 | 0.07 | Elymus repens | 0.27 | 0.35 |
| Stellaria media | 0.04 | 0.05 | Poa pratensis | 0.12 | 0.17 | Festuca arundinacea | 0.06 | 0.07 |
| Taraxacum officinale | 0.21 | 0.25 | Poa trivialis | 0.05 | 0.08 | Galium aparine | 0.04 | 0.05 |
| Taraxacum officinale | 0.16 | 0.22 | Poa pratensis | 0.16 | 0.20 | |||
| Trifolium repens | 0.14 | 0.29 | Urtica dioica | 0.04 | 0.05 | |||
| Sum | 0.82 | 1.00 | Sum | 0.70 | 1.00 | Sum | 0.78 | 1.00 |
| Nb. of species | 6 | Nb. of species | 8 | Nb. of species | 8 | |||
| Block 2 | ||||||||
| Achillea millefolium | 0.06 | 0.07 | Achillea millefolium | 0.04 | 0.07 | Alopecurus pratensis | 0.30 | 0.46 |
| Alopecurus pratensis | 0.18 | 0.23 | Alopecurus pratensis | 0.08 | 0.11 | Arrhenatherum elatius | 0.06 | 0.08 |
| Holcus lanatus | 0.07 | 0.08 | Festuca arundinacea | 0.05 | 0.07 | Elymus repens | 0.04 | 0.07 |
| Lolium perenne | 0.06 | 0.07 | Lolium perenne | 0.09 | 0.13 | Holcus lanatus | 0.04 | 0.07 |
| Poa pratensis | 0.17 | 0.20 | Poa pratensis | 0.12 | 0.18 | Poa pratensis | 0.11 | 0.18 |
| Stellaria media | 0.04 | 0.05 | Taraxacum officinale | 0.13 | 0.18 | Urtica dioica | 0.09 | 0.14 |
| Taraxacum officinale | 0.15 | 0.19 | Trifolium repens | 0.11 | 0.15 | |||
| Trisetum flavescens | 0.08 | 0.10 | Trisetum flavescens | 0.08 | 0.12 | |||
| Sum | 0.80 | 1.00 | Sum | 0.70 | 1.00 | Sum | 0.72 | 1.00 |
| Nb. of species | 8 | Nb. of species | 8 | Nb. of species | 6 | |||
| NPK | Null | Ab | ||||
|---|---|---|---|---|---|---|
| Functional Group | Relative Abundances | Relative Abundances | Relative Abundances | |||
| Obs | Rec | Obs | Rec | Obs | Rec | |
| Block 1 | ||||||
| A | 0.07 | 0.07 | 0.21 | 0.21 | 0.24 | 0.24 |
| B | 0.45 | 0.45 | 0.49 | 0.49 | 0.34 | 0.34 |
| b | 0.48 | 0.48 | 0.30 | 0.30 | 0.42 | 0.42 |
| Sum | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Block 2 | ||||||
| A | 0.47 | 0.47 | 0.39 | 0.40 | 0.59 | 0.61 |
| B | 0.33 | 0.33 | 0.32 | 0.32 | 0.27 | 0.28 |
| b | 0.20 | 0.20 | 0.28 | 0.28 | 0.11 | 0.11 |
| Sum | 1.00 | 1.00 | 0.99 | 1.00 | 0.97 | 1.00 |
| Year | Doy | Observed Biomass | ModVege | ModVege-CoSMo FG | ModVege-CoSMo Species | |||
|---|---|---|---|---|---|---|---|---|
| Simulated Biomass | Difference | Simulated Biomass | Difference | Simulated Biomass | Difference | |||
| Block 1 | ||||||||
| 2006 | 200 | 6742 | 6302 | −440 | 5509 | −1233 | 5503 | −1239 |
| 2007 | 213 | 5781 | 8465 | 2684 | 7581 | 1800 | 7678 | 1897 |
| 2008 | 218 | 8116 | 7955 | −161 | 7049 | −1067 | 7072 | −1044 |
| 2009 | - | - | - | - | - | - | - | - |
| 2010 | 201 | 7734 | 6372 | −1362 | 5445 | −2289 | 5592 | −2142 |
| 2011 | 201 | 5674 | 4736 | −938 | 3848 | −1826 | 3675 | −1999 |
| 2012 | 201 | 8560 | 6934 | −1626 | 5992 | −2568 | 6111 | −2449 |
| 2013 | 198 | 6655 | 6495 | −160 | 5587 | −1068 | 5579 | −1076 |
| 2014 | 203 | 6755 | 6357 | −398 | 5447 | −1308 | 5401 | −1354 |
| 2015 | 202 | 4056 | 3591 | −465 | 2890 | −1166 | 2717 | −1339 |
| 2016 | 202 | 4789 | 6955 | 2166 | 6026 | 1237 | 6051 | 1262 |
| 2017 | 205 | 4498 | 6332 | 1834 | 5376 | 878 | 5370 | 872 |
| 2018 | 204 | 3005 | 6744 | 3739 | 5822 | 2817 | 5758 | 2753 |
| Mean | 6030 | 6437 | 406 | 5548 | −483 | 5542 | −488 | |
| Minimum | 3005 | 3591 | −1626 | 2890 | −2568 | 2717 | −2449 | |
| Maximum | 8560 | 8465 | 3739 | 7581 | 2817 | 7678 | 2753 | |
| Block 2 | ||||||||
| 2006 | 199 | 4576 | 6069 | 1493 | 5240 | 664 | 5124 | 548 |
| 2007 | 213 | 6822 | 8197 | 1375 | 7250 | 428 | 7285 | 463 |
| 2008 | 218 | 5832 | 7810 | 1978 | 6867 | 1035 | 6866 | 1034 |
| 2009 | - | - | - | - | - | - | - | - |
| 2010 | 201 | 8061 | 5982 | −2079 | 5005 | −3056 | 5135 | −2926 |
| 2011 | 201 | 3867 | 5048 | 1181 | 4211 | 344 | 3945 | 78 |
| 2012 | 200 | 8227 | 6576 | −1651 | 5664 | −2563 | 5739 | −2488 |
| 2013 | 198 | 7003 | 6395 | −608 | 5494 | −1509 | 5440 | −1563 |
| 2014 | 203 | 6592 | 7859 | 1267 | 6948 | 152 | 5401 | −1191 |
| 2015 | 202 | 3652 | 3911 | 259 | 3107 | −545 | 2923 | −729 |
| 2016 | 202 | 5131 | 6670 | 1539 | 5738 | 607 | 5786 | 655 |
| 2017 | 205 | 4312 | 6101 | 1789 | 5198 | 886 | 5169 | 857 |
| 2018 | 204 | 3656 | 6631 | 2975 | 5728 | 2072 | 5630 | 1974 |
| Mean | 5661 | 6437 | 776 | 5538 | −124 | 5405 | 548 | |
| Minimum | 3652 | 3911 | −2079 | 3107 | −3056 | 2910 | −2926 | |
| Maximum | 8227 | 8197 | 2975 | 7250 | 2072 | 6989 | 1974 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piseddu, F.; Martin, R.; Movedi, E.; Louault, F.; Confalonieri, R.; Bellocchi, G. Simulation of Multi-Species Plant Communities in Perturbed and Nutrient-Limited Grasslands: Development of the Growth Model ModVege. Agronomy 2022, 12, 2468. https://doi.org/10.3390/agronomy12102468
Piseddu F, Martin R, Movedi E, Louault F, Confalonieri R, Bellocchi G. Simulation of Multi-Species Plant Communities in Perturbed and Nutrient-Limited Grasslands: Development of the Growth Model ModVege. Agronomy. 2022; 12(10):2468. https://doi.org/10.3390/agronomy12102468
Chicago/Turabian StylePiseddu, Francesca, Raphaël Martin, Ermes Movedi, Frédérique Louault, Roberto Confalonieri, and Gianni Bellocchi. 2022. "Simulation of Multi-Species Plant Communities in Perturbed and Nutrient-Limited Grasslands: Development of the Growth Model ModVege" Agronomy 12, no. 10: 2468. https://doi.org/10.3390/agronomy12102468
APA StylePiseddu, F., Martin, R., Movedi, E., Louault, F., Confalonieri, R., & Bellocchi, G. (2022). Simulation of Multi-Species Plant Communities in Perturbed and Nutrient-Limited Grasslands: Development of the Growth Model ModVege. Agronomy, 12(10), 2468. https://doi.org/10.3390/agronomy12102468







