The Key Regulators and Metabolic Intermediates of Lignin Response to Low Temperatures Revealed by Transcript and Targeted Metabolic Profiling Analysis in Poplar
Abstract
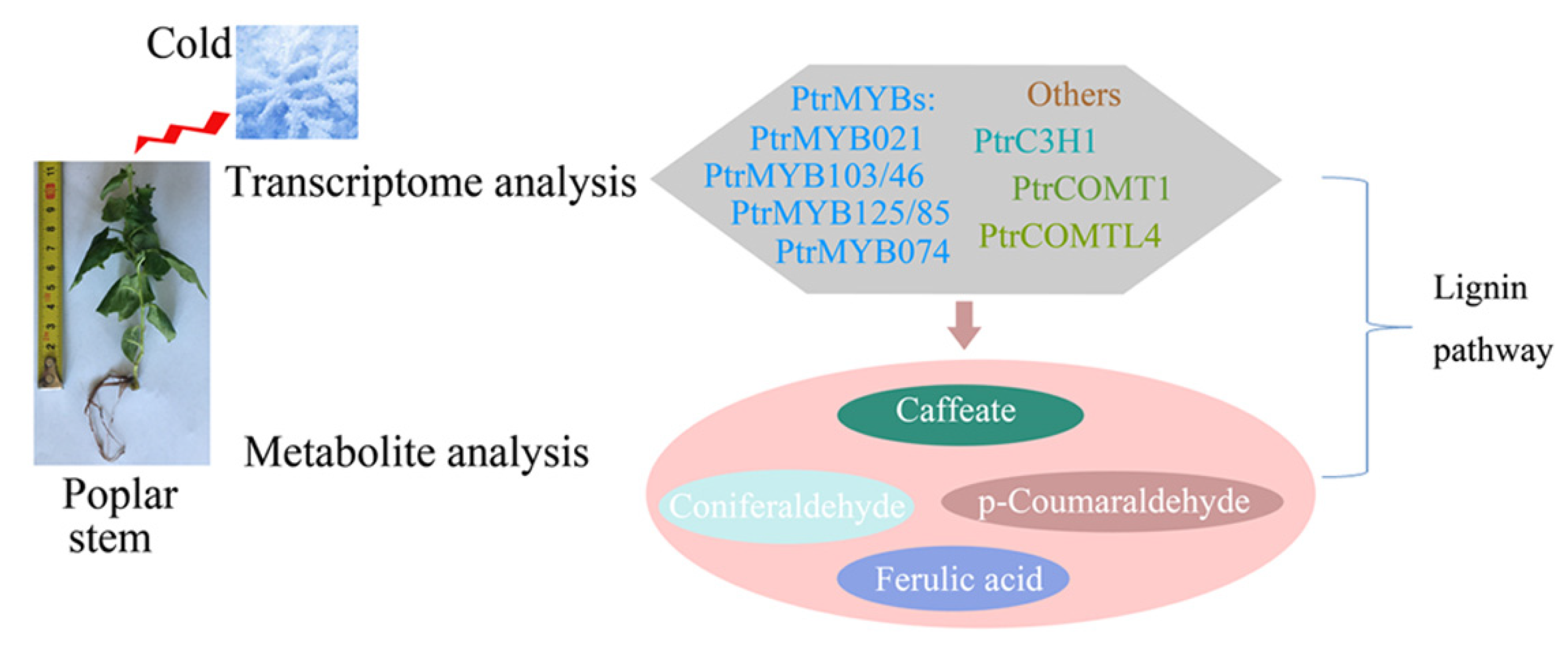
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Treatments
2.2. Extraction of Acid-Soluble and Acid-Insoluble Lignin and the Measurement of Their Contents
2.3. Measurement of the Intermediate Metabolites of Lignin
2.4. Transcriptome Sequencing
2.5. Data Analysis
2.6. RT-qPCR
3. Results
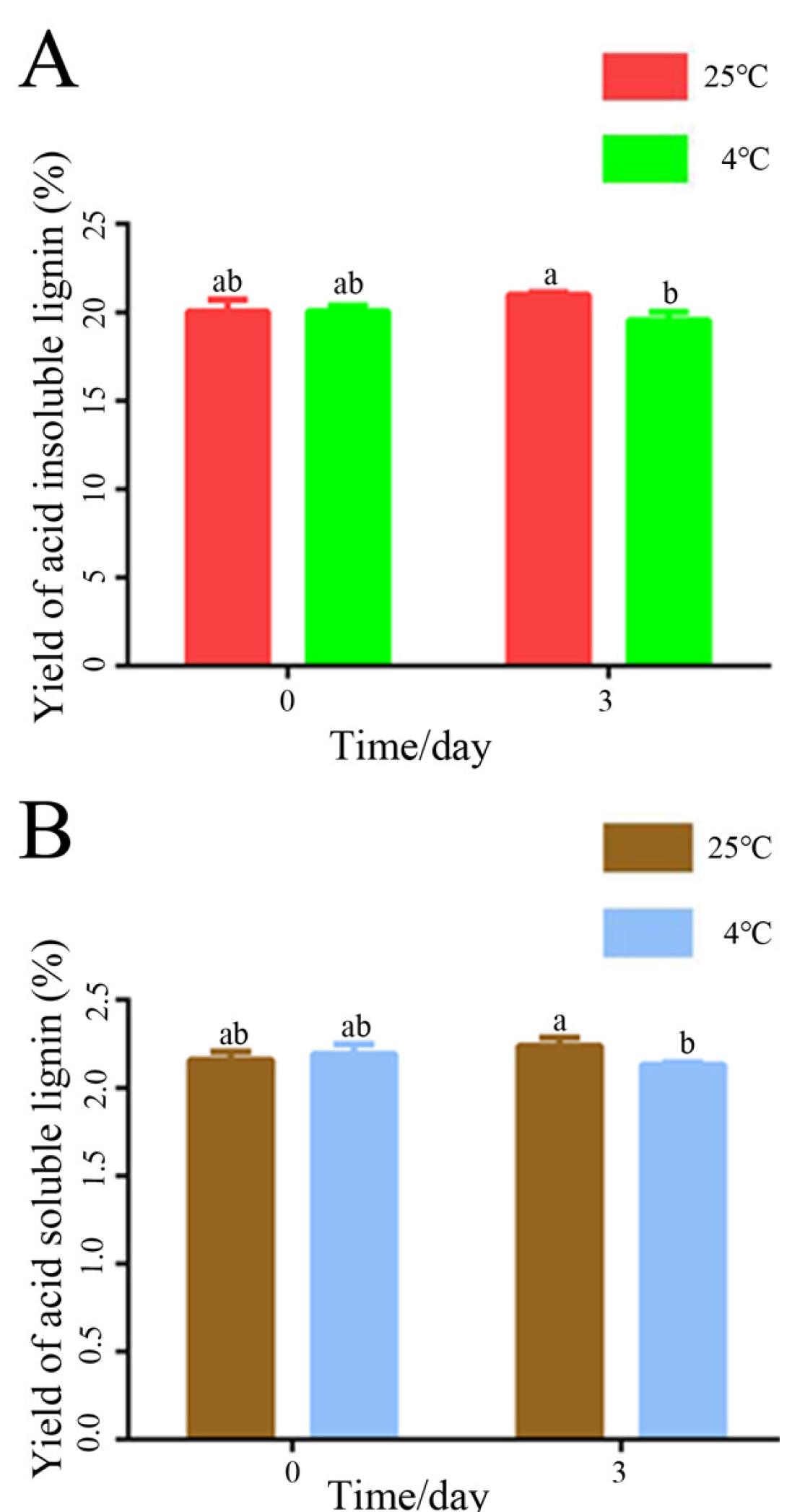
3.1. Effect of Low-Temperature Treatment on the Content of Lignin in Poplar Stems
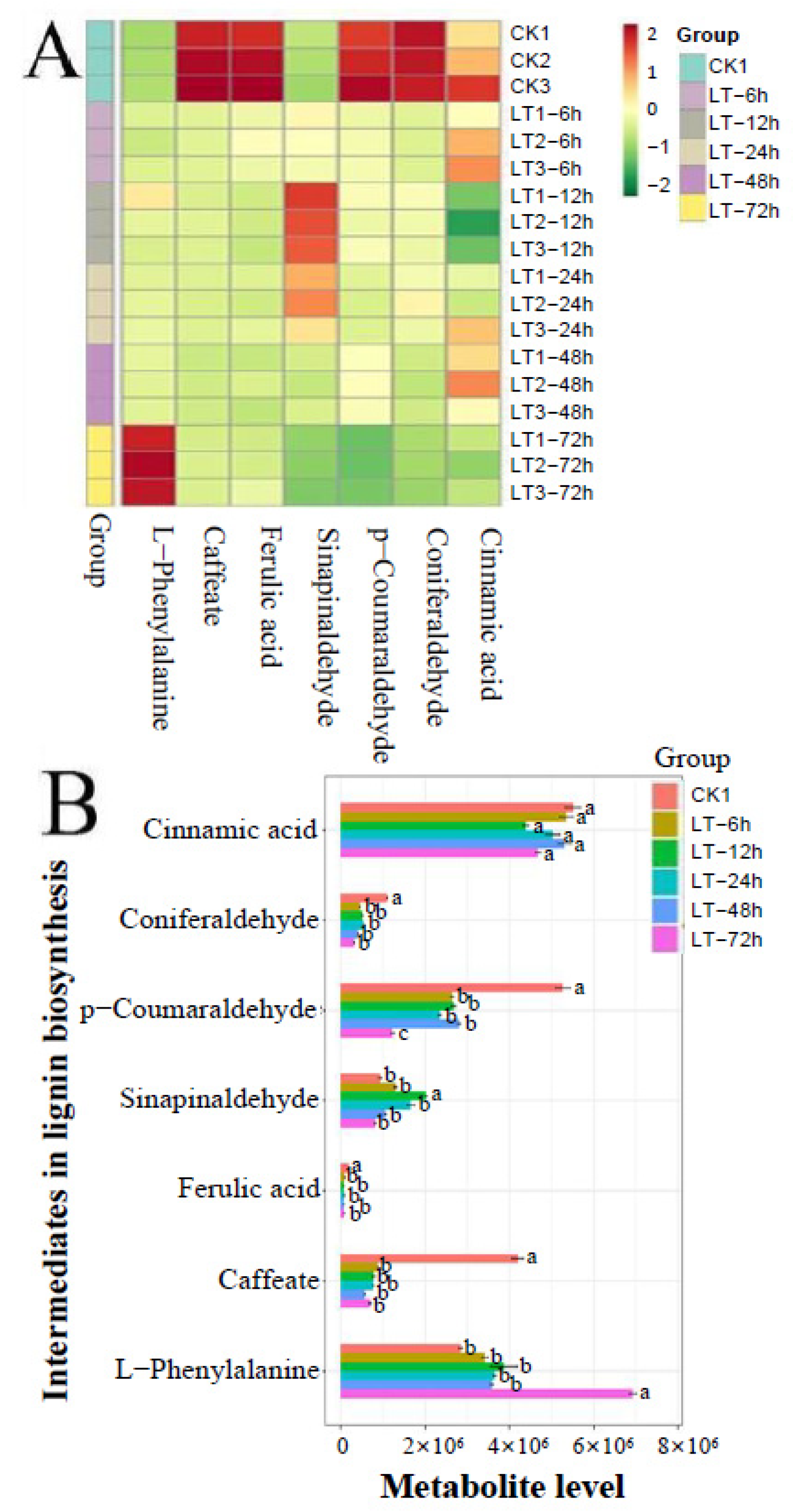
3.2. Response of Lignin Intermediate Metabolites to Low Temperatures
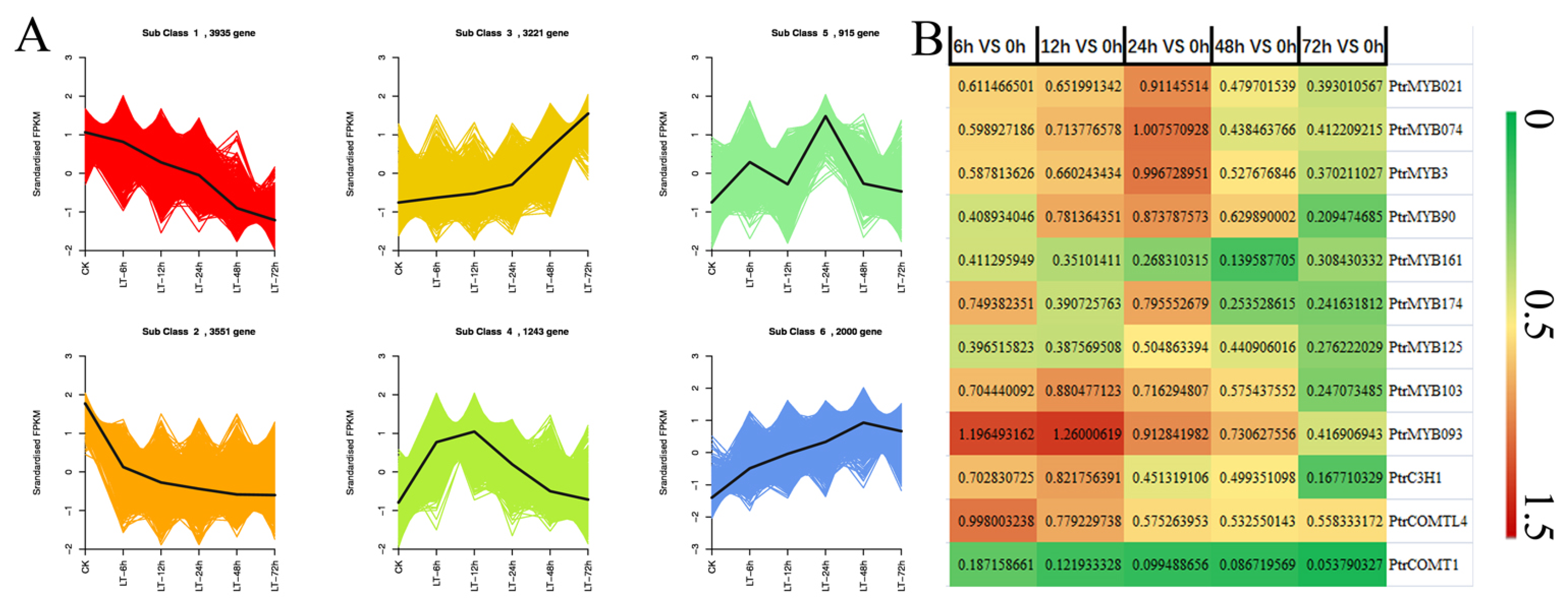
3.3. Differential Expression Profiles Related to Defenses Induced by Cold
3.4. Functional Annotation Analysis of Differentially Expressed Genes (DEGs)
3.5. Identification of the Genes Involved in the Decrease in Lignin Intermediate Metabolites
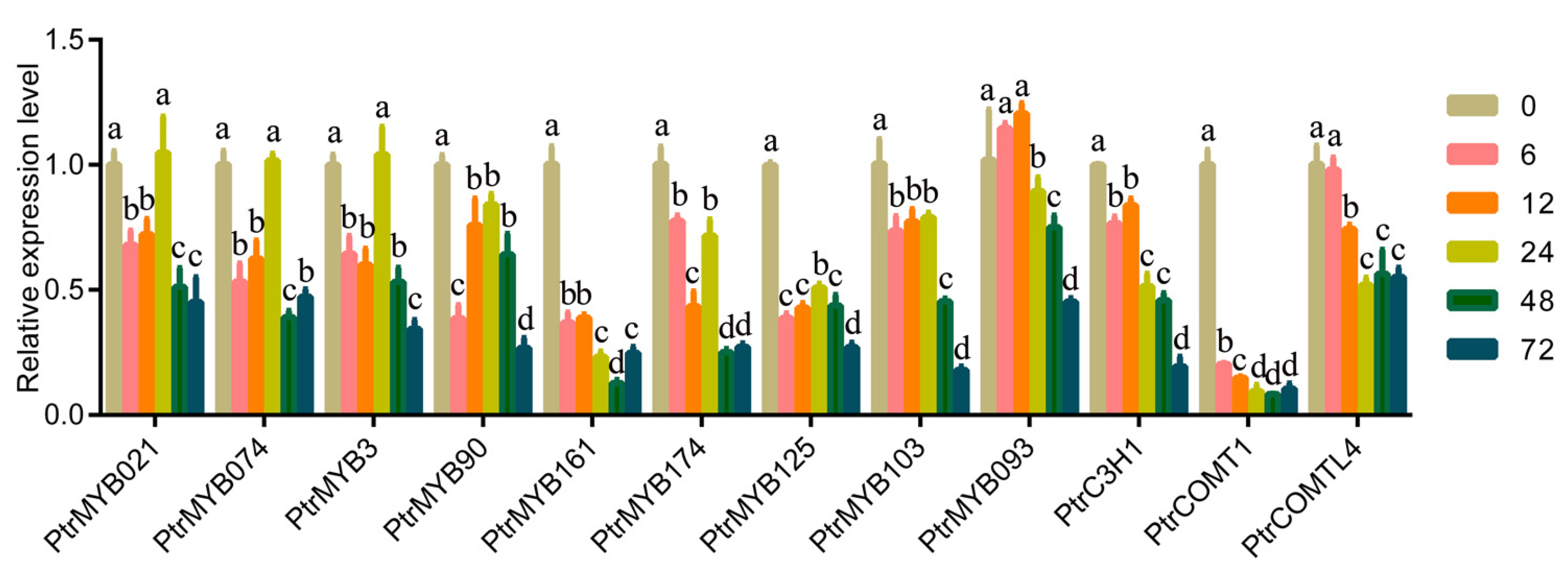
3.6. Quantitative Analysis of the Structural Genes and Transcription Factors Related to Lignin Synthesis and Metabolism in Response to Low-Temperature Stress
4. Discussion
4.1. Low-Temperature Stress Changed the Content of Lignin in Poplar Stems
4.2. PtrC3′H1 Contributed to the Decrease in Caffeate Content in Response to Cold Stress
4.3. The Low-Level Expression of PtrCOMTs upon Cold Stimulus May Result in a Decrease in Coniferaldehyde
4.4. MYB Could Be Involved in the Alteration of Lignin Biosynthesis under Low Temperatures
4.5. Enzymatic Activity Could Contribute to Lignin Reduction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Agarwal, M.; Hao, Y.; Kapoor, A.; Dong, C.H.; Fujii, H.; Zheng, X.; Zhu, J.K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006, 281, 37636–37645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fedoroff, N.V.; Battisti, D.S.; Beachy, R.N.; Cooper, P.J.; Fischhoff, D.A.; Hodges, C.N.; Knauf, V.C.; Lobel, D.; Mazur, B.J.; Molden, D.; et al. Radically rethinking agriculture for the 21st century. Science 2010, 327, 833–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Ding, Y.; Yang, S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Vanholme, R.; De Meester, B.; Ralph, J.; Boerjan, W. Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotech. 2019, 56, 230–239. [Google Scholar] [CrossRef]
- Moura, J.C.M.S.; Bonine, C.A.V.; de Oliveira Fernandes Viana, J.; Dornelas, M.C.; Mazzafera, P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef]
- Behr, M.; Guerriero, G.; Grima-Pettenati, J.; Baucher, M. A molecular blueprint of lignin repression. Trends Plant Sci. 2019, 24, 1052–1064. [Google Scholar] [CrossRef] [Green Version]
- Bryant, N.D.; Pu, Y.; Tschaplinski, T.J.; Tuskan, G.A.; Muchero, W.; Kalluri, U.C.; Yoo, C.G.; Ragauskas, A.J. Transgenic Poplar Designed for Biofuels. Trends Plant Sci. 2020, 25, 881–896. [Google Scholar] [CrossRef]
- Schoch, G.; Goepfert, S.; Morant, M.; Hehn, A.; Meyer, D.; Ullmann, P.; Werck-Reichhart, D. CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J. Biol. Chem. 2001, 276, 36566–36574. [Google Scholar] [CrossRef] [Green Version]
- Franke, R.; Humphreys, J.M.; Hemm, M.R.; Denault, J.W.; Ruegger, M.O.; Cusumano, J.C.; Chapple, C. The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J. 2002, 30, 33–45. [Google Scholar] [CrossRef]
- Coleman, H.D.; Park, J.Y.; Nair, R.; Chapple, C.; Mansfield, S.D. RNAi-mediated suppression of p-coumaroyl-CoA 3′-hydroxylase in hybrid poplar impacts lignin deposition and soluble secondary metabolism. Proc. Natl. Acad. Sci. USA 2008, 105, 4501–4506. [Google Scholar] [CrossRef] [Green Version]
- Bonawitz, N.D.; Im Kim, J.; Tobimatsu, Y.; Ciesielski, P.N.; Anderson, N.A.; Ximenes, E.; Maeda, J.; Ralph, J.; Donohoe, B.S.; Ladisch, M.; et al. Disruption of mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 2014, 509, 376–380. [Google Scholar] [CrossRef]
- Tu, Y.; Rochfort, S.; Liu, Z.; Ran, Y.; Griffith, M.; Badenhorst, P.; Louie, G.V.; Bowman, M.E.; Smith, K.F.; Noel, J.P.; et al. Functional analyses of caffeic acid O-methyltransferase and cinnamoyl-CoA-reductase genes from perennial ryegrass (Lolium perenne). Plant Cell 2010, 22, 3357–3373. [Google Scholar] [CrossRef] [Green Version]
- Daly, P.; McClellan, C.; Maluk, M.; Oakey, H.; Lapierre, C.; Waugh, R.; Stephens, J.; Marshall, D.; Barakate, A.; Tsuji, Y.; et al. RNA i-suppression of barley caffeic acid O-methyltransferase modifies lignin despite redundancy in the gene family. Plant Biotechnol. J. 2020, 17, 594–607. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.Z.; Dixon, R.A. On-off switches for secondary cell wall biosynthesis. Mol. Plant. 2012, 5, 297–303. [Google Scholar] [CrossRef] [Green Version]
- Geng, P.; Zhang, S.; Liu, J.; Zhao, C.; Wu, J.; Cao, Y.; Fu, C.; Han, X.; He, H.; Zhao, Q. MYB20, MYB42, MYB43, and MYB85 regulate phenylalanine and lignin biosynthesis during secondary cell wall formation. Plant Physiol. 2020, 182, 1272–1283. [Google Scholar] [CrossRef] [Green Version]
- Nakano, Y.; Yamaguchi, M.; Endo, H.; Rejab, N.A.; Ohtani, M. NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 2015, 6, 288. [Google Scholar] [CrossRef] [Green Version]
- Kim, W.C.; Ko, J.H.; Kim, J.Y.; Kim, J.; Bae, H.J.; Han, K.H. MYB46 directly regulates the gene expression of secondary wall-associated cellulose synthases in Arabidopsis. Plant J. 2013, 73, 26–36. [Google Scholar] [CrossRef]
- Grima-Pettenati, J.; Soler, M.; Camargo, E.L.O.; Wang, H. Transcriptional regulation of the lignin biosynthetic pathway revisited: New players and insights. Adv. Bot. Res. 2012, 61, 173–218. [Google Scholar]
- Chen, H.; Wang, J.P.; Liu, H.; Li, H.; Lin, Y.C.J.; Shi, R.; Sederoff, R.R. Hierarchical transcription factor and chromatin binding network for wood formation in Populus trichocarpa. Plant Cell. 2019, 31, 602–626. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Xu, Y.; Ma, Q.; Xu, W.; Wang, T.; Xue, Y.; Chong, K. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol. 2007, 143, 1739–1751. [Google Scholar] [CrossRef]
- An, J.P.; Li, R.; Qu, F.J.; You, C.X.; Wang, X.F.; Hao, Y.J. R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanid in accumulation in apple. Plant J. 2018, 96, 562–577. [Google Scholar] [CrossRef] [Green Version]
- Taylor, G. Populus: Arabidopsis for forestry. Do we need a model tree? Ann. Bot. 2002, 90, 681–689. [Google Scholar] [CrossRef] [Green Version]
- Crivellaro, A.; Büntgen, U. New evidence of thermally constrained plant cell wall lignification. Trends Plant Sci. 2020, 25, 322–324. [Google Scholar] [CrossRef]
- Sluiter, A.; Hames, B.; Ruiz, R.; Scarlata, C.; Sluiter, J.; Templeton, D.; Crocker, D. Determination of structural carbohydrates and lignin in biomass. Lab. Anal. Proceed. 2008, 1617, 1–16. [Google Scholar]
- Jiang, C. Efficient extraction of RNA from various Camellia species rich in secondary metabolites for deep transcriptome sequencing and gene expression analysis. Afr. J. Biotechnol. 2019, 10, 16763–16768. [Google Scholar] [CrossRef] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Wang, Z.; Mao, Y.; Guo, Y.; Gao, J.; Liu, X.; Li, S.; Lin, Y.J.; Chen, H.; Wang, J.P.; Chiang, V.L.; et al. MYB Transcription Factor 161 Mediates Feedback Regulation of Secondary wall-associated NAC-Domain 1 Family Genes for Wood Formation. Plant Physiol. 2020, 184, 1389–1406. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Barros, J.; Escamilla-Trevino, L.; Song, L.; Rao, X.; Serrani-Yarce, J.C.; Palacios, M.D.; Engle, N.K.; Choudhury, F.; Tschaplinski, T.J.; Venables, B.J.; et al. 4-Coumarate 3-hydroxylase in the lignin biosynthesis pathway is a cytosolic ascorbate peroxidase. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Chapple, C. Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu. Rev. Plant Biol. 1998, 49, 311–343. [Google Scholar] [CrossRef] [PubMed]
- Ro, D.K.; Mah, N.; Ellis, B.E.; Douglas, C.J. Functional characterization and subcellular localization of poplar (Populus trichocarpa × Populus deltoides) cinnamate 4-hydroxylase. Plant Physiol. 2001, 126, 317–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hausman, J.F.; Evers, D.; Thiellement, H.; Jouve, L. Compared responses of poplar cuttings and in vitro raised shoots to short-term chilling treatments. Plant Cell Rep. 2000, 19, 954–960. [Google Scholar] [CrossRef] [PubMed]
- Olenichenko, N.; Zagoskina, N. Response of winter wheat to cold: Production of phenolic compounds and l-phenylalanine ammonia lyase activity. Appl. Biochem. Microbiol. 2005, 41, 600–603. [Google Scholar] [CrossRef]
- Zeng, J.K.; Li, X.; Zhang, J.; Ge, H.; Yin, X.R.; Chen, K.S. Regulation of loquat fruit low temperature response and lignification involves interaction of heat shock factors and genes associated with lignin biosynthesis. Plant Cell Environ. 2016, 39, 1780–1789. [Google Scholar] [CrossRef] [PubMed]
- Cabane, M.; Afif, D.; Hawkins, S. Lignins and abiotic stresses. Adv. Bot. Res. 2012, 61, 219–262. [Google Scholar]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and biological functions in plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef] [Green Version]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef]
- Alvarez, S.; Marsh, E.L.; Schroeder, S.G.; Schachtman, D.P. Metabolomic and proteomic changes in the xylem sap of maize under drought. Plant Cell Environ. 2008, 31, 325–340. [Google Scholar] [CrossRef]
- Reddy, M.S.; Chen, F.; Shadle, G.; Jackson, L.; Aljoe, H.; Dixon, R.A. Targeted down-regulation of cytochrome P450 enzymes for forage quality improvement in alfalfa (Medicago sativa L.). Proc. Natl. Acad. Sci. USA 2005, 102, 16573–16578. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, M.; Blaschek, L.; Subbotina, E.; Kajita, S.; Pesquet, E. Importance of lignin coniferaldehyde residues for plant properties and sustainable uses. ChemSuschem 2020, 13, 4400. [Google Scholar] [CrossRef]
- Van Acker, R.; Déjardin, A.; Desmet, S.; Hoengenaert, L.; Vanholme, R.; Morreel, K.; Laurans, F.; Kim, H.; Santoro, N.; Foster, C.; et al. Different metabolic routes for coniferaldehyde and sinapaldehyde with Cinnamyl alcohol dehydrogenase1 deficiency. Plant Physiol. 2017, 175, 1018–1039. [Google Scholar] [CrossRef]
- Zhang, W.; Wei, R.; Chen, S.; Jiang, J.; Li, H.; Huang, H.; Yang, G.; Wang, S.; Wei, H.; Liu, G. Functional characterization of CCR in birch (Betula platyphylla × Betula pendula) through overexpression and suppression analysis. Physiol. Plantarum. 2015, 154, 283–296. [Google Scholar] [CrossRef]
- De Meester, B.; Madariaga Calderón, B.; de Vries, L.; Pollier, J.; Goeminne, G.; Doorsselaere, J.V.; Chen, M.; Ralph, J.; Vanholme, R.; Boerjan, W. Tailoring poplar lignin without yield penalty by combining a null and haploinsufficient CINNAMOYL-CoA REDUCTASE2 allele. Nat. Commun. 2020, 11, 5020. [Google Scholar] [CrossRef]
- Bout, S.; Vermerris, W. A candidate-gene approach to clone the sorghum Brown midrib gene encoding caffeic acid O-methyltransferase. Mol. Genet. Genomics 2003, 269, 205–214. [Google Scholar] [CrossRef]
- Bhuiyan, N.H.; Liu, W.; Liu, G.; Selvaraj, G.; Wei, Y.; King, J. Transcriptional regulation of genes involved in the pathways of biosynthesis and supply of methyl units in response to powdery mildew attack and abiotic stresses in wheat. Plant Mol. Biol. 2007, 64, 305–318. [Google Scholar] [CrossRef]
- Hu, Y.; Li, W.C.; Xu, Y.Q.; Li, G.J.; Liao, Y.; Fu, F.L. Differential expression of candidate genes for lignin biosynthesis under drought stress in maize leaves. J. Appl. Genet. 2009, 50, 213–223. [Google Scholar] [CrossRef]
- Gui, J.; Lam, P.Y.; Tobimatsu, Y.; Sun, J.; Huang, C.; Cao, S.; Zhong, Y.; Umezawa, T.; Li, L. Fibre-specific regulation of lignin biosynthesis improves biomass quality in Populus. New Phytol. 2020, 226, 1074–1087. [Google Scholar] [CrossRef] [Green Version]
- Ohtani, M.; Demura, T. The quest for transcriptional hubs of lignin biosynthesis: Beyond the NAC-MYB-gene regulatory network model. Curr. Opin. Biotech. 2019, 56, 82–87. [Google Scholar] [CrossRef]
- Zhong, R.; Lee, C.; Ye, Z.H. Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol. Plant. 2010, 3, 1087–1103. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.H.; Jeon, H.W.; Kim, W.C.; Kim, J.Y.; Han, K.H. The MYB46/MYB83-mediated transcriptional regulatory programme is a gatekeeper of secondary wall biosynthesis. Ann. Bot. 2014, 114, 1099–1107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarthy, R.L.; Zhong, R.; Fowler, S.; Lyskowski, D.; Piyasena, H.; Carleton, K.; Spicer, C.; Ye, Z.H. The poplar MYB transcription factors, PtrMYB3 and PtrMYB20, are involved in the regulation of secondary wall biosynthesis. Plant Cell Physiol. 2010, 51, 1084–1090. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, P.; Das, A.; Datta, S.; Banerjee, I.; Tripathy, S.; Chaudhuri, S. Understanding the early cold response mechanism in IR64 indica rice variety through comparative transcriptome analysis. BMC Genomics 2020, 21, 425. [Google Scholar] [CrossRef] [PubMed]
- Mitiouchkina, T.; Mishin, A.S.; Somermeyer, L.G.; Markina, N.M.; Chepurnyh, T.V.; Guglya, E.B.; Karataeva, T.A.; Palkina, K.A.; Shakhova, E.S.; Fakhranurova, L.I.; et al. Plants with genetically encoded autoluminescence. Nat. Biotechnol. 2020, 38, 944–946. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.; Dixon, R.A. Current models for transcriptional regulation of secondary cell wall biosynthesis in grasses. Front. Plant Sci. 2018, 9, 399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, C.N., Jr.; Lenaghan, S.C. Lighting the Way: Advances in Engineering Autoluminescent Plants. Trends Plant Sci. 2020, 25, 1176–1179. [Google Scholar]
- Grabber, J.H.; Ralph, J.; Hatfield, R.D. Severe inhibition of maize wall degradation by synthetic lignins formed with coniferaldehyde. J. Sci. Food Agr. 1998, 78, 81–87. [Google Scholar] [CrossRef]
- Blaschek, L.; Champagne, A.; Dimotakis, C.; Decou, R.; Hishiyama, S.; Kratzer, S.; Kajita, S.; Pesquet, E. Cellular and genetic regulation of coniferaldehyde incorporation in lignin of herbaceous and woody plants by quantitative wiesner staining. Front. Plant Sci. 2020, 11, 109. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Li, P.; Li, C.; Xia, T. The Key Regulators and Metabolic Intermediates of Lignin Response to Low Temperatures Revealed by Transcript and Targeted Metabolic Profiling Analysis in Poplar. Agronomy 2022, 12, 2506. https://doi.org/10.3390/agronomy12102506
Zhao X, Li P, Li C, Xia T. The Key Regulators and Metabolic Intermediates of Lignin Response to Low Temperatures Revealed by Transcript and Targeted Metabolic Profiling Analysis in Poplar. Agronomy. 2022; 12(10):2506. https://doi.org/10.3390/agronomy12102506
Chicago/Turabian StyleZhao, Xianyan, Panpan Li, Can Li, and Tao Xia. 2022. "The Key Regulators and Metabolic Intermediates of Lignin Response to Low Temperatures Revealed by Transcript and Targeted Metabolic Profiling Analysis in Poplar" Agronomy 12, no. 10: 2506. https://doi.org/10.3390/agronomy12102506
APA StyleZhao, X., Li, P., Li, C., & Xia, T. (2022). The Key Regulators and Metabolic Intermediates of Lignin Response to Low Temperatures Revealed by Transcript and Targeted Metabolic Profiling Analysis in Poplar. Agronomy, 12(10), 2506. https://doi.org/10.3390/agronomy12102506




