Genetic Relationships and Diversity of Common Buckwheat Accessions in Bosnia and Herzegovina
Abstract
1. Introduction
2. Materials and Methods
2.1. SSR Analyses
2.2. Morphological Analyses
2.3. Biostatistical Analyses
3. Results and Discussion
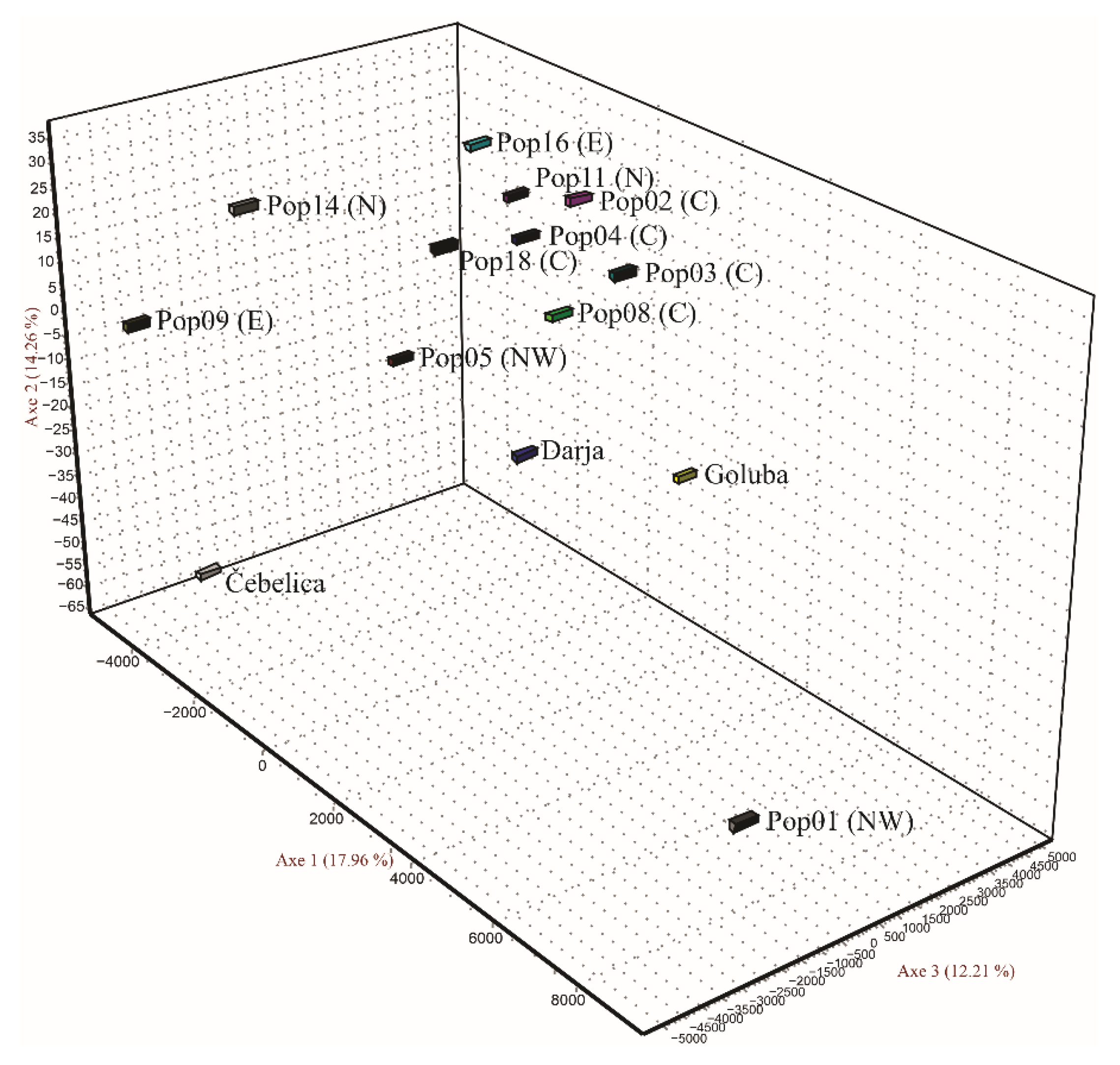
3.1. SSR Polymorphism and Genetic Relationships among the Analyzed Accessions
3.2. Phenotypic Data on Common Buckwheat Accessions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gao, X.D.; Kim, J.H.; Park, C.H.; Hong, S.K. Studies on genetic diversity of buckwheat germplasms. Korean J. Plant Res. 2010, 23, 214–222. [Google Scholar]
- Javornik, B.; Eggum, B.O.; Kreft, I. Studies on protein fractions and protein quality of buckwheat. Genetika 1981, 13, 115–121. [Google Scholar]
- Holasova, M.; Fiedlerova, V.; Smrcinova, H.; Orsak, M.; Lachman, J.; Vavreinova, S. Buckwheat—the source of antioxidant activity in functional foods. Food Res. Int. 2002, 35, 207–211. [Google Scholar] [CrossRef]
- Kreft, I.; Fabjan, N.; Yasumoto, K. Rutin content in buckwheat (Fagopyrum esculentum Moench) food materials and products. Food Chem. 2006, 98, 508–512. [Google Scholar] [CrossRef]
- Iwata, H.; Imon, K.; Tsumura, Y.; Ohsawa, R. Genetic diversity amongJapanese indigenous common buckwheat (Fagopyrum esculentum) cultivars as determined from amplified fragment length polymorphism and simple sequence repeat markers and quantitative agronomic traits. Genome 2005, 48, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.H.; Kim, N.S.; Lee, G.A.; Lee, S.Y.; Lee, J.K.; Yi, J.Y.; Park, Y.J.; Kim, T.S.; Gwag, J.G.; Kwon, S.J. Development of SSR markers for studies of diversity in the genus Fagopyrum. TAG 2009, 119, 1247–1254. [Google Scholar] [CrossRef]
- Chrungoo, N.K.; Chettry, U. Buckwheat: A critical approach towards assessment of its potential as a super crop. Indian J. Genet. Plant Breed. 2021, 81, 1–23. [Google Scholar] [CrossRef]
- Ohnishi, O. Search for the wild ancestor of buckwheat III: The wild ancestor of cultivated common buckwheat, and of Tatary buckwheat. Econ. Bot. 1998, 52, 123–133. [Google Scholar] [CrossRef]
- Ikeda, K. Buckwheat: Composition, chemistry, and processing. Adv. Food Nutr. Res. 2002, 44, 395–434. [Google Scholar]
- Hadžibegović, I. Historijske Monografije—Knjiga 1: Bosanskohercegovački Gradovi na Razmeđu 19. i 20. Stoljeća; Institut za istoriju: Sarajevo, Bosna i Hercegovina, 2004; pp. 104–105. [Google Scholar]
- Gadžo, D.; Đikić, M.; Oručević-Žuljević, S.; Gavrić, T.; Grahić, J. Proizvodnja heljde u brdsko-planinskim područjima—dosadašnja iskustva i budući izazovi. In Proceedings of the Simpozij unapređenje poljoprivrede, šumarstva i vodoprivrede u kraškim, brdskim i planinskim područjima—Racionalno korištenje i zaštita, Sarajevo, Bosnia and Herzegovina, 23 June 2016; pp. 51–59. [Google Scholar]
- Grahić, J.; Đikić, M.; Gadžo, D.; Kurtović, M.; Šimon, S.; Lazarević, B.; Vranac, A.; Gaši, F. Ispitivanje utjecaja organskih peletiranih đubriva na hemijski sastav i parametre prinosa heljde. Work. Fac. Agric. Food Sci. 2016, 66, 31–47. [Google Scholar]
- Nikolić, L.J.; Latković, D.; Berenji, J.; Sikora, V. Morphological characteristics of various buckwheat varieties (Fagopyrum esculentum Moench.). Bull. Altern. Plant Species 2010, 42, 53–59. [Google Scholar]
- Bavec, F.; Pušnik, S.; Rajčan, I. Yield performance of two buckwheat genotypes grown as a full-season and stubble-crop. Rostl. Výroba 2002, 48, 351–355. [Google Scholar] [CrossRef]
- Grahić, J.; Đikić, M.; Gadžo, D.; Uzunović, M.; Okić, A.; Kurtović, M.; Gaši, F. Analysis of agronomic practices of buckwheat producers in Bosnia and Herzegovina. Work. Fac. Agric. Food Sci. 2016, 66, 21–30. [Google Scholar]
- Song, Y.; Cheng, Z.; Dong, Y.; Liu, D.; Bai, K.; Jarvis, D.; Feng, J.; Long, C. Diversity of Tartary buckwheat (Fagopyrum tataricum) landraces from Liangshan, Southwest China: Evidence from morphology and SSR markers. Agronomy 2022, 12, 1022. [Google Scholar] [CrossRef]
- Bajracharya, J.; Brown, A.H.D.; Joshi, B.K.; Panday, D.; Baniya, B.K.; Sthapit, B.R.; Jarvis, D.I. Traditional seed management and genetic diversity in barley varieties in high-hill agro-ecosystems of Nepal. Genet. Resour. Crop Evol. 2012, 59, 389–398. [Google Scholar] [CrossRef]
- Sabreena; Nazir, M.; Mahajan, R.; Hashim, M.J.; Iqbal, J.; Alyemeni, M.N.; Ganai, B.A.; Zargar, S.M. Deciphering allelic variability and population structure in buckwheat: An analogy between the efficiency of ISSR and SSR markers. Saudi J. Biol. Sci. 2021, 28, 6050–6056. [Google Scholar] [CrossRef] [PubMed]
- Bashir, E.; Mahajan, R.; Mir, R.A.; Dar, W.A.; Zargar, S.M. Unravelling the genetic variability and population structure of buckwheat (Fagopyrum spp.): A collection of north western Himalayas. Nucleus 2020, 64, 93–101. [Google Scholar] [CrossRef]
- Facho, Z.; Farhatullah, A.; Tao, W. Species divergence and diversity in buckwheat landraces collected from the western himalayan region of pakistan. Pak. J. Bot. 2019, 51, 2215–2224. [Google Scholar] [CrossRef]
- Grahić, J.; Đikić, M.; Gadžo, D.; Šimon, S.; Kurtović, M.; Pejić, I.; Gaši, F. Assessment of genetic relationships among common buckwheat (Fagopyrum esculentum Moench) varieties from Western Balkans using morphological and SSR molecular markers. Genetika 2018, 50, 791–802. [Google Scholar] [CrossRef]
- Song, Y.J.; Lee, G.A.; Yoon, M.S.; Ma, K.H.; Choi, Y.M.; Lee, J.R.; Jung, Y.; Park, H.J.; Kim, C.K.; Lee, M.C. Analysis of genetic diversity and population structure of buckwheat (Fagopyrum esculentum Moench) landraces of Korea using SSR markers. Korean J. Plant Res. 2011, 24, 702–711. [Google Scholar] [CrossRef][Green Version]
- Kishore, G.; Gupta, S.; Pandey, A. Assessment of population genetic diversity of Fagopyrum tataricum using SSR molecular marker. Biochem. Sys. Ecol. 2012, 43, 32–41. [Google Scholar] [CrossRef]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotech. 2000, 18, 233–234. [Google Scholar] [CrossRef] [PubMed]
- IPGRI. Descriptors for Buckwheat (Fagopyrum spp.); International Plant Genetic Resources Institute: Rome, Italy, 1994. [Google Scholar]
- Hardy, O.J.; Vekemans, X. A versatile computer program to analyse spatial genetic structure at the individual or population level. Mol. Ecol. Notes 2002, 2, 618–620. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Belkhir, K.; Borsa, P.; Chikhi, L.; Raufast, N.; Bonhomme, F. GENETIX 4.02, Logiciel Sous Windows TM Pour Lagénétique des Populations; Laboratoire Génome, Populations, Interactions, CNRS UMR 5000, Université deMontpellier II: Montpellier, France, 2001. [Google Scholar]
- Excoffier, L.; Smouse, P.E.; Quattro, J.M. Analysis of molecular variance interfered from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 1992, 131, 479–491. [Google Scholar] [CrossRef]
- Ohta, T.; Kimura, M. A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a finite population. Genet Res. 1973, 22, 201–204. [Google Scholar] [CrossRef]
- Meirmans, P.; Van Tienderen, P. GenoType and GenoDive: Two programs for the analysis of genetic diversity of asexual organisms. Mol. Ecol. Notes 2004, 4, 792–794. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Earl, D.A.; Von Holdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Vigouroux, Y.; Glaubitz, J.C.; Matsuoka, Y.; Goodman, M.M.; Sánchez, G.J.; Doebley, J. Population structure and genetic diversity of New World maize races assessed by DNA microsatellites. Am. J. Bot. 2008, 95, 1240–1253. [Google Scholar] [CrossRef] [PubMed]
- Pagès, J. Multiple Factor Analysis by Example Using R, 1st ed.; Chapman and Hall: New York, NY, USA, 2014. [Google Scholar]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Grahić, A.; Grahić, J. MADC—Marker Analysis Data Compiler User’s Manual. 2017. Available online: www.divisionagro.ba/apps/docs/madc-marker-analysis-data-compiler/user-manual (accessed on 18 June 2022).
- Tsukada, M.; Sugita, S.; Tsukada, Y. Oldest primitive agriculture and vegetational environments in Japan. Nature 1986, 322, 632–634. [Google Scholar] [CrossRef]
- Grahić, J.; Kurtović, M.; Đikić, M.; Šimon, S.; Gaši, F. Genetic purity assessment of common buckwheat variety ‘Darja’ with the use of SSR molecular markers. G&A 2017, 1, 8–13. [Google Scholar]
- Aubert, L.; Decamps, C.; Jacquemin, G.; Quinet, M. Comparison of plant morphology, yield and nutritional quality of Fagopyrum esculentum and Fagopyrum tataricum grown under field conditions in Belgium. Plants 2021, 10, 258. [Google Scholar] [CrossRef]
- Murayama, S.; Miyamoto, K.; Yanokuchi, Y. Effects of variety, amount of fertilizer application and seeding rate on lodging of common buckwheat. Hokuriku Crop Sci. 2004, 40, 78–81. [Google Scholar]
- Morishita, T.; Suzuki, T.; Mukasa, Y. Characteristics of a novel ‘semidwarf materials’ in common buckwheat. Fagopyrum 2015, 32, 9–14. [Google Scholar]
- Morishita, T.; Tetsuka, T. Year-to-year variation and varietal difference of agronomic characters of common buckwheat in the kyushu area. Jpn. J. Crop Sci. 2001, 70, 379–386. [Google Scholar] [CrossRef]
- Tetsuka, T.; Morishita, T. Relationships between lodging and characteristics of Japanese landraces of buckwheat. Rep. Kyusyu Br. Crop Sci. Soc. Japan 1999, 65, 79–80. [Google Scholar]
- Malhotra, N.; Sharma, P.; Sood, H.; Chandora, R.; Arya, M.; Rana, J.C.; Singh, M. Agro-morphological characterization and nutritional profiling of traditional Himalayan crop landraces for their promotion toward mainstream agriculture. Front. Plant Sci. 2022, 13, 898220. [Google Scholar] [CrossRef]
- Klykov, A.G.; Moiseenko, L.M.; Barsukova, Y.N. Biological resources and selection value of species of Fagopyrum Mill. genus in the far east of Russia. In Molecular Breeding and Nutritional Aspects of Buckwheat; Zhou, M., Kreft, I., Woo, S.H., Chrungoo, N., Wieslander, G., Eds.; Elsevier Science: London, UK, 2016; pp. 51–60. [Google Scholar]
- Matsuri, K.; Eguchi, K.; Tetsuka, T. A novel gene that diverts the anthocyanin biosynthetic pathway towards the production of pro-anthocyanidins in common buckwheat (Fagopyrum esculentum). Breed. Sci. 2008, 58, 143–148. [Google Scholar] [CrossRef][Green Version]

| Loci | Number of Alleles | HO | HE | PIC |
|---|---|---|---|---|
| GB-FE-043 | 3.0 | 0.698 | 0.526 | 0.414 |
| GB-FE-191 | 5.0 | 0.333 | 0.386 | 0.348 |
| Fem1303 | 16.0 | 0.590 | 0.637 | 0.614 |
| Fem1840 | 4.0 | 0.682 | 0.632 | 0.554 |
| Fes1094 | 5.0 | 0.302 | 0.270 | 0.255 |
| Fes1368 | 13.0 | 0.627 | 0.677 | 0.654 |
| Fes1497 | 12.0 | 0.911 | 0.729 | 0.691 |
| Mean | 8.3 | 0.592 | 0.551 | 0.504 |
| Accession | Plant Height | Stem Color | Stem Length | Stem Diameter | Thickness of Stem Tissue |
| Goluba | 97.83 ± 6.201 | pink | 97.70 ± 6.228 | 0.40 ± 0.046 | 0.10 ± 0.002 |
| Darja | 83.61 ± 3.086 | green | 80.51 ± 3.063 | 0.40 ± 0.019 | 0.16 ± 0.006 |
| Čebelica | 129.24 ± 2.822 | red | 127.80 ± 2.878 | 0.53 ± 0.035 | 0.12 ± 0.005 |
| Pop01 | 83.16 ± 2.262 | red | 82.70 ± 2.293 | 0.52 ± 0.023 | 0.20 ± 0.029 |
| Pop02 | 71.05 ± 1.597 | pink | 69.08 ± 1.667 | 0.51 ± 0.021 | 0.12 ± 0.011 |
| Pop03 | 85.31 ± 2.098 | pink | 82.81 ± 2.196 | 0.51 ± 0.021 | 0.23 ± 0.025 |
| Pop04 | 73.21 ± 2.348 | green | 70.78 ± 2.027 | 0.32 ± 0.022 | 0.12 ± 0.014 |
| Pop05 | 86.01 ± 3.030 | pink | 84.94 ± 2.933 | 0.38 ± 0.020 | 0.10 ± 0.000 |
| Pop08 | 74.16 ± 2.034 | red | 73.61 ± 2.106 | 0.31 ± 0.025 | 0.10 ± 0.005 |
| Pop09 | 101.86 ± 1.854 | red | 99.50 ± 2.004 | 0.54 ± 0.019 | 0.11 ± 0.018 |
| Pop11 | 73.4 ± 2.369 | pink | 73.40 ± 2.369 | 0.36 ± 0.034 | 0.27 ± 0.030 |
| Pop14 | 77.28 ± 1.815 | red | 76.50 ± 1.833 | 0.30 ± 0.020 | 0.23 ± 0.026 |
| Pop16 | 102.61 ± 3.137 | red | 103.78 ± 3.206 | 0.51 ± 0.019 | 0.12 ± 0.007 |
| Pop18 | 94.35 ± 3.167 | green | 94.83 ± 3.214 | 0.50 ± 0.020 | 0.12 ± 0.008 |
| Accession | Number of Seeds per Cyme | Seed Shape | Seed Color | Seed Surface | Seed Weight |
| Goluba | 4.40 ± 0.587 | ovate | brown | smooth | 0.026 ± 0.0008 |
| Darja | 8.77 ± 0.735 | triangular | brown | smooth | 0.019 ± 0.0013 |
| Čebelica | 4.91 ± 0.444 | triangular | brown | smooth | 0.023 ± 0.0011 |
| Pop01 | 4.55 ± 0.563 | triangular | brown | smooth | 0.026 ± 0.0006 |
| Pop02 | 7.45 ± 0.671 | ovate | brown | smooth | 0.023 ± 0.0014 |
| Pop03 | 9.47 ± 1.099 | triangular | brown | smooth | 0.018 ± 0.0010 |
| Pop04 | 8.70 ± 0.925 | ovate | brown | smooth | 0.021 ± 0.0013 |
| Pop05 | 8.62 ± 0.908 | ovate | brown | smooth | 0.025 ± 0.0012 |
| Pop08 | 6.84 ± 0.718 | triangular | brown | smooth | 0.022 ± 0.0014 |
| Pop09 | 5.75 ± 0.481 | triangular | brown | smooth | 0.020 ± 0.0021 |
| Pop11 | 5.56 ± 0.363 | ovate | black | smooth | 0.019 ± 0.0014 |
| Pop14 | 3.78 ± 0.280 | ovate | brown | smooth | 0.016 ± 0.0013 |
| Pop16 | 4.06 ± 0.250 | triangular | brown | smooth | 0.019 ± 0.0013 |
| Pop18 | 5.75 ± 0.595 | ovate | black | wrinkled | 0.032 ± 0.0078 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grahić, J.; Okić, A.; Šimon, S.; Djikić, M.; Gadžo, D.; Pejić, I.; Gaši, F. Genetic Relationships and Diversity of Common Buckwheat Accessions in Bosnia and Herzegovina. Agronomy 2022, 12, 2676. https://doi.org/10.3390/agronomy12112676
Grahić J, Okić A, Šimon S, Djikić M, Gadžo D, Pejić I, Gaši F. Genetic Relationships and Diversity of Common Buckwheat Accessions in Bosnia and Herzegovina. Agronomy. 2022; 12(11):2676. https://doi.org/10.3390/agronomy12112676
Chicago/Turabian StyleGrahić, Jasmin, Arnela Okić, Silvio Šimon, Mirha Djikić, Drena Gadžo, Ivan Pejić, and Fuad Gaši. 2022. "Genetic Relationships and Diversity of Common Buckwheat Accessions in Bosnia and Herzegovina" Agronomy 12, no. 11: 2676. https://doi.org/10.3390/agronomy12112676
APA StyleGrahić, J., Okić, A., Šimon, S., Djikić, M., Gadžo, D., Pejić, I., & Gaši, F. (2022). Genetic Relationships and Diversity of Common Buckwheat Accessions in Bosnia and Herzegovina. Agronomy, 12(11), 2676. https://doi.org/10.3390/agronomy12112676







