High-Throughput Plant Phenotyping (HTPP) in Resource-Constrained Research Programs: A Working Example in Ghana
Abstract
1. Introduction
1.1. Background
1.2. The Importance of Groundnut and Its Yield Reduction Factors in Ghana
1.3. The Scenario of Field Crop Research in Ghana
1.4. Why Practise High-Throughput Plant Phenotyping in Ghana?
2. Materials and Methods
2.1. Study Area, Characteristics of Genotypes and Field Design
2.2. Conventional (Manual) Data Collection
2.3. HTPP (Drone-Based) Data Capture
2.3.1. Tools Used
2.3.2. Flight Planning and Image Acquisition
2.3.3. Image Alignment and Orthomosaic Generation
2.3.4. Data Extraction
2.4. Statistical Analysis
3. Results
3.1. Early and Late Leaf Spot Disease Tracking Using Conventional and HTPP
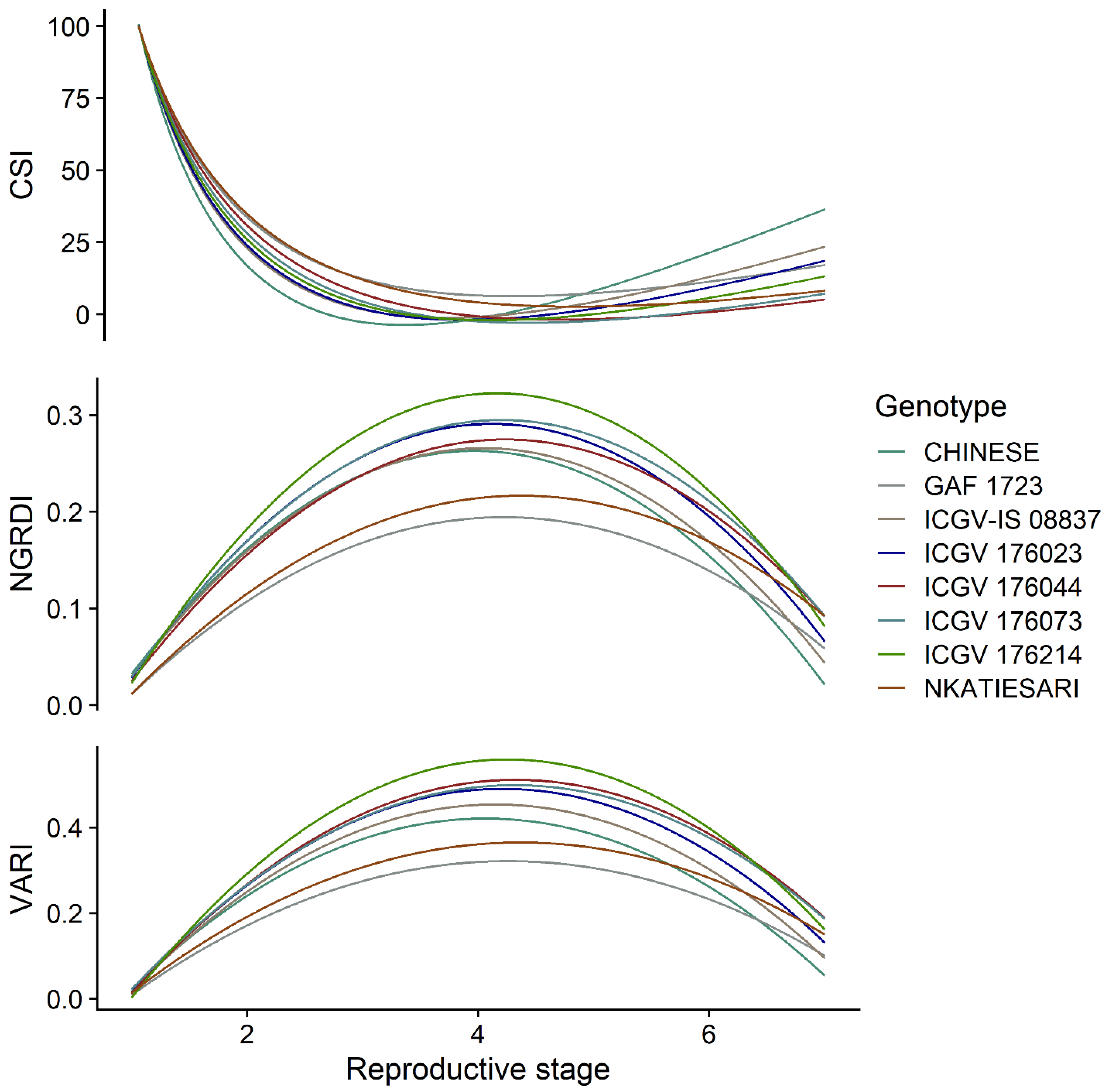
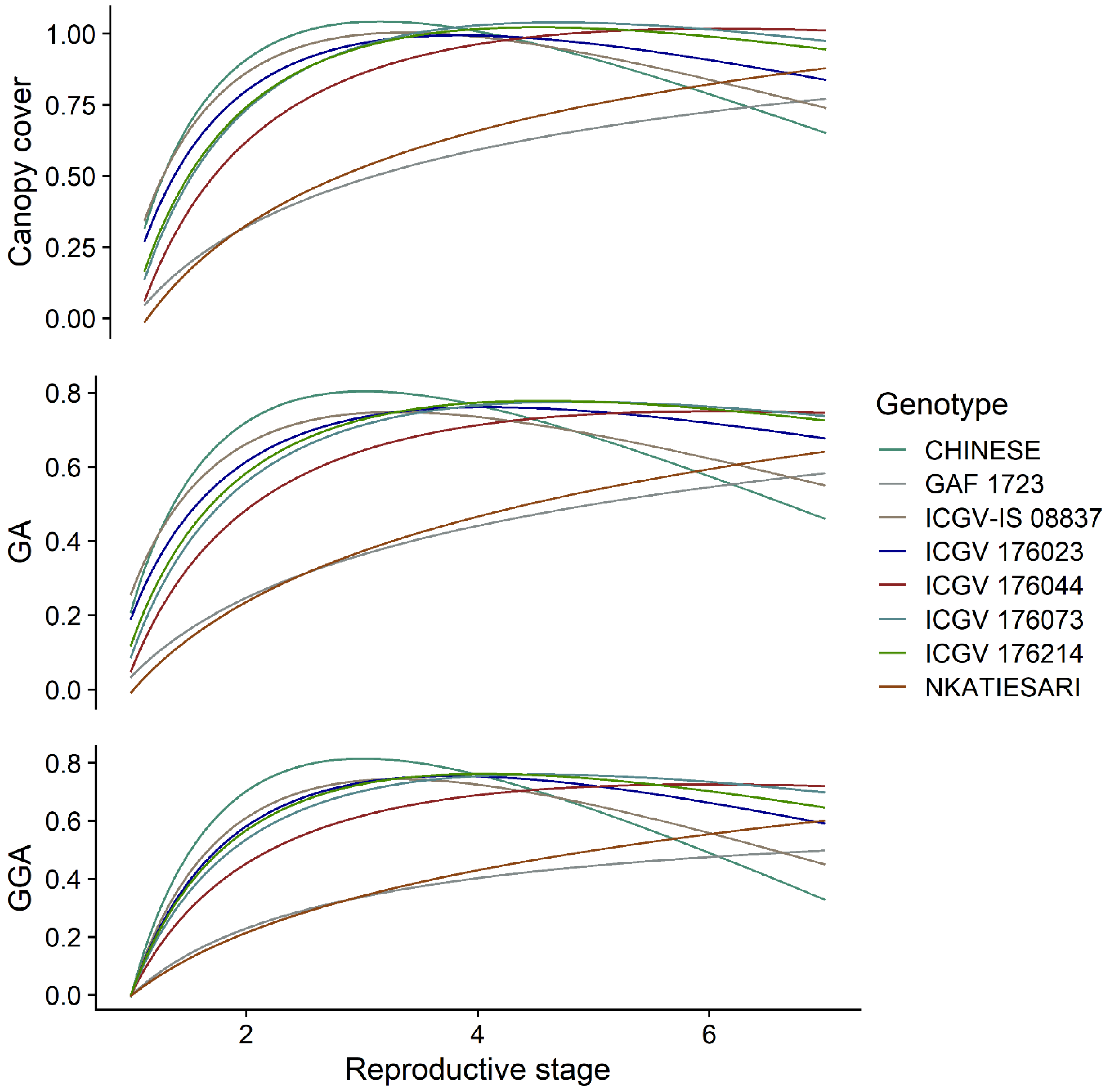
3.2. Temporal Dynamics of the Vegetation Indices among the Genotypes
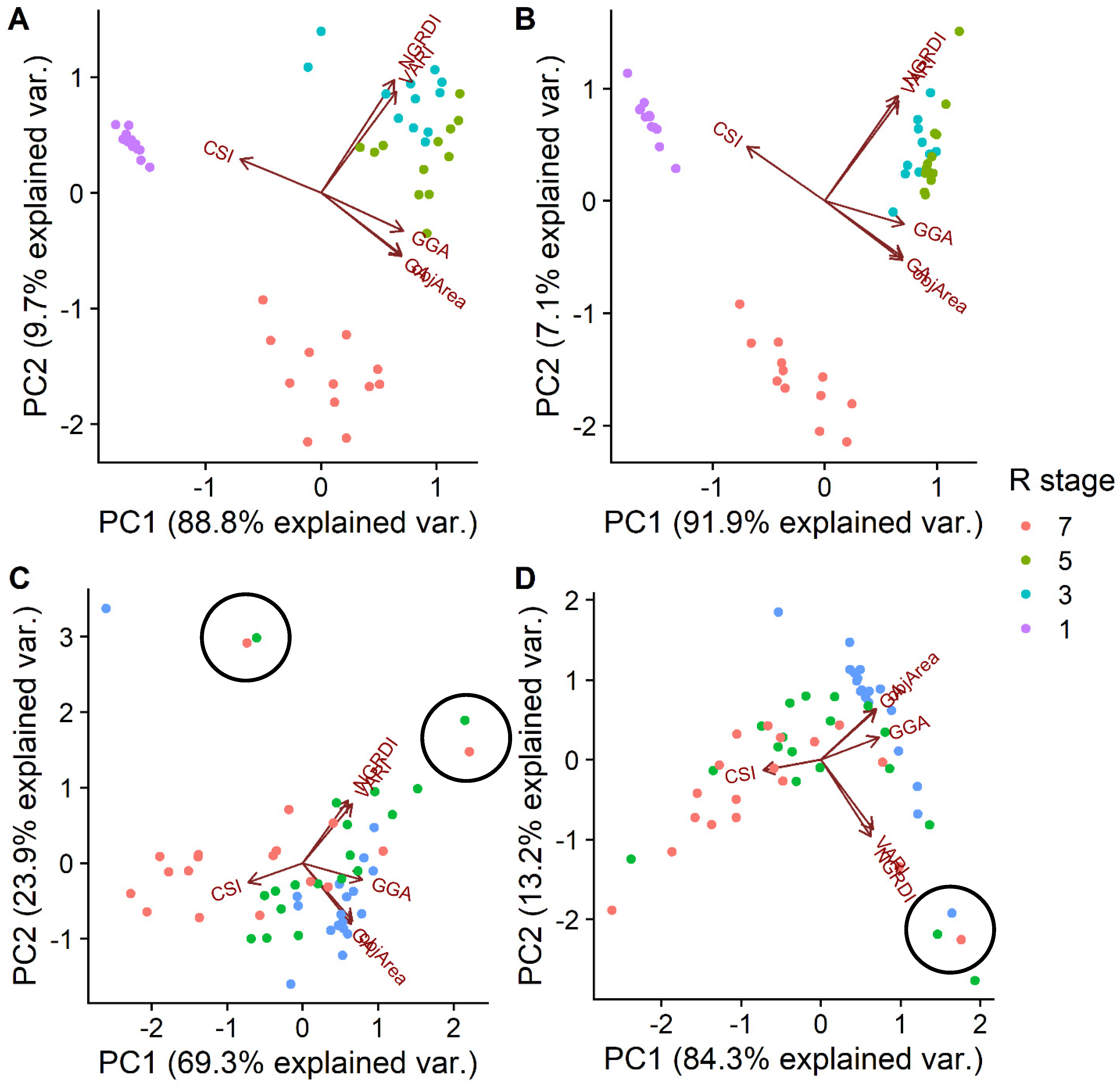
3.3. The Effect of Genotype-By-Environment Interaction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Addis Ababa: Organization of African Unity. Lagos Plan of Action for the Economic Development of Africa, 1980–2000; Organization of African Unity: Addis Ababa, Ethiopia, 1982.
- Mêgnigbêto, E. International Collaboration in Scientific Publishing: The Case of West Africa (2001–2010). Scientometrics 2013, 96, 761–783. [Google Scholar] [CrossRef]
- Vadez, V.; Berger, J.D.; Warkentin, T.; Asseng, S.; Ratnakumar, P.; Rao, K.P.C.; Gaur, P.M.; Munier-Jolain, N.; Larmure, A.; Voisin, A.-S.; et al. Adaptation of Grain Legumes to Climate Change: A Review. Agron. Sustain. Dev. 2012, 32, 31–44. [Google Scholar] [CrossRef]
- Salekdeh, G.H.; Reynolds, M.; Bennett, J.; Boyer, J. Conceptual Framework for Drought Phenotyping during Molecular Breeding. Trends Plant Sci. 2009, 14, 488–496. [Google Scholar] [CrossRef]
- Fiorani, F.; Schurr, U. Future Scenarios for Plant Phenotyping. Annu. Rev. Plant Biol. 2013, 64, 267–291. [Google Scholar] [CrossRef]
- Adzawla, W.; Kissidue, I.N.; Martey, E.; Etwire, P.M.; Atakora, W.K.; Gouzaye, A.; Bindraban, P.S. Baseline Study on Fertilizer Use and Food/Nutrition Security in the Sudan, Guinea Savanna and TranGhananal Zones of Ghana. In IFDC FERARI Report 5; IFDC: Accra, Ghana, 2021. [Google Scholar]
- Prasad, P.V.V.; Kakani, V.G.; Upadhyaya, H.D. Growth and Production of Groundnut. In UNESCO Encyclopedia; Encyclopedia of Life Support Systems (EOLSS); UNESCO: Oxford, UK, 2010; pp. 1–26. [Google Scholar]
- Oteng-Frimpong, R.; Dakora, F.D. Multienvironment Testing for Trait Stability and G × E Interaction on N2 Fixation, Plant Development, and Water-Use Efficiency of 21 Elite Groundnut (Arachis hypogaea L.) Genotypes in the Guinea Savanna. Front. Plant Sci. 2019, 10, 1070. [Google Scholar] [CrossRef]
- Pasupuleti, J.; Nigam, S.N.; Pandey, M.K.; Nagesh, P.; Varshney, R.K. Groundnut Improvement: Use of Genetic and Genomic Tools. Front. Plant Sci. 2013, 4, 23. [Google Scholar]
- Danful, R.; Kassim, Y.B.; Puozaa, D.K.; Oteng-Frimpong, R.; Rasheed, M.A.; Wireko-Kena, A.; Akromah, R. Genetics of Stay-Green Trait and Its Association with Leaf Spot Tolerance and Pod Yield in Groundnut. Int. J. Agron. 2019, 2019, 3064026. [Google Scholar] [CrossRef]
- Oteng-Frimpong, R.; Kassim, Y.B.; Danful, R.; Akromah, R.; Wireko-Kena, A.; Forson, S. Modeling Groundnut (Arachis hypogaea L.) Performance under Drought Conditions. J. Crop Improv. 2019, 33, 125–144. [Google Scholar] [CrossRef]
- Oteng-Frimpong, R.; Kassim, Y.B.; Puozaa, D.K.; Nboyine, J.A.; Issah, A.-R.; Rasheed, M.A.; Adjebeng-Danquah, J.; Kusi, F. Characterization of Groundnut (Arachis hypogaea L.) Test Locations Using Consensus Representative Environments with Farmer Preferred Traits. Front. Plant Sci. 2021, 12, 291. [Google Scholar] [CrossRef]
- National Variety Release and Registration Committee (N.V.R.R.C). Catalogue of Crop Varieties Released and Registered in Ghana. 2019. Available online: https://nastag.org/docx/resources/2019%20NATIONAL%20CROP%20VARIETY%20CATALOGUE.pdf (accessed on 1 November 2022).
- Chiteka, Z.A.; Gorbet, D.W.; Shokes, F.M.; Kucharek, T.A.; Knauft, D.A. Components of Resistance to Late Leafspot in Peanut. I. Levels and Variability-Implications for Selection 1. Peanut Sci. 1988, 15, 25–30. [Google Scholar] [CrossRef]
- Chiteka, Z.A.; Gorbet, D.W.; Knauft, D.A.; Shokes, F.M.; Kucharek, T.A. Components of Resistance to Late Leafspot in Peanut. II. Correlations Among Components and Their Significance in Breeding for Resistance 1. Peanut Sci. 1988, 15, 76–81. [Google Scholar] [CrossRef]
- Padi, F.K. Genotype × Environment Interaction for Yield and Reaction to Leaf Spot Infections in Groundnut in Semiarid West Africa. Euphytica 2008, 164, 143–161. [Google Scholar] [CrossRef]
- Subrahmanyam, P.; McDonald, D.; Waliyar, F.; Reddy, L.J.; Nigam, S.N.; Gibbons, R.W.; Rao, V.R.; Singh, A.K.; Pande, S.; Reddy, P.M.; et al. Screening Methods and Sources of Resistance to Rust and Late Leaf Spot of Groundnut. Information Bulletin no. 47. Available online: http://oar.icrisat.org/3477/ (accessed on 28 May 2019).
- Varshney, R.K.; Mohan, S.M.; Gaur, P.M.; Gangarao, N.; Pandey, M.K.; Bohra, A.; Sawargaonkar, S.L.; Chitikineni, A.; Kimurto, P.K.; Janila, P.; et al. Achievements and Prospects of Genomics-Assisted Breeding in Three Legume Crops of the Semi-Arid Tropics. Biotechnol. Adv. 2013, 31, 1120–1134. [Google Scholar] [CrossRef]
- Pasupuleti, J.; Pandey, M.K.; Shasidhar, Y.; Variath, M.T.; Sriswathi, M.; Khera, P.; Manohar, S.S.; Nagesh, P.; Vishwakarma, M.K.; Mishra, G.P.; et al. Molecular Breeding for Introgression of Fatty Acid Desaturase Mutant Alleles (AhFAD2A and AhFAD2B) Enhances Oil Quality in High and Low Oil Containing Peanut Genotypes. Plant Sci. 2016, 242, 203–213. [Google Scholar] [CrossRef]
- Pasupuleti, J.; Variath, M.T.; Pandey, M.K.; Desmae, H.; Motagi, B.N.; Okori, P.; Manohar, S.S.; Rathnakumar, A.L.; Radhakrishnan, T.; Liao, B.; et al. Genomic Tools in Groundnut Breeding Program: Status and Perspectives. Front. Plant Sci. 2016, 7, 289. [Google Scholar]
- Varshney, R.K. Exciting Journey of 10 Years from Genomes to Fields and Markets: Some Success Stories of Genomics-Assisted Breeding in Chickpea, Pigeonpea and Groundnut. Plant Sci. 2016, 242, 98–107. [Google Scholar] [CrossRef]
- Burow, M.D.; Starr, J.L.; Park, C.-H.; Simpson, C.E.; Paterson, A.H. Introgression of Homeologous Quantitative Trait Loci (QTLs) for Resistance to the Root-Knot Nematode [Meloidogyne Arenaria (Neal) Chitwood] in an Advanced Backcross-QTL Population of Peanut (Arachis hypogaea L.). Mol. Breed. 2014, 34, 393–406. [Google Scholar] [CrossRef]
- Simpson, C.E.; Starr, J.L.; Church, G.T.; Burow, M.D.; Paterson, A.H. Registration of ‘NemaTAM’ Peanut. Crop Sci. 2003, 43, 1561. [Google Scholar] [CrossRef]
- Eberius, M.; Lima-Guerra, J. High-Throughput Plant Phenotyping—Data Acquisition, Transformation, and Analysis. In Bioinformatics: Tools and Applications; Edwards, D., Stajich, J., Hansen, D., Eds.; Springer: New York, NY, USA, 2009; pp. 259–278. [Google Scholar] [CrossRef]
- Koltes, J.E.; Cole, J.B.; Clemmens, R.; Dilger, R.N.; Kramer, L.M.; Lunney, J.K.; McCue, M.E.; McKay, S.D.; Mateescu, R.G.; Murdoch, B.M.; et al. A Vision for Development and Utilization of High-Throughput Phenotyping and Big Data Analytics in Livestock. Front. Genet. 2019, 10, 1197. [Google Scholar] [CrossRef]
- Araus, J.L.; Kefauver, S.C.; Zaman-Allah, M.; Olsen, M.S.; Cairns, J.E. Translating High-Throughput Phenotyping into Genetic Gain. Trends Plant Sci. 2018, 23, 451–466. [Google Scholar] [CrossRef]
- Barker, J.; Zhang, N.; Sharon, J.; Steeves, R.; Wang, X.; Wei, Y.; Poland, J. Development of a Field-Based High-Throughput Mobile Phenotyping Platform. Comput. Electron. Agric. 2016, 122, 74–85. [Google Scholar] [CrossRef]
- Jordan, B.S.; Branch, W.D.; Coffin, A.W.; Smith, C.M.; Culbreath, A.K. Comparison of Trimble GreenSeeker and Crop Circle (Model ACS-210) Reflectance Meters for Assessment of Severity of Late Leaf Spot. Peanut Sci. 2019, 46, 110–117. [Google Scholar] [CrossRef]
- Luis, J.M.; Ozias-Akins, P.; Holbrook, C.C.; Kemerait, R.C., Jr.; Snider, J.L.; Liakos, V. Phenotyping Peanut Genotypes for Drought Tolerance. Peanut Sci. 2016, 43, 36–48. [Google Scholar] [CrossRef]
- Pallottino, F.; Figorilli, S.; Cecchini, C.; Costa, C. Light Drones for Basic In-Field Phenotyping and Precision Farming Applications: RGB Tools Based on Image Analysis. In Crop Breeding: Genetic Improvement Methods; Tripodi, P., Ed.; Springer: New York, NY, USA, 2021; pp. 269–278. [Google Scholar] [CrossRef]
- Pourazar, H.; Samadzadegan, F.; Javan, F.D. Aerial Multispectral Imagery for Plant Disease Detection: Radiometric Calibration Necessity Assessment. Eur. J. Remote Sens. 2019, 52 (Suppl. S3), 17–31. [Google Scholar] [CrossRef]
- Chapu, I.; Okello, D.K.; Okello, R.C.O.; Odong, T.L.; Sarkar, S.; Balota, M. Exploration of Alternative Approaches to Phenotyping of Late Leaf Spot and Groundnut Rosette Virus Disease for Groundnut Breeding. Front. Plant Sci. 2022, 13, 912332. [Google Scholar] [CrossRef]
- Sie, E.K.; Oteng-Frimpong, R.; Kassim, Y.B.; Puozaa, D.K.; Adjebeng-Danquah, J.; Masawudu, A.R.; Ofori, K.; Danquah, A.; Cazenave, A.B.; Hoisington, D.; et al. RGB-Image Method Enables Indirect Selection for Leaf Spot Resistance and Yield Estimation in a Groundnut Breeding Program in Western Africa. Front. Plant Sci. 2022, 13, 957061. [Google Scholar] [CrossRef]
- Boote, K.J. Growth Stages of Peanut (Arachis hypogaea L.) 1. Peanut Sci. 1982, 9, 35–40. [Google Scholar] [CrossRef]
- Rife, T.W.; Poland, J.A. Field Book: An Open-Source Application for Field Data Collection on Android. Crop Sci. 2014, 54, 1624–1627. [Google Scholar] [CrossRef]
- Vacca, G. WEB Open Drone Map (WebODM) a Software Open Source to Photogrammetry Process; Smart Surveyors for Land and Water Management: Enschede, The Netherlands, 2020; Volume 9. [Google Scholar]
- Anderson, S.L.; Murray, S.C. R/UAStools::Plotshpcreate: Create Multi-Polygon Shapefiles for Extraction of Research Plot Scale Agriculture Remote Sensing Data. Front. Plant Sci. 2020, 11, 511768. [Google Scholar] [CrossRef]
- Matias, F.I.; Caraza-Harter, M.V.; Endelman, J.B. FIELDimageR: An R Package to Analyze Orthomosaic Images from Agricultural Field Trials. Plant Phenome J. 2020, 3, e20005. [Google Scholar] [CrossRef]
- Escadafal, R.; Belghit, A.; Ben-Moussa, A. Indices Spectraux Pour La Télédétection de La Dégradation Des Milieux Naturels En Tunisie Aride. In Proceedings of the 6th International Symposium on Physical Measurements and Signatures in Remote Sensing, Val D’Isere, France, 17–21 January 1994; pp. 17–21. [Google Scholar]
- Tucker, C.J. Red and Photographic Infrared Linear Combinations for Monitoring Vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Stark, R.; Rundquist, D. Novel Algorithms for Remote Estimation of Vegetation Fraction. Remote Sens. Environ. 2002, 80, 76–87. [Google Scholar] [CrossRef]
- Casadesús, J.; Kaya, Y.; Bort, J.; Nachit, M.M.; Araus, J.L.; Amor, S.; Ferrazzano, G.; Maalouf, F.; Maccaferri, M.; Martos, V.; et al. Using Vegetation Indices Derived from Conventional Digital Cameras as Selection Criteria for Wheat Breeding in Water-Limited Environments. Ann. Appl. Biol. 2007, 150, 227–236. [Google Scholar] [CrossRef]
- Casadesús, J.; Villegas, D. Conventional Digital Cameras as a Tool for Assessing Leaf Area Index and Biomass for Cereal Breeding. J. Integr. Plant Biol. 2014, 56, 7–14. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 1 November 2022).
- Lenth, R.; Singmann, H.; Love, J.; Buerkner, P.; Herve, M. Emmeans: Estimated Marginal Means, Aka Least-Squares Means. 2019. Available online: https://cran.r-project.org/web/packages/emmeans/emmeans.pdf (accessed on 1 November 2022).
- de Mendiburu, F. Agricolae: Statistical Procedures for Agricultural Research. 2019. Available online: https://cran.r-project.org/web/packages/agricolae/index.html (accessed on 1 November 2022).
- Kassambara, A. Ggpubr: “ggplot2” Based Publication Ready Plots. 2020. Available online: https://cran.r-project.org/web/packages/ggpubr/index.html (accessed on 1 November 2022).
- Wickham, H.; Chang, W.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K.; Studio, R. Ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. 2019. Available online: https://ggplot2.tidyverse.org/reference/ggplot2-package.html (accessed on 1 November 2022).
- Wilke, C.O. Cowplot: Streamlined Plot Theme and Plot Annotations for “Ggplot2”. 2020. Available online: https://wilkelab.org/cowplot/ (accessed on 1 November 2022).
- Gauch, H.G. A Simple Protocol for AMMI Analysis of Yield Trials. Crop Sci. 2013, 53, 1860–1869. [Google Scholar] [CrossRef]
- Piepho, H.P. Robustness of Statistical Tests for Multiplicative Terms in the Additive Main Effects and Multiplicative Interaction Model for Cultivar Trials. Theoret. Appl. Genet. 1995, 90, 438–443. [Google Scholar] [CrossRef]
- Oteng-Frimpong, R.; Danful, R.; Kassim, Y.B.; Denwar, N.N.; Akromah, R. Stay-Green Trait and Its Association with Leaf Spot Disease Tolerance in Groundnut. In Crop Breeding and Genomics; WACCI: Accra, Ghana, 2018; pp. 68–69. [Google Scholar]
- Oteng-Frimpong, R.; Danful, R.; Kassim, Y.B.; Denwar, N.N.; Puozaa, D.K.; Adombila, R.; Masawudu, A.R.; Issah, A.R. Relationship between Stay-Green Trait and Leaf Spot Tolerance in Groundnut. In 2017 Annual Report; CSIR-Savanna Agricultural Research Institute: Accra, Ghana, 2017; pp. 40–46. [Google Scholar]

| Medium Duration (MD) | Short Duration (SD) | ||||
|---|---|---|---|---|---|
| No. | Genotype | Years | No. | Genotype | Years |
| 1 | ICGV 176044 | 2020; 2021 | 1 | ICGV 176217 | 2020; 2021 |
| 2 | ICGV 176107 | 2020; 2021 | 2 | ICGV 176166 | 2020; 2021 |
| 3 | ICGV 176084 | 2020; 2021 | 3 | ICGV 176222 | 2020; 2021 |
| 4 | ICGV 176067 | 2020; 2021 | 4 | ICGV 176160 | 2020; 2021 |
| 5 | ICGV 176129 | 2020; 2021 | 5 | ICGV 176214 | 2020; 2021 |
| 6 | ICGV 176073 | 2020; 2021 | 6 | ICGV 176151 | 2020; 2021 |
| 7 | ICGV 176033 | 2020; 2021 | 7 | ICGV 176023 | 2020; 2021 |
| 8 | ICGV 176225 | 2020; 2021 | 8 | ICGV 176156 | 2020; 2021 |
| 9 | ICGV 176124 | 2020; 2021 | 9 | ICGV 176019 | 2020; 2021 |
| 10 | ICGV 176203 | 2020; 2021 | 10 | ICGV 176049 | 2020; 2021 |
| 11 | NKATIESARI | 2020; 2021 | 11 | CHINESE | 2020; 2021 |
| 12 | GAF 1723 | 2020; 2021 | 12 | ICGV-IS 08837 | 2020; 2021 |
| 13 | GAF 1665 | 2021 | 13 | ICGV 15403 | 2021 |
| 14 | ICGV 176051 | 2021 | 14 | ICGV 176004 | 2021 |
| 15 | ICGV 176112 | 2021 | 15 | ICGV 176010 | 2021 |
| 16 | ICGV 176124b | 2021 | 16 | ICGV 176053 | 2021 |
| 17 | ICGV 176192 | 2021 | 17 | ICGV 176154 | 2021 |
| 18 | ICGV-SM 10523 | 2021 | 18 | YENYAWOSO | 2021 |
| No. | Tool | Purpose | Commercial/Open Source |
|---|---|---|---|
| Hardware tools | |||
| 1 | UAV quadcopter with an RGB camera (DJI, California, USA) | Take images | Commercial |
| 2 | Smart phone (Apple Inc., Accra, Ghana) | Augment drone | Commercial/personal |
| 3 | Laptop (Dell, Accra, Ghana) | Process and analyze images/data | Commercial/personal |
| Software tools | |||
| 1 | UAV Forecast (version 2.6.3) | Monitor weather | Open source |
| 2 | Pix4Dcapture (version 4.10.0) | Mission planning | Open source |
| 3 | WebODM (version 1.9.11) | Generate orthomosaics | Open source |
| 4 | QGIS (version 3.16.14) | Georeference orthomosaics | Open source |
| 5 | R statistical software (version 4.2.0) | Platform for image analysis packages | Open source |
| 6 | UASTools (version 0.4.0) | Generate shapefile for microplots | Open source |
| 7 | FieldImageR (version 0.3.3) | Analyze images | Open source |
| ELS | LLS | |||||
|---|---|---|---|---|---|---|
| Median | Min | Max | Median | Min | Max | |
| 2020 | ||||||
| Medium duration | 35.88 | 12.83 | 56.19 | 46.13 | 15.39 | 56.12 |
| Short duration | 61.48 | 41.39 | 72.60 | 62.09 | 41.84 | 70.53 |
| 2021 | ||||||
| Medium duration | 67.78 | 34.68 | 90.86 | 67.05 | 30.85 | 88.29 |
| Short duration | 61.66 | 41.50 | 78.75 | 69.20 | 42.94 | 85.60 |
| MD | SD | |||||||
|---|---|---|---|---|---|---|---|---|
| Reproductive Stage | ELS AUDPC | LLS AUDPC | ELS AUDPC | LLS AUDPC | ||||
| 2020 | 2021 | 2020 | 2021 | 2020 | 2021 | 2020 | 2021 | |
| R3 | ||||||||
| NGRDI | 0.67 * | −0.22 ns | 0.78 ** | −0.50 * | −0.18 ns | −0.43 ns | −0.32 ns | −0.81 *** |
| VARI | 0.56 ns | −0.28 ns | 0.64 * | −0.54 * | −0.25 ns | −0.42 ns | −0.33 ns | −0.82 *** |
| Canopy cover | 0.73 ** | 0.28 ns | 0.91 *** | 0.51 * | 0.20 ns | −0.54 * | 0.01 ns | −0.74 *** |
| GA | 0.73 ** | 0.27 ns | 0.90 *** | 0.53 * | 0.25 ns | −0.49 * | −0.02 ns | −0.70 ** |
| GGA | 0.72 ** | 0.22 ns | 0.90 *** | 0.50 * | 0.20 ns | −0.38 ns | −0.02 ns | −0.70 ** |
| CSI | −0.64 * | −0.06 ns | −0.83 *** | −0.29 ns | −0.03 ns | 0.37 ns | 0.00 ns | 0.69 ** |
| R5 | ||||||||
| NGRDI | 0.03 ns | −0.60 ** | 0.32 ns | −0.80 *** | −0.65 * | −0.62 ** | −0.80 ** | −0.87 *** |
| VARI | 0.10 ns | −0.61 ** | 0.36 ns | −0.79 *** | −0.58 ns | −0.63 ** | −0.62 * | −0.88 *** |
| Canopy cover | 0.46 ns | 0.10 ns | 0.75 ** | 0.27 ns | 0.05 ns | −0.43 ns | 0.15 ns | −0.70 ** |
| GA | 0.40 ns | 0.08 ns | 0.77 ** | 0.30 ns | −0.42 ns | −0.46 ns | −0.43 ns | −0.71 *** |
| GGA | 0.40 ns | −0.41 ns | 0.77 ** | −0.42 ns | −0.44 ns | −0.55 * | −0.45 ns | −0.83 *** |
| CSI | −0.37 ns | 0.56 * | −0.71 ** | 0.72 *** | 0.35 ns | 0.55 * | 0.34 ns | 0.84 *** |
| R7 | ||||||||
| NGRDI | −0.74 ** | −0.63 ** | −0.38 ns | −0.86 *** | −0.88 *** | −0.62 ** | −0.64 * | −0.86 *** |
| VARI | −0.66 * | −0.64 ** | −0.28 ns | −0.86 *** | −0.88 *** | −0.62 ** | −0.63 * | −0.86 *** |
| Canopy cover | −0.33 ns | −0.37 ns | 0.12 ns | −0.37 ns | −0.69 * | −0.54 * | −0.69 * | −0.86 *** |
| GA | −0.35 ns | −0.33 ns | 0.15 ns | −0.28 ns | −0.77 ** | −0.55 * | −0.71 * | −0.86 *** |
| GGA | −0.51 ns | −0.54 * | 0.00 ns | −0.68 ** | −0.83 *** | −0.61 ** | −0.56 ns | −0.88 *** |
| CSI | 0.67 * | 0.58 * | 0.21 ns | 0.81 *** | 0.77 ** | 0.63 ** | 0.38 ns | 0.86 *** |
| Variation Source | df | Sum of Squares | |||
|---|---|---|---|---|---|
| ELS AUDPC | LLS AUDPC | Haulm Yield (kgha−1) | Pod Yield (kgha−1) | ||
| MD in 2020 | |||||
| Environment Ghana | 3 | 9905.4 *** | 14,938.8 *** | 441,412,003 *** | 32,875,500 *** |
| Genotype (G) | 11 | 3923.1 *** | 11,262.3 *** | 16,931,646 ** | 23,250,254 *** |
| Genotype × Environment | 33 | 4699.6 *** | 5372.9 * | 23,674,776 ns | 12,776,519 *** |
| G × E signal | 0.7581 | 0.4496 | 0.2733 | 0.7691 | |
| SD in 2020 | |||||
| EnvironmeGhana(E) | 3 | 35,052 *** | 14,005.6 *** | 171,352,883 *** | 16,265,746 *** |
| Genotype (G) | 11 | 2799 *** | 7702.7 *** | 33,553,632 *** | 19,915,906 *** |
| Genotype × Environment | 33 | 1911 ** | 3095.6 *** | 10,956,981 * | 15,445,714 *** |
| G × E signal | 0.5238 | 0.7165 | 0.3965 | 0.7738 | |
| MD in 2021 | |||||
| EnviroGhanant (E) | 3 | 178,981 *** | 154,147 *** | 156,608,615 *** | 264,852,830 *** |
| Genotype (G) | 17 | 21,916 *** | 17,784 *** | 49,675,436 *** | 77,008,509 *** |
| Genotype × Environment | 51 | 33,535 *** | 32,443 *** | 115,776,686 *** | 38,860,002 *** |
| G × E signal | 0.9302 | 0.8380 | 0.8175 | 0.8258 | |
| SD in 2021 | |||||
| EnvGhananment (E) | 3 | 146,356 *** | 195,735 *** | 664,657,480 *** | 284,254,679 *** |
| Genotype (G) | 17 | 11,577 *** | 15,269 *** | 69,544,867 ** | 47,883,935 *** |
| Genotype × Environment | 51 | 14,675 *** | 11,339 *** | 116,259,584 ** | 26,920,315 *** |
| G × E signal | 0.8592 | 0.6924 | 0.5232 | 0.8008 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kassim, Y.B.; Oteng-Frimpong, R.; Puozaa, D.K.; Sie, E.K.; Abdul Rasheed, M.; Abdul Rashid, I.; Danquah, A.; Akogo, D.A.; Rhoads, J.; Hoisington, D.; et al. High-Throughput Plant Phenotyping (HTPP) in Resource-Constrained Research Programs: A Working Example in Ghana. Agronomy 2022, 12, 2733. https://doi.org/10.3390/agronomy12112733
Kassim YB, Oteng-Frimpong R, Puozaa DK, Sie EK, Abdul Rasheed M, Abdul Rashid I, Danquah A, Akogo DA, Rhoads J, Hoisington D, et al. High-Throughput Plant Phenotyping (HTPP) in Resource-Constrained Research Programs: A Working Example in Ghana. Agronomy. 2022; 12(11):2733. https://doi.org/10.3390/agronomy12112733
Chicago/Turabian StyleKassim, Yussif Baba, Richard Oteng-Frimpong, Doris Kanvenaa Puozaa, Emmanuel Kofi Sie, Masawudu Abdul Rasheed, Issah Abdul Rashid, Agyemang Danquah, Darlington A. Akogo, James Rhoads, David Hoisington, and et al. 2022. "High-Throughput Plant Phenotyping (HTPP) in Resource-Constrained Research Programs: A Working Example in Ghana" Agronomy 12, no. 11: 2733. https://doi.org/10.3390/agronomy12112733
APA StyleKassim, Y. B., Oteng-Frimpong, R., Puozaa, D. K., Sie, E. K., Abdul Rasheed, M., Abdul Rashid, I., Danquah, A., Akogo, D. A., Rhoads, J., Hoisington, D., Burow, M. D., & Balota, M. (2022). High-Throughput Plant Phenotyping (HTPP) in Resource-Constrained Research Programs: A Working Example in Ghana. Agronomy, 12(11), 2733. https://doi.org/10.3390/agronomy12112733








