Genome-Wide Identification and Expression Analysis of BrATGs and Their Different Roles in Response to Abiotic Stresses in Chinese Cabbage
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Identification of ATGs in Chinese Cabbage
2.3. Physicochemical Property Analysis and Subcellular Localization of BrATGs
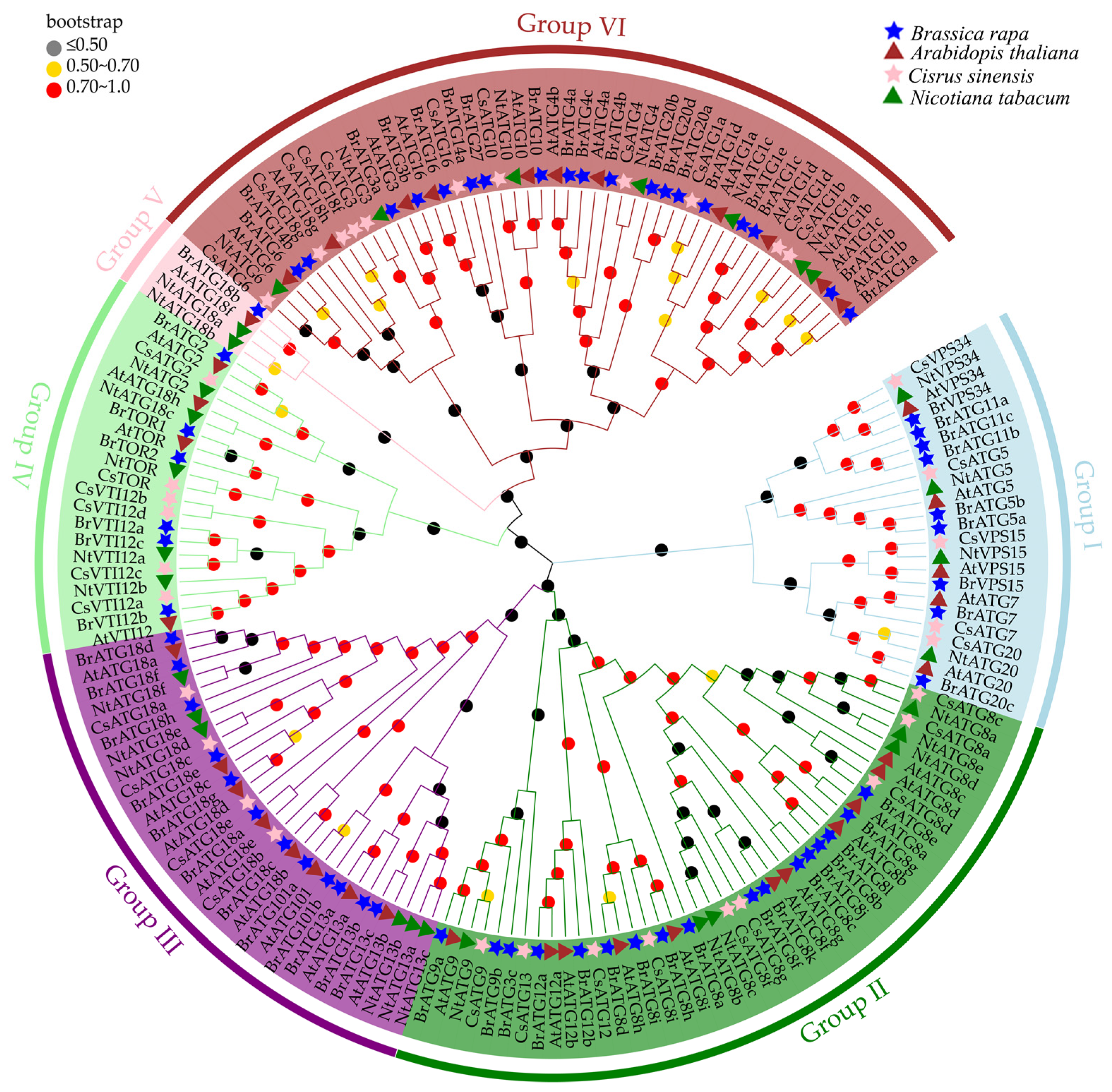
2.4. Phylogenetic Tree Analysis of BrATGs
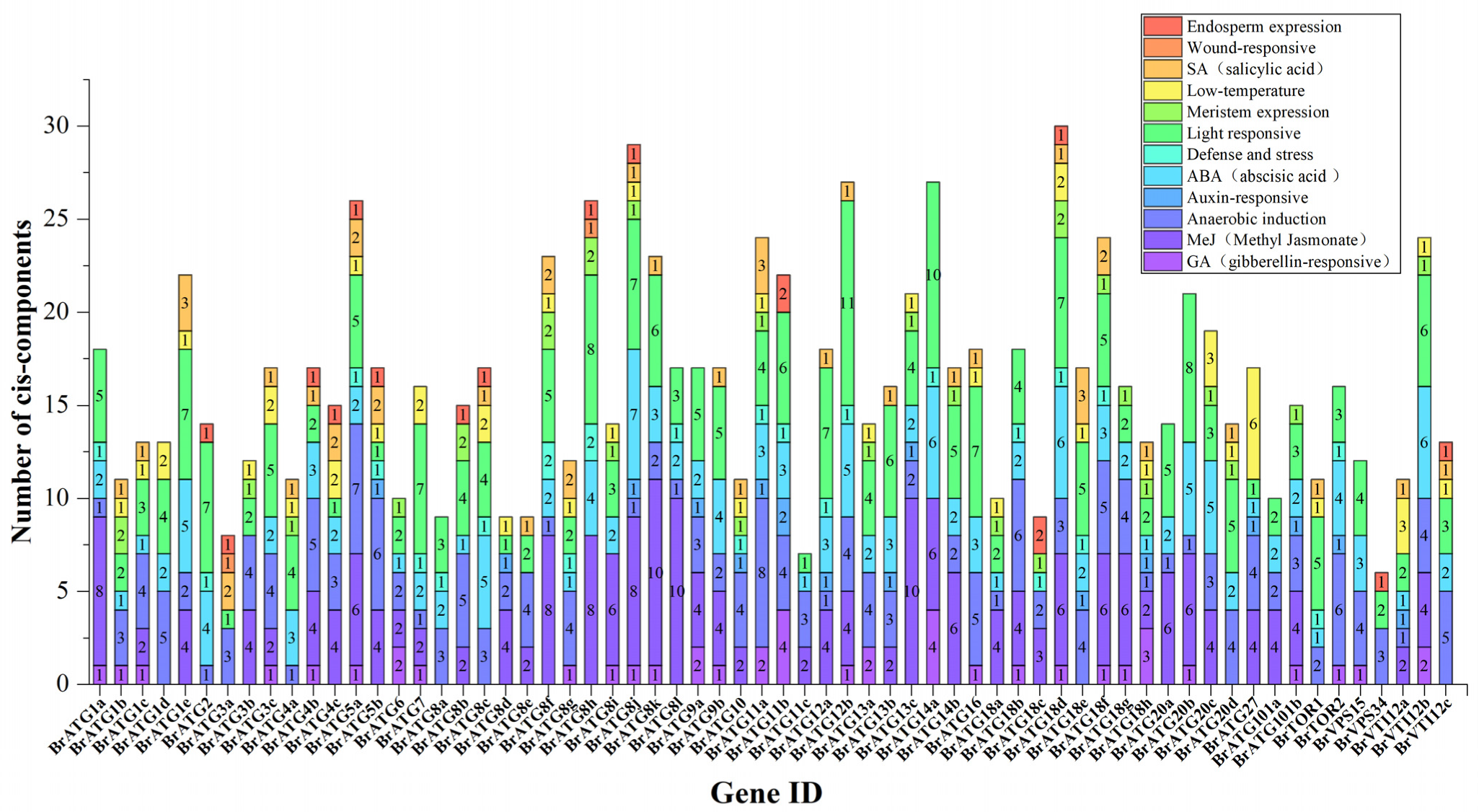
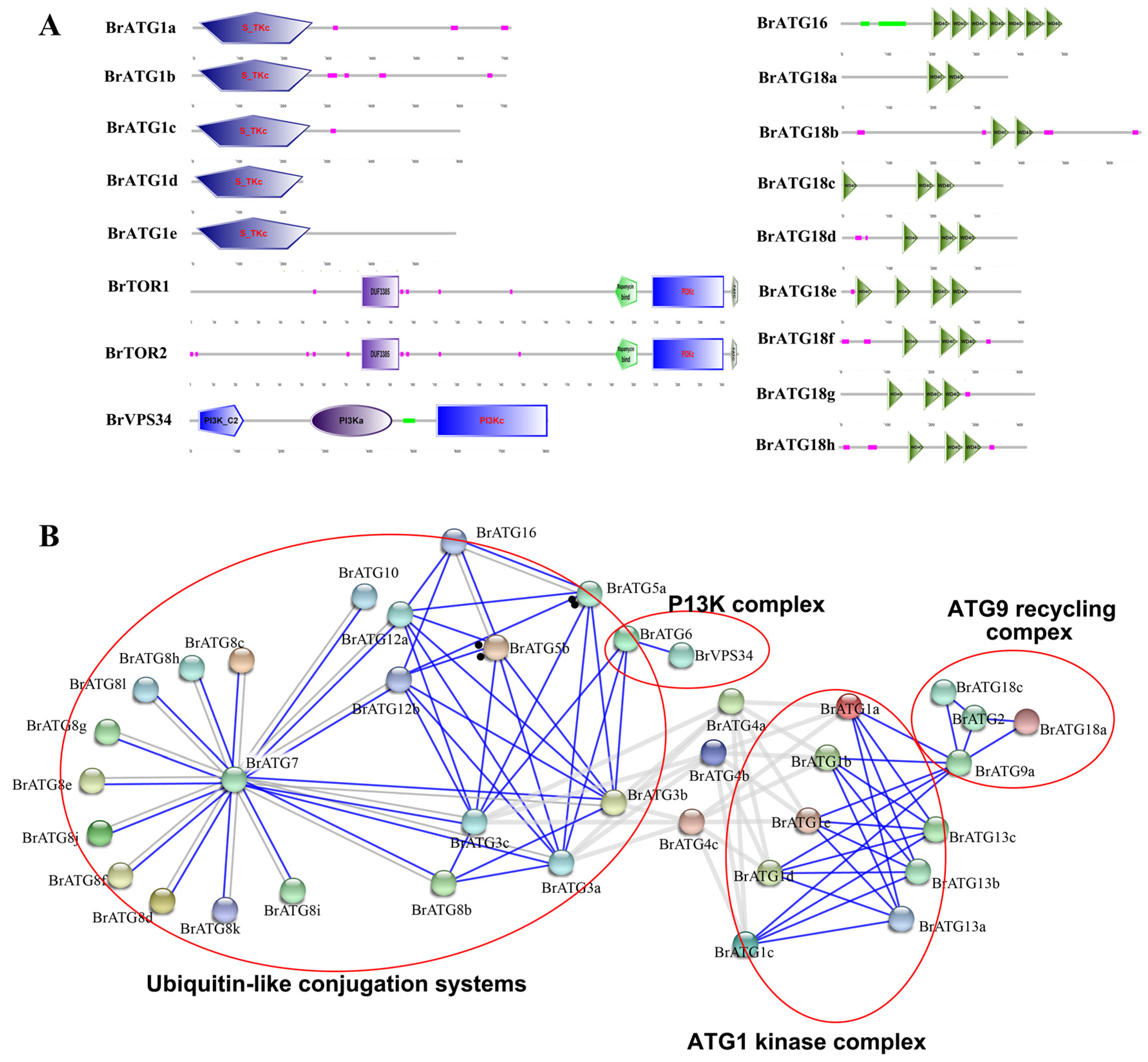
2.5. Analysis of BrATGs Gene Structure, Protein Structural Domain Distribution, Cis-Acting Elements, and Protein Interaction Networks
2.6. Chromosome Distribution and Gene Duplication Analysis of BrATGs
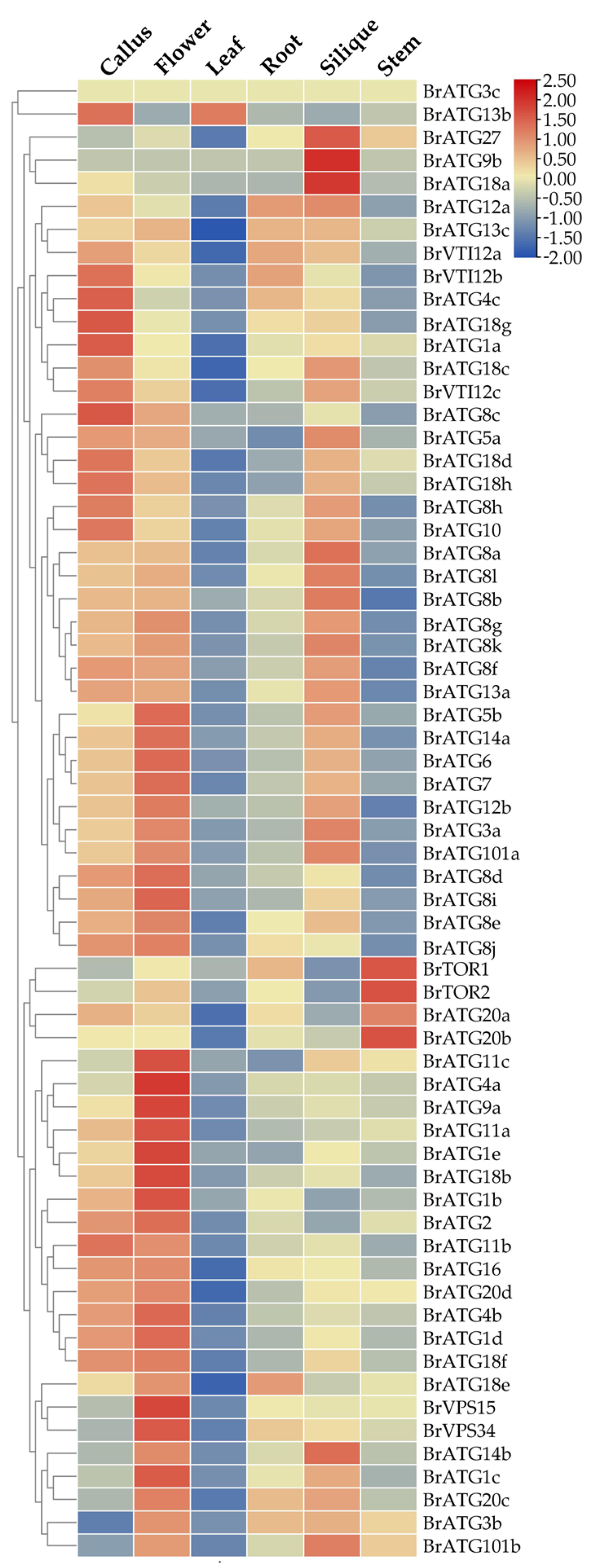
2.7. Analysis of Expression Patterns of BrATGs Genes in Different Tissues
2.8. Analysis of Expression Specificity in Response to Abiotic Stress by qRT–PCR
3. Results
3.1. Identification of BrATGs in the Chinese Cabbage Genome
3.2. Phylogenetic Analysis of BrATGs
3.3. Analysis of Gene Structure and Protein Structural Domain Distribution of BrATGs
3.4. Analysis of Cis-Acting Elements and Protein Interactions of BrATGs
3.5. Analysis of Conserved Structural Domains and Protein-Protein Interaction Networks of BrATG Proteins
3.6. Chromosome Distribution and Gene Duplication Analysis of BrATGs
3.7. Analysis of the Expression Pattern of BrATG in Various Tissues
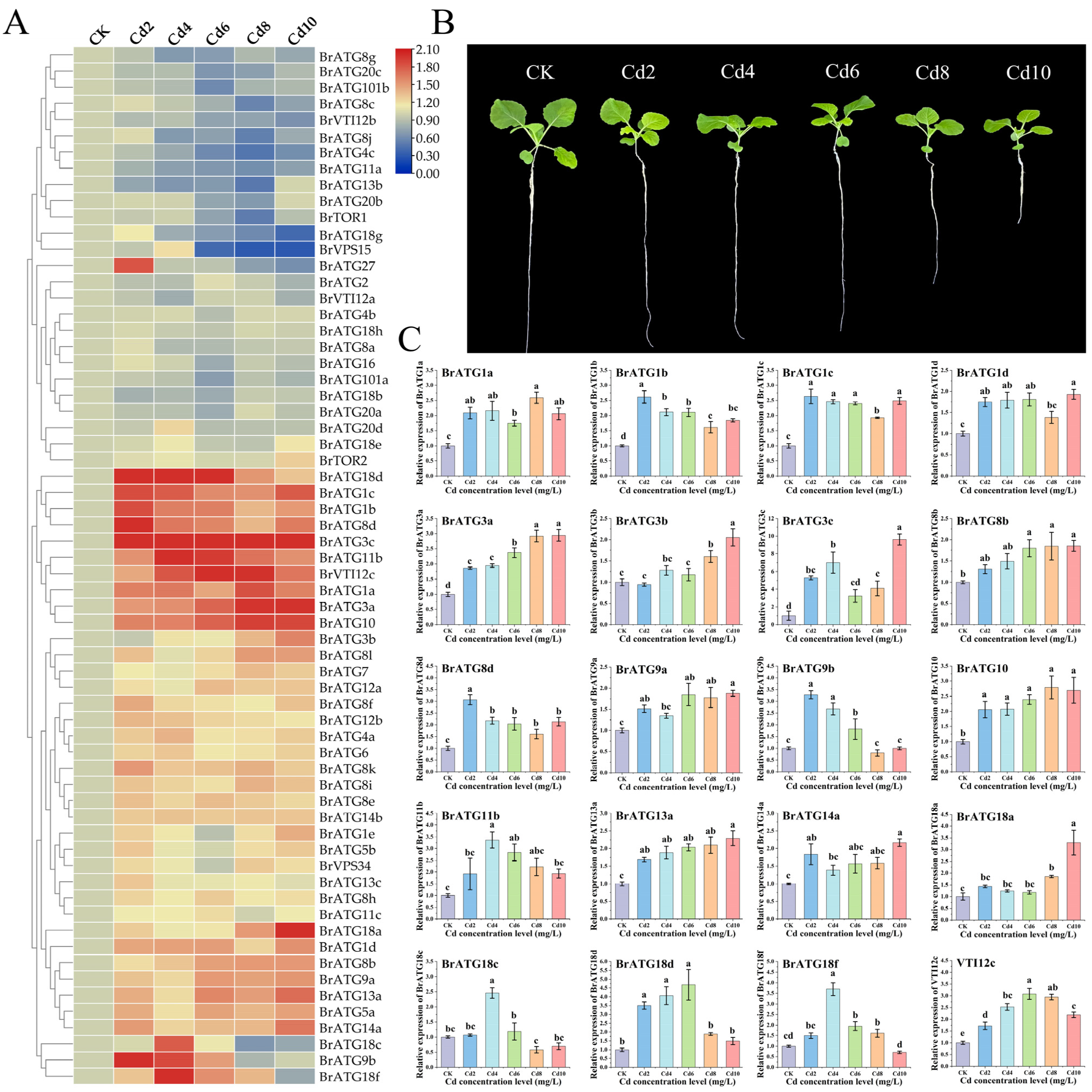
3.8. Analysis of the Expression Pattern of BrATGs under Cadmium (Cd) Stress
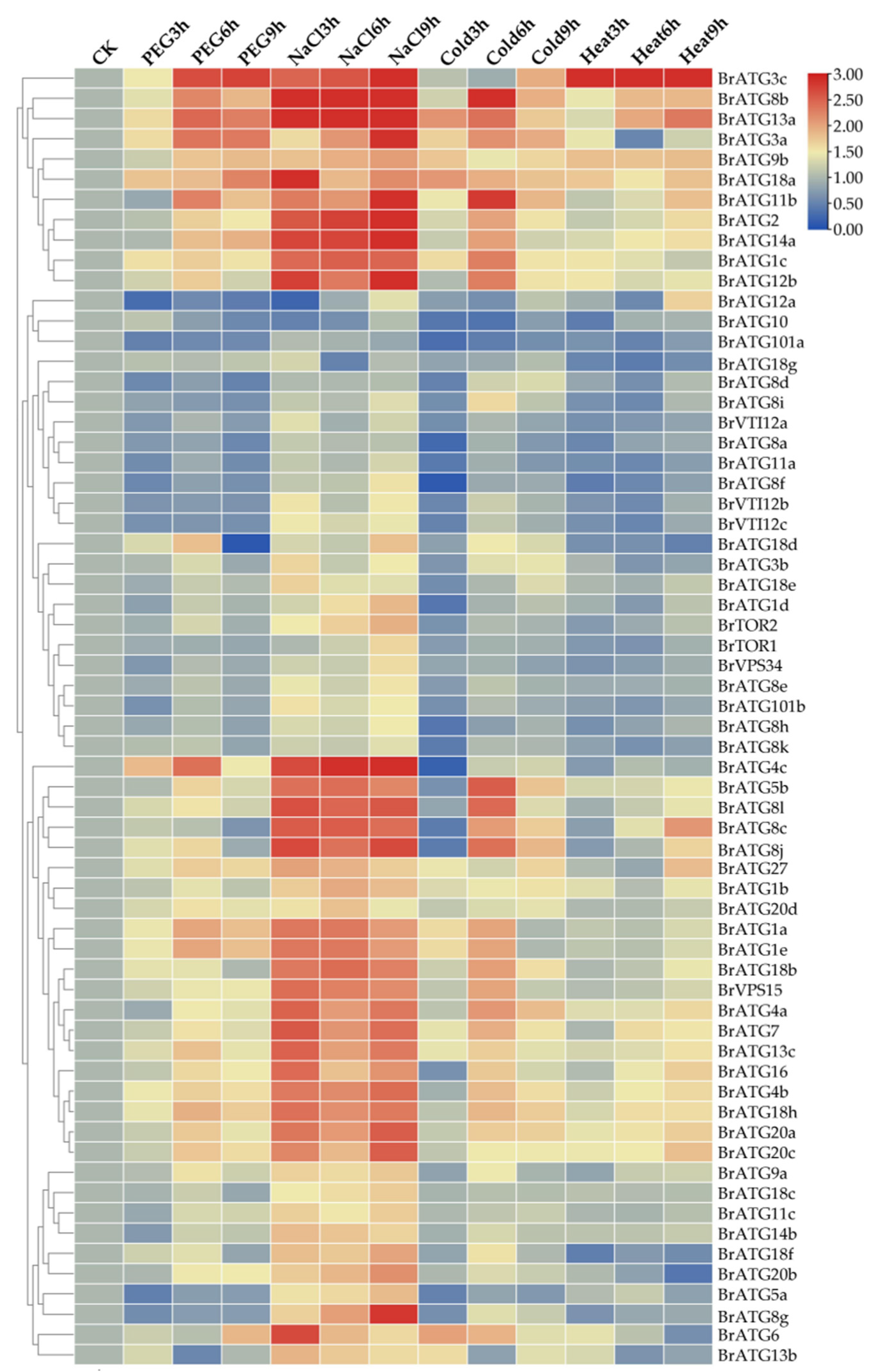
3.9. Expression Patterns of BrATGs under Other Abiotic Stresses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, X.; Zhao, P.; Wang, W.; Zou, J.; Cheng, T.; Peng, X.; Sun, M. A comprehensive, genome-wide analysis of autophagy-related genes identified in tobacco suggests a central role of autophagy in plant response to various environmental cues. DNA Res. 2015, 22, 245–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bozhkov, P.V. Plant autophagy: Mechanisms and functions. J. Exp. Bot. 2018, 69, 1281–1285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.H.; Kwon, C.; Lee, J.H.; Chung, T. Genes for plant autophagy: Functions and interactions. Mol. Cells 2012, 34, 413–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reggiori, F.; Klionsky, D.J. Autophagy in the eukaryotic cell. Eukaryot. Cell 2002, 1, 11–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sridhar, S.; Botbol, Y.; Macian, F.; Cuervo, A.M. Autophagy and disease: Always two sides to a problem. J. Pathol. 2012, 226, 255–273. [Google Scholar] [CrossRef] [Green Version]
- Nakatogawa, H.; Suzuki, K.; Kamada, Y.; Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat. Rev. Mol. Cell Biol. 2009, 10, 458–467. [Google Scholar] [CrossRef] [Green Version]
- Yoshimoto, K.; Takano, Y.; Sakai, Y. Autophagy in plants and phytopathogens. FEBS Lett. 2010, 584, 1350–1358. [Google Scholar] [CrossRef] [Green Version]
- Xia, K.; Liu, T.; Ouyang, J.; Wang, R.; Fan, T.; Zhang, M. Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.). DNA Res. 2011, 18, 363–377. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Fu, X.Z.; Zhou, X.; Xu, Y.Y.; Hui, Q.L.; Chun, C.P.; Ling, L.L.; Peng, L.Z. Comprehensive Analysis of Autophagy-Related Genes in Sweet Orange (Citrus sinensis) Highlights Their Roles in Response to Abiotic Stresses. Int. J. Mol. Sci. 2020, 21, 2699. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Chung, T.; Pennington, J.G.; Federico, M.L.; Kaeppler, H.F.; Kaeppler, S.M.; Otegui, M.S.; Vierstra, R.D. Autophagic Recycling Plays a Central Role in Maize Nitrogen Remobilization. Plant Cell 2015, 27, 1389–1408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, W.; Nie, X.; Cui, L.; Zhi, Y.; Zhang, T.; Du, X.; Song, W. Genome-wide sequence and expressional analysis of autophagy Gene family in bread wheat (Triticum aestivum L.). J. Plant Physiol. 2018, 229, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xiang, Y.; Niu, Y. An Overview of the Molecular Mechanisms and Functions of Autophagic Pathways in Plants. Plant Signal. Behav. 2021, 16, 1977527. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Kubota, Y.; Sekito, T.; Ohsumi, Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 2007, 12, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Suttangkakul, A.; Li, F.; Chung, T.; Vierstra, R.D. The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 2011, 23, 3761–3779. [Google Scholar] [CrossRef] [Green Version]
- Funderburk, S.F.; Wang, Q.J.; Yue, Z. The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond. Trends Cell Biol. 2010, 20, 355–362. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Liu, X.; Zhang, X.; Qin, N.; Xu, K.; Yin, W.; Zheng, Y.; Song, Y.; Zeng, R.; Liu, J. Phosphoinositide 3-Kinase Promotes Oxidative Burst, Stomatal Closure and Plant Immunity in Bacterial Invasion. Front. Plant Sci. 2019, 10, 1740. [Google Scholar] [CrossRef]
- Xu, P.; Damschroder, D.; Zhang, M.; Ryall, K.A.; Adler, P.N.; Saucerman, J.J.; Wessells, R.J.; Yan, Z. Atg2, Atg9 and Atg18 in mitochondrial integrity, cardiac function and healthspan in Drosophila. J. Mol. Cell. Cardiol. 2019, 127, 116–124. [Google Scholar] [CrossRef]
- Suzuki, K.; Ohsumi, Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007, 581, 2156–2161. [Google Scholar] [CrossRef] [Green Version]
- Noda, N.N.; Fujioka, Y.; Hanada, T.; Ohsumi, Y.; Inagaki, F. Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation. EMBO Rep. 2013, 14, 206–211. [Google Scholar] [CrossRef]
- Nakatogawa, H. Two ubiquitin-like conjugation systems that mediate membrane formation during autophagy. Essays Biochem. 2013, 55, 39–50. [Google Scholar] [PubMed]
- Wang, P.; Mugume, Y.; Bassham, D.C. New advances in autophagy in plants: Regulation, selectivity and function. Semin. Cell Dev. Biol. 2018, 80, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Li, X.; Yang, M.; Shao, Q.; Zhao, Y.; Ma, C.; Wang, P. Autophagy: An Intracellular Degradation Pathway Regulating Plant Survival and Stress Response. Front. Plant Sci. 2020, 11, 164. [Google Scholar] [CrossRef] [Green Version]
- Ustun, S.; Hofius, D. Anti- and pro-microbial roles of autophagy in plant-bacteria interactions. Autophagy 2018, 14, 1465–1466. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Zhang, P.; Zhu, R.; Fu, J.; Su, J.; Zheng, J.; Wang, Z.; Wang, D.; Gong, Q. Autophagy Is Rapidly Induced by Salt Stress and Is Required for Salt Tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 1459. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Xiong, Y.; Bassham, D.C. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 2009, 5, 954–963. [Google Scholar] [CrossRef] [Green Version]
- Chiarelli, R.; Roccheri, M.C. Heavy metals and metalloids as autophagy inducing agents: Focus on cadmium and arsenic. Cells 2012, 1, 597–616. [Google Scholar] [CrossRef] [Green Version]
- Taheri, S.H.; Bazgir, E. Thermopriming-Induced Autophagy in Shoot Apical Meristem of Arabidopsis. Iran. J. Biotechnol. 2021, 19, e2901. [Google Scholar]
- Masclaux-Daubresse, C.; Chen, Q.; Have, M. Regulation of nutrient recycling via autophagy. Curr. Opin. Plant Biol. 2017, 39, 8–17. [Google Scholar] [CrossRef]
- Minina, E.A.; Moschou, P.N.; Vetukuri, R.R.; Sanchez-Vera, V.; Cardoso, C.; Liu, Q.; Elander, P.H.; Dalman, K.; Beganovic, M.; Lindberg, Y.J.; et al. Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness. J. Exp. Bot. 2018, 69, 1415–1432. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, W.; Yue, J.; Liu, Y.; Pei, D.; Wang, H. The Responses of Wheat Autophagy and ATG8 Family Genes to Biotic and Abiotic Stresses. J. Plant Growth Regul. 2020, 39, 867–876. [Google Scholar] [CrossRef]
- Li, F.; Vierstra, R.D. Arabidopsis ATG11, a scaffold that links the ATG1-ATG13 kinase complex to general autophagy and selective mitophagy. Autophagy 2014, 10, 1466–1467. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Contento, A.L.; Bassham, D.C. AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 2005, 42, 535–546. [Google Scholar] [CrossRef]
- Wang, Y.; Nishimura, M.T.; Zhao, T.; Tang, D. ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J. 2011, 68, 74–87. [Google Scholar] [CrossRef]
- Song, X.M.; Liu, T.K.; Duan, W.K.; Ma, Q.H.; Ren, J.; Wang, Z.; Li, Y.; Hou, X.L. Genome-wide analysis of the GRAS gene family in Chinese cabbage (Brassica rapa ssp. pekinensis). Genomics 2014, 103, 135–146. [Google Scholar] [CrossRef] [Green Version]
- Duan, W.; Song, X.; Liu, T.; Huang, Z.; Ren, J.; Hou, X.; Li, Y. Genome-wide analysis of the MADS-box gene family in Brassica rapa (Chinese cabbage). Mol. Genet. Genom. 2015, 290, 239–255. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.H.; Bancroft, I.; Cheng, F.; et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Cai, X.; Wu, J.; Liu, M.; Grob, S.; Cheng, F.; Liang, J.; Cai, C.; Liu, Z.; Liu, B.; et al. Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic. Res. 2018, 5, 50. [Google Scholar] [CrossRef] [Green Version]
- Han, B.; Xu, H.; Feng, Y.; Xu, W.; Cui, Q.; Liu, A. Genomic Characterization and Expressional Profiles of Autophagy-Related Genes (ATGs) in Oilseed Crop Castor Bean (Ricinus communis L.). Int. J. Mol. Sci. 2020, 21, 562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klionsky, D.J.; Schulman, B.A. Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat. Struct. Mol. Biol. 2014, 21, 336–345. [Google Scholar] [CrossRef]
- Popelka, H.; Damasio, A.; Hinshaw, J.E.; Klionsky, D.J.; Ragusa, M.J. Structure and function of yeast Atg20, a sorting nexin that facilitates autophagy induction. Proc. Natl. Acad. Sci. USA 2017, 114, E10112–E10121. [Google Scholar] [CrossRef]
- Chang, Y.Y.; Neufeld, T.P. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol. Biol. Cell 2009, 20, 2004–2014. [Google Scholar] [CrossRef] [Green Version]
- Yuan, H.X.; Russell, R.C.; Guan, K.L. Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy 2013, 9, 1983–1995. [Google Scholar] [CrossRef] [Green Version]
- Meijer, W.H.; van der Klei, I.J.; Veenhuis, M.; Kiel, J.A.K.W. ATG Genes Involved in Non-Selective Autophagy are Conserved from Yeast to Man, but the Selective Cvt and Pexophagy Pathways also Require Organism-Specific Genes. Autophagy 2007, 3, 106–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, X.; Pu, X.; Qin, G.; Zhu, T.; Lin, H. The roles of autophagy in development and stress responses in Arabidopsis thaliana. Apoptosis 2014, 19, 905–921. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, S.I.; Park, O.K. Autophagy in plants. J. Plant Biol. 2008, 51, 313–320. [Google Scholar] [CrossRef]
- Wang, H.; Ding, Z.; Gou, M.; Hu, J.; Wang, Y.; Wang, L.; Wang, Y.; Di, T.; Zhang, X.; Hao, X.; et al. Genome-wide identification, characterization, and expression analysis of tea plant autophagy-related genes (CsARGs) demonstrates that they play diverse roles during development and under abiotic stress. BMC Genom. 2021, 22, 121. [Google Scholar] [CrossRef] [PubMed]
- Yen, W.L.; Klionsky, D.J. Atg27 is a second transmembrane cycling protein. Autophagy 2007, 3, 254–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yen, W.L.; Legakis, J.E.; Nair, U.; Klionsky, D.J. Atg27 is required for autophagy-dependent cycling of Atg9. Mol. Biol. Cell 2007, 18, 581–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segarra, V.A.; Boettner, D.R.; Lemmon, S.K. Atg27 tyrosine sorting motif is important for its trafficking and Atg9 localization. Traffic 2015, 16, 365–378. [Google Scholar] [CrossRef]
- Wang, Q.; Hou, S. The emerging roles of ATG1/ATG13 kinase complex in plants. J. Plant Physiol. 2022, 271, 153653. [Google Scholar] [CrossRef]
- Fletcher, K.; Ulferts, R.; Jacquin, E.; Veith, T.; Gammoh, N.; Arasteh, J.M.; Mayer, U.; Carding, S.R.; Wileman, T.; Beale, R.; et al. The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. Embo J. 2018, 37, e97840. [Google Scholar] [CrossRef] [PubMed]
- Quezada-Rodriguez, E.H.; Gomez-Velasco, H.; Arthikala, M.K.; Lara, M.; Hernandez-Lopez, A.; Nanjareddy, K. Exploration of Autophagy Families in Legumes and Dissection of the ATG18 Family with a Special Focus on Phaseolus vulgaris. Plants 2021, 10, 2619. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Sun, X.; Yue, Z.; Liang, D.; Wang, N.; Ma, F. Isolation and characterization of MdATG18a, a WD40-repeat AuTophaGy-related gene responsive to leaf senescence and abiotic stress in Malus. Sci. Hortic. 2014, 165, 51–61. [Google Scholar] [CrossRef]
- Jung, C.H.; Ro, S.H.; Cao, J.; Otto, N.M.; Kim, D.H. mTOR regulation of autophagy. FEBS Lett. 2010, 584, 1287–1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Son, O.; Kim, S.; Kim, D.; Hur, Y.S.; Kim, J.; Cheon, C.I. Involvement of TOR signaling motif in the regulation of plant autophagy. Biochem. Biophys. Res. Commun. 2018, 501, 643–647. [Google Scholar] [CrossRef] [PubMed]
- Kurotani, K.; Tabata, R.; Kawakatsu, Y.; Sugita, R.; Okayasu, K.; Tanoi, K.; Notaguchi, M. Autophagy Is Induced during Plant Grafting for Wound Healing; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2020. [Google Scholar]
- Wang, P.; Nolan, T.M.; Yin, Y.; Bassham, D.C. Identification of transcription factors that regulate ATG8 expression and autophagy in Arabidopsis. Autophagy 2020, 16, 123–139. [Google Scholar] [CrossRef]
- Dong, Z.; Wang, L.; Xu, J.; Li, Y.; Zhang, Y.; Zhang, S.; Miao, J. Promotion of autophagy and inhibition of apoptosis by low concentrations of cadmium in vascular endothelial cells. Toxicol. Vitr. 2009, 23, 105–110. [Google Scholar] [CrossRef]
- Huo, L.; Guo, Z.; Wang, Q.; Jia, X.; Sun, X.; Ma, F. The protective role of MdATG10-mediated autophagy in apple plant under cadmium stress. Ecotoxicol. Environ. Saf. 2022, 234, 113398. [Google Scholar] [CrossRef] [PubMed]
- Li, W.W.; Chen, M.; Zhong, L.; Liu, J.M.; Xu, Z.S.; Li, L.C.; Zhou, Y.B.; Guo, C.H.; Ma, Y.Z. Overexpression of the autophagy-related gene SiATG8a from foxtail millet (Setaria italica L.) confers tolerance to both nitrogen starvation and drought stress in Arabidopsis. Biochem. Biophys. Res. Commun. 2015, 468, 800–806. [Google Scholar] [CrossRef] [PubMed]
- Zhen, X.; Xu, F.; Zhang, W.; Li, N.; Li, X. Overexpression of rice gene OsATG8b confers tolerance to nitrogen starvation and increases yield and nitrogen use efficiency (NUE) in Arabidopsis. PLoS ONE 2019, 14, e223011. [Google Scholar] [CrossRef] [Green Version]
- Huo, L.; Guo, Z.; Jia, X.; Sun, X.; Wang, P.; Gong, X.; Ma, F. Increased autophagic activity in roots caused by overexpression of the autophagy-related gene MdATG10 in apple enhances salt tolerance. Plant Sci. 2020, 294, 110444. [Google Scholar] [CrossRef] [PubMed]
- Huo, L.; Sun, X.; Guo, Z.; Jia, X.; Che, R.; Sun, Y.; Zhu, Y.; Wang, P.; Gong, X.; Ma, F. MdATG18a overexpression improves basal thermotolerance in transgenic apple by decreasing damage to chloroplasts. Hortic. Res. 2020, 7, 21. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Gene ID | pI | MW(kDa) | AA | Instability Index | Aliphatic Index | Location |
|---|---|---|---|---|---|---|---|
| BrATG1a | BraA03g019750.3C | 6.31 | 80.08 | 722 | Unstable | 84.57 | Chloroplast |
| BrATG1b | BraA04g005840.3C | 8.32 | 79.24 | 713 | Unstable | 85.76 | Golgi apparatus |
| BrATG1c | BraA07g024950.3C | 6.31 | 67.61 | 609 | Unstable | 86.54 | Peroxisome |
| BrATG1d | BraA08g004000.3C | 7.13 | 28.69 | 253 | Unstable | 100.16 | Cytoplasm |
| BrATG1e | BraA09g051460.3C | 6.19 | 66.78 | 599 | Unstable | 86.94 | Peroxisome |
| BrATG2 | BraA05g028170.3C | 5.16 | 20.64 | 1878 | Unstable | 87.16 | Nucleus |
| BrATG3a | BraA02g002420.3C | 4.63 | 34.47 | 306 | Unstable | 77.06 | Cytoplasm |
| BrATG3b | BraA03g044500.3C | 4.63 | 35.09 | 309 | Unstable | 70.97 | Nucleus |
| BrATG3c | BraA09g020260.3C | 8.75 | 40.23 | 357 | Stable | 77.84 | Cytoplasm |
| BrATG4a | BraA04g002160.3C | 4.77 | 45.79 | 419 | Unstable | 82.17 | Nucleus |
| BrATG4b | BraA05g004100.3C | 5.1 | 51.72 | 466 | Unstable | 78.61 | Nucleus |
| BrATG4c | BraA09g049830.3C | 4.82 | 48.58 | 443 | Unstable | 50.29 | Nucleus |
| BrATG5a | BraA02g006950.3C | 5.21 | 39.15 | 342 | Unstable | 87.86 | Nucleus |
| BrATG5b | BraA10g022680.3C | 5 | 38.07 | 335 | Unstable | 92.51 | Cytoplasm |
| BrATG6 | BraA07g024790.3C | 5.92 | 58.86 | 521 | Stable | 71.52 | Nucleus |
| BrATG7 | BraA02g032310.3C | 5.03 | 76.59 | 697 | Unstable | 84.25 | Nucleus |
| BrATG8a | BraA01g012690.3C | 5.84 | 37.13 | 327 | Stable | 67.68 | Nucleus |
| BrATG8b | BraA02g027610.3C | 8.78 | 14.12 | 126 | Stable | 86.67 | Cytoplasm |
| BrATG8c | BraA03g023610.3C | 8.69 | 13.83 | 122 | Stable | 83.85 | Nucleus |
| BrATG8d | BraA03g032690.3C | 6.1 | 13.75 | 119 | Unstable | 76.13 | Nucleus |
| BrATG8e | BraA03g042050.3C | 7.9 | 13.72 | 119 | Unstable | 88.49 | Cytoplasm |
| BrATG8f | BraA03g046990.3C | 6.74 | 13.58 | 119 | Stable | 84.37 | Cytoplasm |
| BrATG8g | BraA03g049890.3C | 8.61 | 15.46 | 137 | Stable | 84.01 | Cytoplasm |
| BrATG8h | BraA05g031480.3C | 6.59 | 13.23 | 115 | Unstable | 86.52 | Cytoplasm |
| BrATG8i | BraA05g039000.3C | 6.9 | 13.72 | 119 | Unstable | 68.74 | Nucleus |
| BrATG8j | BraA07g023170.3C | 6.32 | 13.86 | 122 | Stable | 87.05 | Nucleus |
| BrATG8k | BraA08g011950.3C | 7.84 | 13.52 | 118 | Stable | 85.93 | Cytoplasm |
| BrATG8l | BraA09g025640.3C | 7.78 | 13.63 | 120 | Stable | 80.42 | Cytoplasm |
| BrATG9a | BraA04g022550.3C | 5.75 | 98.10 | 857 | Unstable | 85.43 | Plasma membrane |
| BrATG9b | BraA05g013480.3C | 7.02 | 81.16 | 700 | Unstable | 84.8 | Plasma membrane |
| BrATG10 | BraA05g038170.3C | 4.76 | 25.53 | 223 | Stable | 96.5 | Cytoplasm |
| BrATG11a | BraA01g006990.3C | 5.57 | 126.69 | 1128 | Unstable | 78 | Nucleus |
| BrATG11b | BraA03g056180.3C | 5.56 | 123.84 | 1100 | Unstable | 78.64 | Nucleus |
| BrATG11c | BraA08g017920.3C | 5.63 | 124.06 | 1100 | Unstable | 77.94 | Nucleus |
| BrATG12a | BraA05g033070.3C | 9.21 | 9.81 | 89 | Stable | 93.03 | Cytoplasm |
| BrATG12b | BraA06g000790.3C | 9.1 | 9.50 | 85 | Stable | 83.65 | Cytoplasm |
| BrATG13a | BraA01g034190.3C | 9.09 | 65.04 | 590 | Unstable | 69.76 | Chloroplast |
| BrATG13b | BraA05g028550.3C | 8.61 | 66.70 | 599 | Unstable | 62.99 | Nucleus |
| BrATG13c | BraA06g025150.3C | 9.58 | 62.22 | 563 | Unstable | 77.51 | Nucleus |
| BrATG14a | BraA07g041070.3C | 8.85 | 50.81 | 449 | Unstable | 78.95 | Mito |
| BrATG14b | BraA09g029210.3C | 9.34 | 53.22 | 474 | Unstable | 78.02 | Chloroplast |
| BrATG16 | BraA10g008580.3C | 6.19 | 55.38 | 502 | Unstable | 87.43 | Cytoplasm |
| BrATG18a | BraA03g002010.3C | 8.81 | 41.82 | 377 | Stable | 90.66 | Cytoplasm |
| BrATG18b | BraA03g013760.3C | 5.57 | 82.52 | 759 | Unstable | 73.95 | Nucleus |
| BrATG18c | BraA03g055830.3C | 8.7 | 39.68 | 366 | Unstable | 91.37 | Plasma membrane |
| BrATG18d | BraA04g000590.3C | 6.63 | 43.73 | 397 | Unstable | 81.99 | Endoplasmic reticulum |
| BrATG18e | BraA04g028920.3C | 6.13 | 45.04 | 408 | Unstable | 81.45 | Nucleus |
| BrATG18f | BraA07g025220.3C | 7.63 | 45.65 | 416 | Unstable | 77.04 | Cytoplasm |
| BrATG18g | BraA09g047160.3C | 6.39 | 48.63 | 442 | Unstable | 89.32 | Nucleus |
| BrATG18h | BraA09g052060.3C | 8.33 | 47.57 | 430 | Unstable | 78.4 | Endoplasmic reticulum |
| BrATG20a | BraA03g002860.3C | 5.44 | 57.44 | 514 | Unstable | 77.2 | Cytoplasm |
| BrATG20b | BraA10g016310.3C | 5.17 | 61.82 | 556 | Unstable | 77.16 | Cytoplasm |
| BrATG20c | BraA10g028870.3C | 6.72 | 46.31 | 402 | Unstable | 78.88 | Chloroplast |
| BrATG20d | BraA10g030100.3C | 5.06 | 59.94 | 537 | Unstable | 75.74 | Nucleus |
| BrATG27 | BraA05g005810.3C | 9.34 | 41.00 | 374 | Unstable | 81.07 | Plasma membrane |
| BrATG101a | BraA07g016990.3C | 7.67 | 33.11 | 281 | Unstable | 82.46 | Nucleus |
| BrATG10b | BraA09g009280.3C | 5.86 | 25.30 | 217 | Unstable | 96.04 | Nucleus |
| BrTOR1 | BraA05g019460.3C | 6.3 | 278.83 | 2480 | Unstable | 99.34 | Chloroplast |
| BrTOR2 | BraA06g003510.3C | 6.29 | 279.29 | 2486 | Unstable | 100 | Chloroplast |
| BrVPS15 | BraA01g008260.3C | 8.18 | 216.56 | 1926 | Unstable | 86.63 | Nucleus |
| BrVPS34 | BraA09g017180.3C | 6.34 | 93.23 | 813 | Unstable | 91.01 | Nucleus |
| BrVTI12a | BraA04g011910.3C | 9.72 | 24.74 | 221 | Stable | 94.52 | Golgi apparatus |
| BrVTI12b | BraA07g013170.3C | 8.37 | 25.14 | 222 | Unstable | 93.11 | Golgi apparatus |
| BrVTI12c | BraA09g003760.3C | 9.56 | 24.97 | 221 | Stable | 94.43 | Golgi apparatus |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Y.; Zhang, M.; Yin, F.; Cao, X.; Fan, S.; Wu, C.; Xiao, X. Genome-Wide Identification and Expression Analysis of BrATGs and Their Different Roles in Response to Abiotic Stresses in Chinese Cabbage. Agronomy 2022, 12, 2976. https://doi.org/10.3390/agronomy12122976
Hu Y, Zhang M, Yin F, Cao X, Fan S, Wu C, Xiao X. Genome-Wide Identification and Expression Analysis of BrATGs and Their Different Roles in Response to Abiotic Stresses in Chinese Cabbage. Agronomy. 2022; 12(12):2976. https://doi.org/10.3390/agronomy12122976
Chicago/Turabian StyleHu, Yuanfeng, Ming Zhang, Fengrui Yin, Xiaoqun Cao, Shuying Fan, Caijun Wu, and Xufeng Xiao. 2022. "Genome-Wide Identification and Expression Analysis of BrATGs and Their Different Roles in Response to Abiotic Stresses in Chinese Cabbage" Agronomy 12, no. 12: 2976. https://doi.org/10.3390/agronomy12122976




