Genetic Diversity and Population Differentiation of Dongxiang Wild Rice (Oryza rufipogon Griff.) Based on SNP Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Investigation of Root Characteristics
2.3. Genome Resequencing, SNP, and InDel Calling
- (1)
- The genomic DNA was fragmented using Ultra II FS enzyme mixed solution and repaired at the end, and an A tail was added to it.
- (2)
- The processed products were connected to specific sequencing joints.
- (3)
- AMPure XP beads in specific proportions were selected for target fragment selection in accordance with the expected library size, and purification was performed to remove joint contamination.
- (4)
- With polymerase chain reaction (PCR) enrichment, purification was performed using AMPure XP beads to prepare the sequencing library.
- (1)
- Preliminary quantification was performed using Qubit 3.0.
- (2)
- The insert size of the library was detected using Agilent 2100, and the next step of the experiment began only when the insert size met the expectation, and joint contamination was not found.
- (3)
- The effective concentration of the library was accurately quantified using a German ANALYTIKJENA QTOWER real-time fluorescent quantitative PCR analyzer, and the library was qualified when the effective concentration was >2 nM.
- (1)
- The Cutadapt software (version 1.13) [8] was used to remove adapter sequences from the reads.
- (2)
- The Trimmomatic software (version 0.36) [9] was used to remove low-quality nucleobases in reads (the average quantity was calculated with a 4 bp sliding window, and all the following nucleobases were removed if they were lower than 15).
- (3)
- The length of the reads should be greater than 50 bp.
- (1)
- The sequencing depth of samples should not be less than 3.
- (2)
- The missing genotype proportions of all samples should not exceed 50%.
- (3)
- A relatively low allele frequency should not be less than 5%.
- (4)
- The hybridization percentage should not exceed 55%.
2.4. Analysis of Population Structure
3. Results
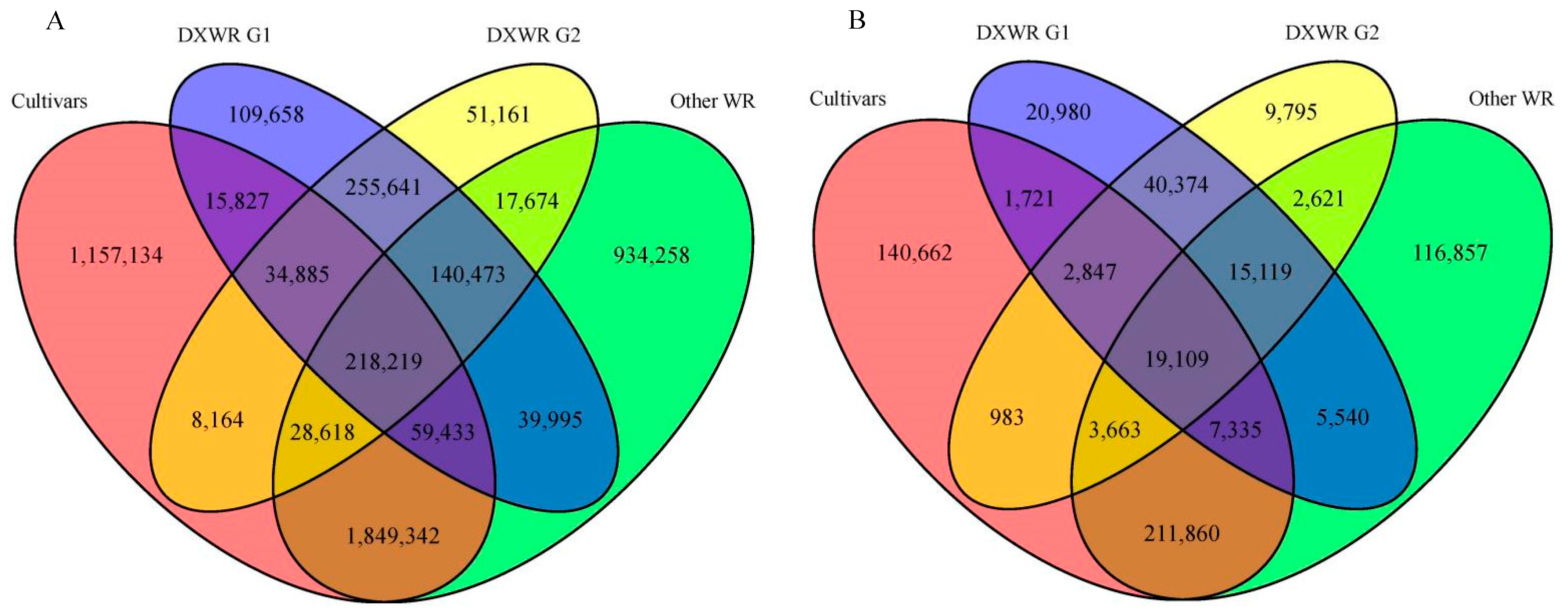
3.1. Genetic Diversity of DXWR
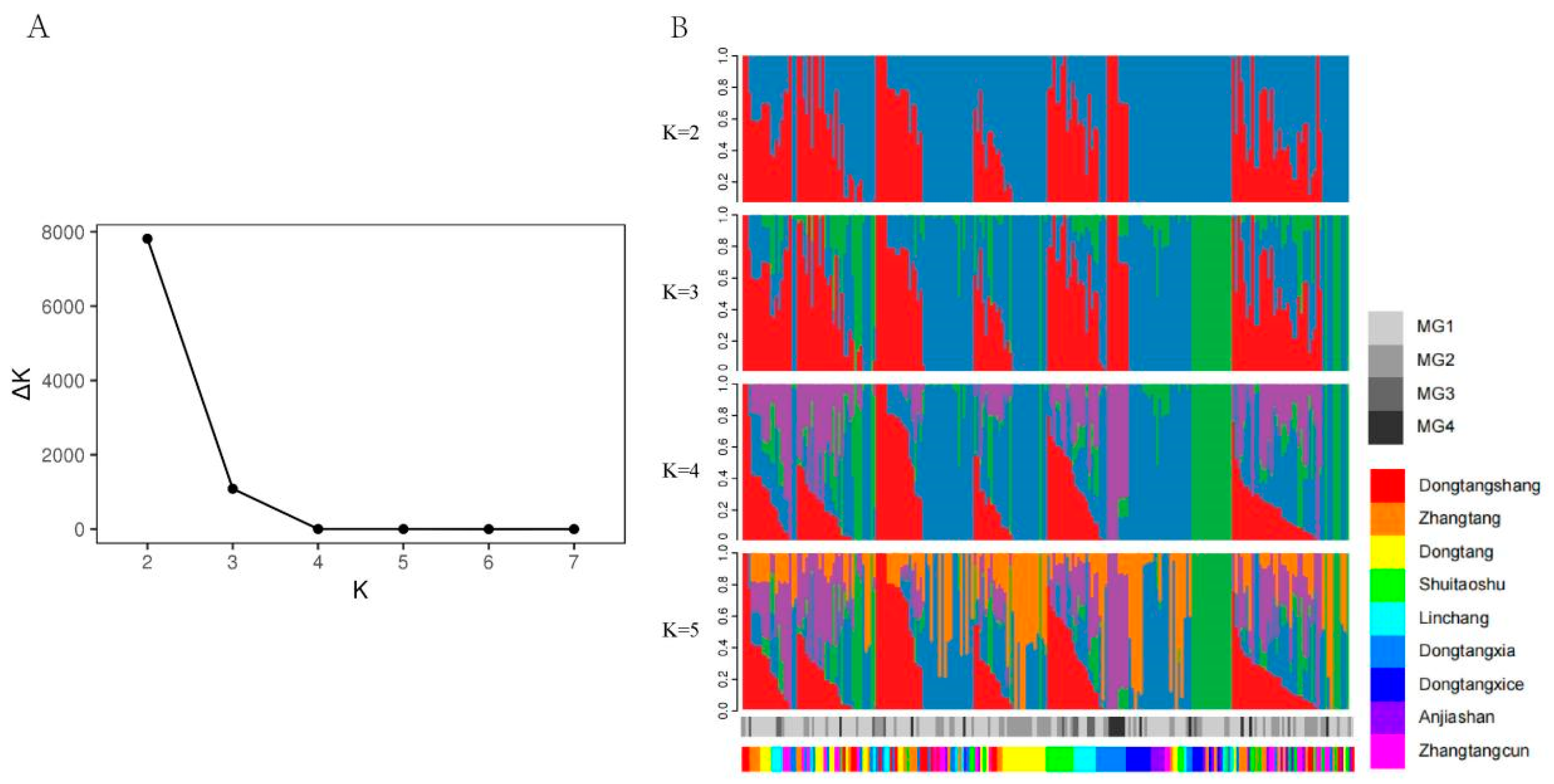
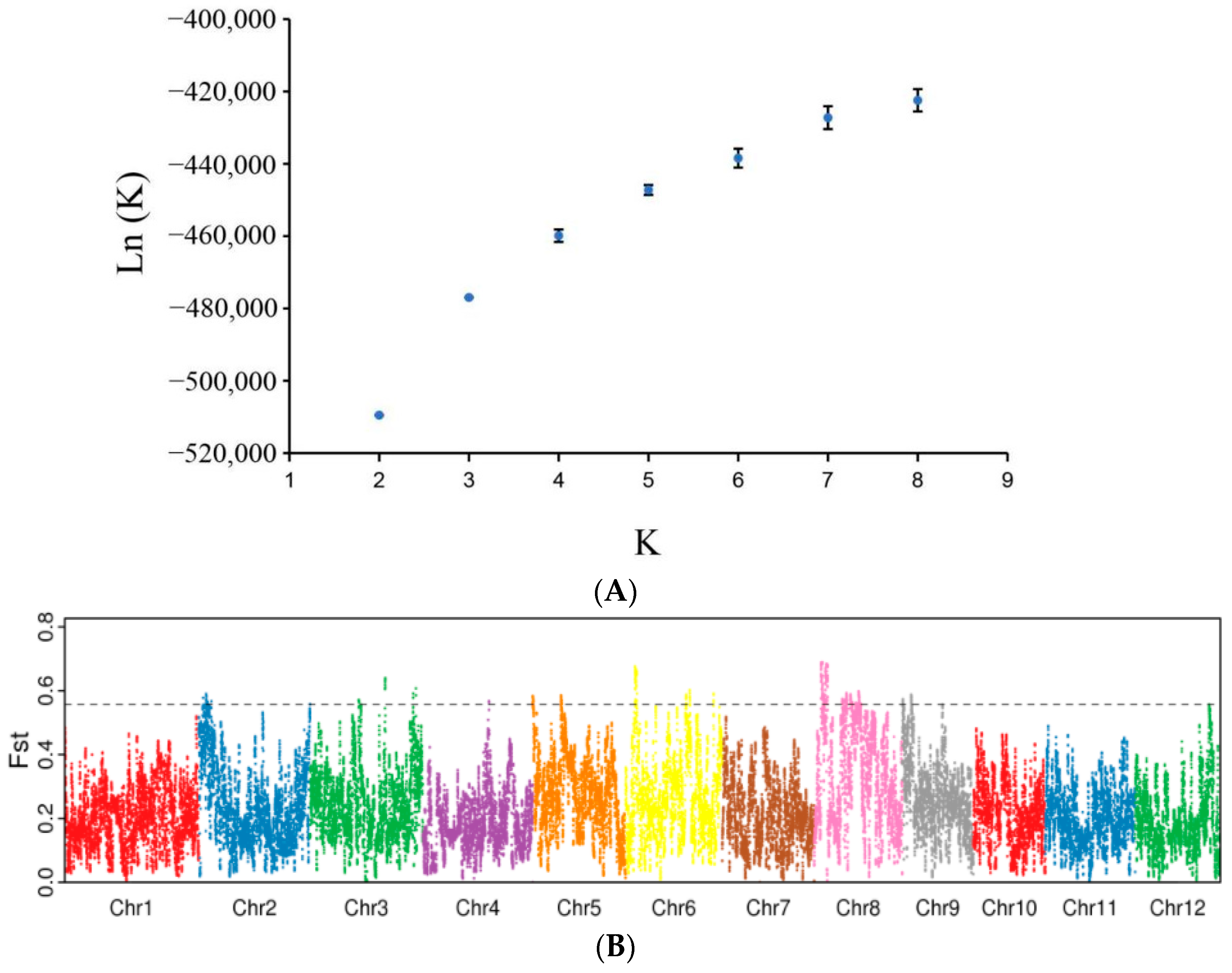
3.2. Population Structure of DXWR
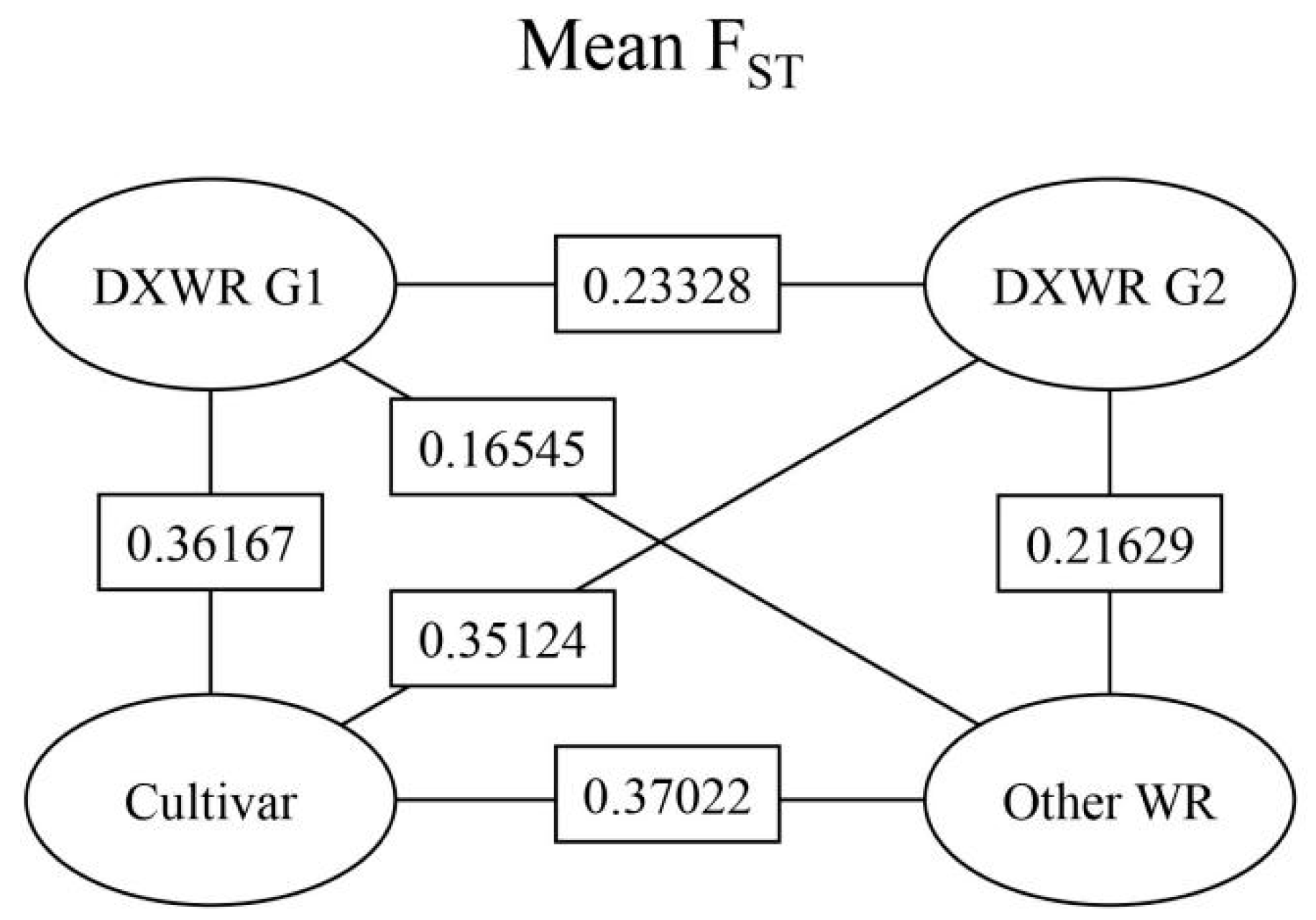
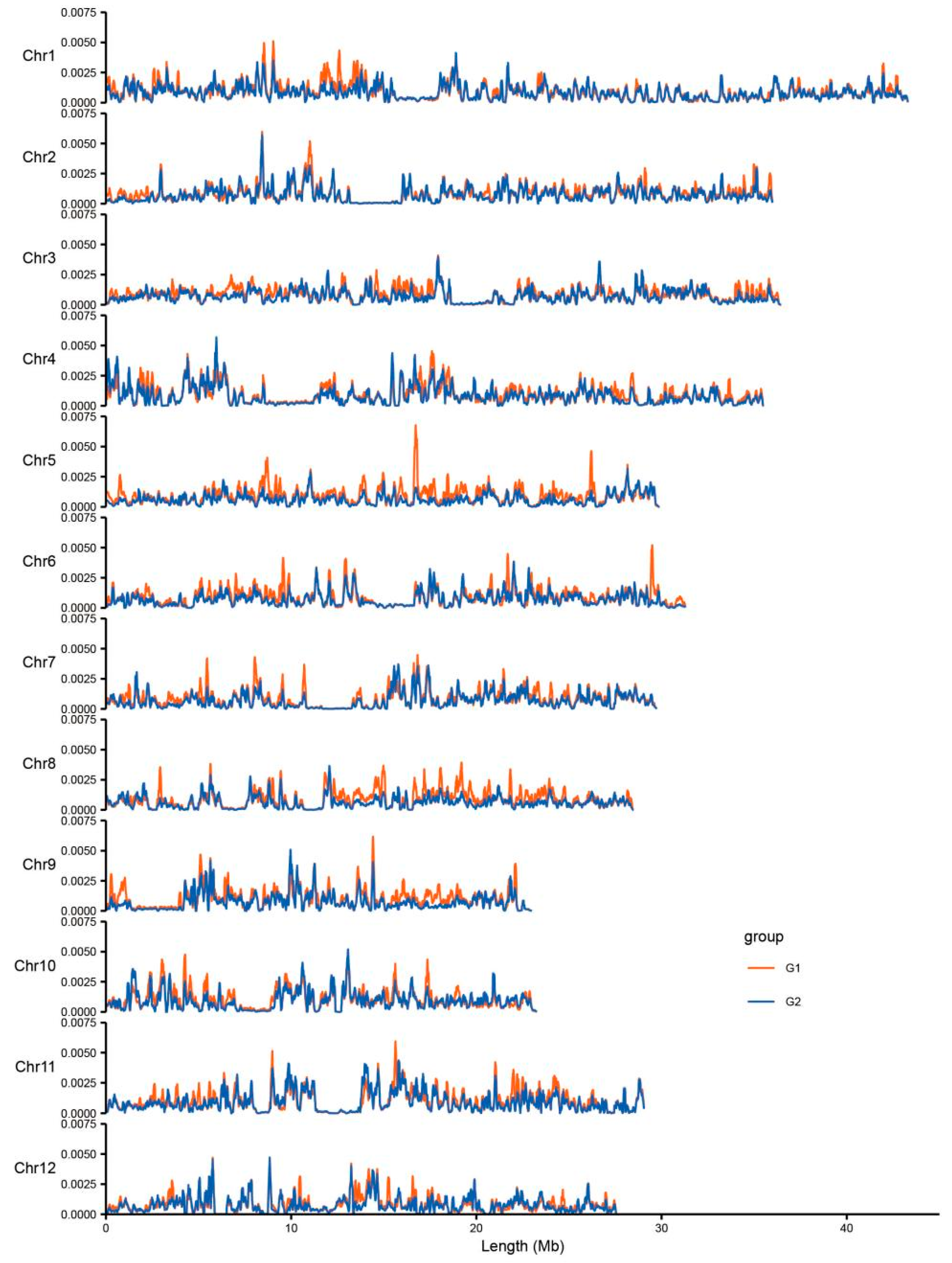
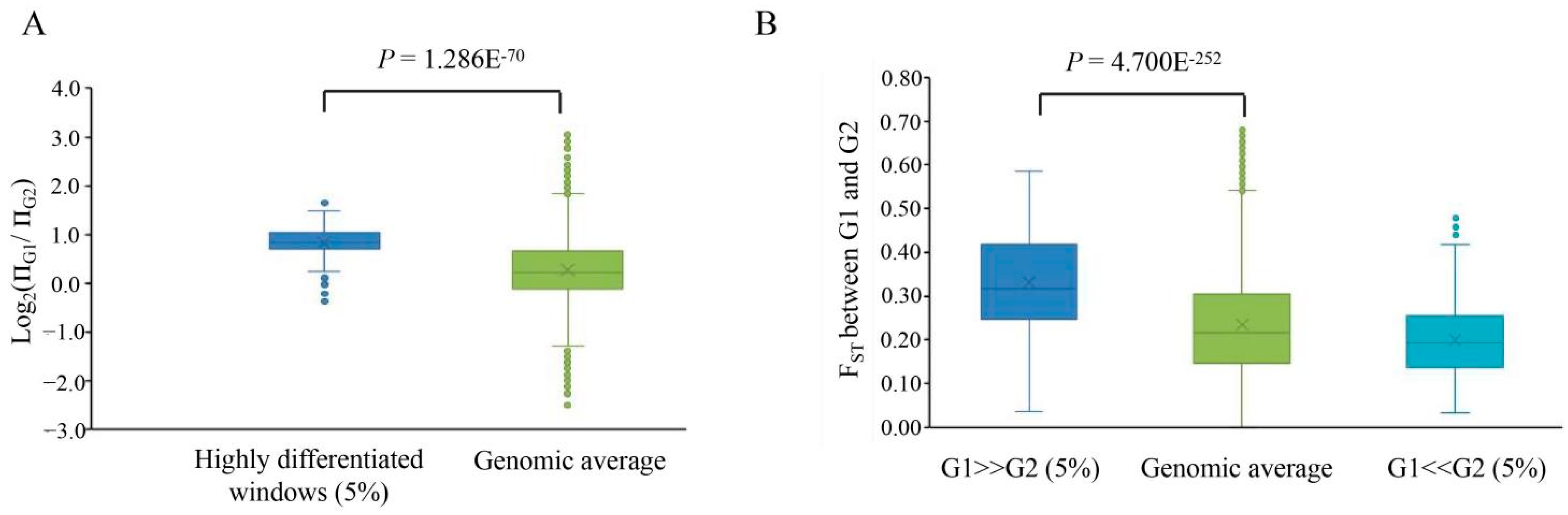
3.3. Genetic Differentiation between Two Subpopulations of DXWR
3.4. Differences in Deep Root Trait between the Two Subpopulations of DXWR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Nass, L.L.; Sigrist, M.S.; Ribeiro, C.; Reifschneider, F. Genetic resources: The basis for sustainable and competitive plant breeding. Crop Breed. Appl. Biot. 2012, 12, 75–86. [Google Scholar] [CrossRef]
- E, Z.G.; Wang, L. Discovery and utilization of favorable genes in wild rice. Hereditas 2008, 30, 1397–1405. [Google Scholar] [CrossRef]
- Xie, J.; Agrama, H.A.; Kong, D.; Zhuang, J.; Hu, B.; Wan, Y.; Yan, W. Genetic diversity associated with conservation of endangered Dongxiang wild rice (Oryza rufipogon). Genet. Resour. Crop Evol. 2010, 57, 597–609. [Google Scholar] [CrossRef]
- Luo, L. Breeding for water-saving and drought-resistance rice (WDR) in China. J. Exp. Bot. 2010, 61, 3509–3517. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.Q.; Wang, X.K.; Yoshimura, A.; Iwata, N. A study of the genetic diversity of common wild rice (O. rufipogon Griff.) and cultivated rice (O. sativa L.) by RFLP analysis. Acta Genet. Sin. 2000, 27, 227–234. [Google Scholar]
- Uga, Y.; Okuno, K.; Yano, M. Dro1, a major QTL involved in deep rooting of rice under upland field conditions. J. Exp. Bot. 2011, 62, 2485–2494. [Google Scholar] [CrossRef] [PubMed]
- Scott, O.R.; Bendich, A.J. Extraction of DNA from plant tissue. Plant Mol. Biol. Manual. 1988, A6, 73–83. [Google Scholar]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997v2. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Mckenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample groupinformation. Mol. Ecol. Res. 2009, 9, 1322–1332. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.C.; Cam, T.L.; Shashaank, V.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2015, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Smith, D.; Zhu, H.; Guan, Y.; Lam, T.T.Y. ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.; Lunter, G.; Marth, G.; Sherry, S.T.; et al. The variant call format and vcftools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- You, J.; Zong, W.; Hu, H.H.; Li, X.H.; Xiao, J.H.; Xiong, L.Z. A STRESS-RESPONSIVE NAC1-Regulated Protein Phosphatase Gene Rice Protein Phosphatase18 Modulates Drought and Oxidative Stress Tolerance through Abscisic Acid-Independent Reactive Oxygen Species Scavenging in Rice. Plant. Physiol. 2014, 166, 2100–2114. [Google Scholar] [CrossRef]
- Kim, S.-K.; You, Y.N.; Park, J.C.; Joung, Y.; Kim, B.-G.; Ahn, J.C.; Cho, H.S. The rice thylakoid lumenal cyclophilin OsCYP20-2 confers enhanced environmental stress tolerance in tobacco and Arabidopsis. Plant Cell Rep. 2012, 31, 417–426. [Google Scholar] [CrossRef]
- Bang, S.W.; Lee, D.K.; Jung, H.; Chung, P.J.; Kim, Y.S.; Choi, Y.D.; Suh, J.W.; Kim, J.K. Overexpression of OsTF1L, a rice HD-Zip transcription factor, promotes lignin biosynthesis and stomatal closure that improves drought tolerance. Plant Biotechnol. J. 2019, 17, 118–131. [Google Scholar] [CrossRef]
- Nie, Y.; Xia, H.; Ma, X.; Lou, Q.; Liu, Y.; Zhang, A.; Chen, L.; Yan, L.; Luo, L. Dissecting genetic Basis of Deep Rooting in Dongxiang Wild Rice. Rice Sci. 2022, 29, 277–287. [Google Scholar]
- Zhang, F.; Luo, X.; Zhou, Y.; Xie, J. Genome-wide identification of conserved microRNA and their response to drought stress in Dongxiang wild rice (Oryza rufipogon Griff.). Biotechnol. Lett. 2016, 38, 711–721. [Google Scholar] [CrossRef]
- Li, Z.; Liu, G.; Yu, W. Genetic heterogeneity of wild rice in Dongxiang wild rice. J. Southwest Agric. Univ. 1989, 3, 285–287. [Google Scholar]
- Qiu, B.; Xiong, Y.; Yu, L.; Hu, B.; Zhang, Z.; Xie, J. Cluster analysis of plant morphological characters in population of Dongxiang wild rice. Acta Agric. Jiangxi 2006, 18, 1–5. [Google Scholar]
- Yu, L.; Xu, Q.; Qiu, B.; Xiong, Y.; Rao, S. Comparative Studies on the Main Agronomic Characteristics between In-situ and Ex-situ Conserved Wild Rice Populations in Dongxiang. J. Plant Genet. Res. 2007, 8, 99–101. [Google Scholar]
- Huang, Y.; Li, G.; Chen, D.; Shi, Q.; Xiao, Y.; Wang, F.; Yu, L.; Xiao, Y. A preliminary study on the genetic diversity of Allozymic loci of Dongxiang wild rice (Oryza rufipogon Griff.). Acta Agric. Univ. Jiangxiensis 2002, 24, 1–5. [Google Scholar]
- Xie, J.; Chen, D.; Xiao, Y.; Luo, S.; Zheng, K.; Zhuang, J. Genetic diversity of Dongxiang wild rice detected by SSLP markers. Sci Agric Sin 2003, 36, 873–878. [Google Scholar]
- Kong, D.; Xie, J.; Zhuang, J.; Fan, Y.; Wan, Y. Genetic diversity of in situ conserved Dongxiang wild rice detected by SSR. J. Agric. Biotech. 2006, 14, 742–746. [Google Scholar]
- Yang, Q.; Yu, L.; Zhang, W.; Shi, J.; Ren, J.; Miao, H. The Genetic Differentiation of Dongxiang Wild Rice (Oryza rufipogon Griff.) and Its Implications for In-Situ Conservation. Sci. Agric. Sin. 2007, 40, 1085–1093. [Google Scholar]
- Xie, J.; Hu, B.; Wan, Y.; Zhang, T.; Li, X.; Liu, R.; Huang, Y.; Dai, L.; Luo, X. Comparison of the drought resistance characters at Seedling Stage between Dongxiang common wild rice (Oryza rufipogon Griff.) and cultivars (Oryza sativa L.). Acta Ecol. Sin. 2010, 30, 1665–1674. [Google Scholar]
- Gao, L.; Ge, S.; Hong, D.; Lin, R.; Tao, G.; Xu, Z. Allozyme variation and conservation genetics of common wild rice (Oryza rufipogon Griff.) in Yunnan, China. Euphytica 2002, 124, 273–281. [Google Scholar] [CrossRef]
- Song, Z.; Xu, X.; Wang, B.; Chen, J.; Lu, B. Gentic diversity in the northernmost Oryza rufipogon populations estimated by SSR markers. Theor. Appl. Genet. 2003, 107, 1492–1499. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Zhang, Q.; Wang, M. Ribosomal DNA polymorphisms of the common wild rice from China. Acta Genet. Sin. 1998, 25, 531–537. [Google Scholar]
- Sun, C.; Wang, X.; Li, Z.; Yoshimura, A. Comparison of the genetic diversity of common wild rice (Oryza rufipogon) and cultivated rice (O. Sativa) using RFLP markers. Theor. Appl. Genet. 2001, 102, 157–162. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, H.; Wang, H.; Lu, J.; Yuan, X.; Peng, S.; Wei, X. Comparative Assessment of SSR Allelic Diversity in Wild and Cultivated Rice in China. Acta Agron. Sin. 2008, 34, 591–597. [Google Scholar] [CrossRef]
- Duan, S.; Li, S.; Li, S.; Zhu, Y. Genetic Diversity of Wild Rice and Cultivated Rice. Acta Agron. Sin. 2009, 35, 467–474. [Google Scholar] [CrossRef]
- Zhu, Z.; Sun, C.; Fu, Y.; Zhang, P.; Wang, X. Comparison of the genetic diversity of common wild rice and cultivated rice using SSR markers. Sci. Agric. Sin. 2002, 35, 1437–1441. [Google Scholar]
- Zhang, J.; Xu, B.; Li, M. Genetic diversity of populations of an endangered medicinal plant species (Glycyrrhiza uralensis) in different environments of North China. J. Med. Plant Res. 2010, 4, 830–836. [Google Scholar]
- Shen, X.H.; Yan, S.; Huang, R.L.; Zhu, S.; Xiong, H.L.; Shen, L.G. Development of novel cytoplasmic male sterile source from Dongxiang Wild Rice (Oryza rufipogon). Rice Sci. 2013, 20, 379–382. [Google Scholar] [CrossRef]
- Cao, Z.B.; Tang, H.W.; Cai, Y.H.; Zeng, B.H.; Zhao, J.L.; Tang, X.Y.; Lu, M.; Wang, H.M.; Zhu, X.J.; Wu, X.F.; et al. Natural variation of HTH5 from wild rice, Oryza rufipogon Griff., is involved in conferring high-temperature tolerance at the heading stage. Plant Biotechnol. J. 2022, 20, 1591–1605. [Google Scholar] [CrossRef]
- Yang, K.S.; Chen, X.R.; Fu, J.R.; Peng, X.S.; Zhu, C.I.; He, X.P.; He, H.H. Survey of the Biologic Characteristic on the Nutritional Stress in the Dongxiang Wild Rice. J. Plant Genet. Res. 2006, 4, 427–433. [Google Scholar]
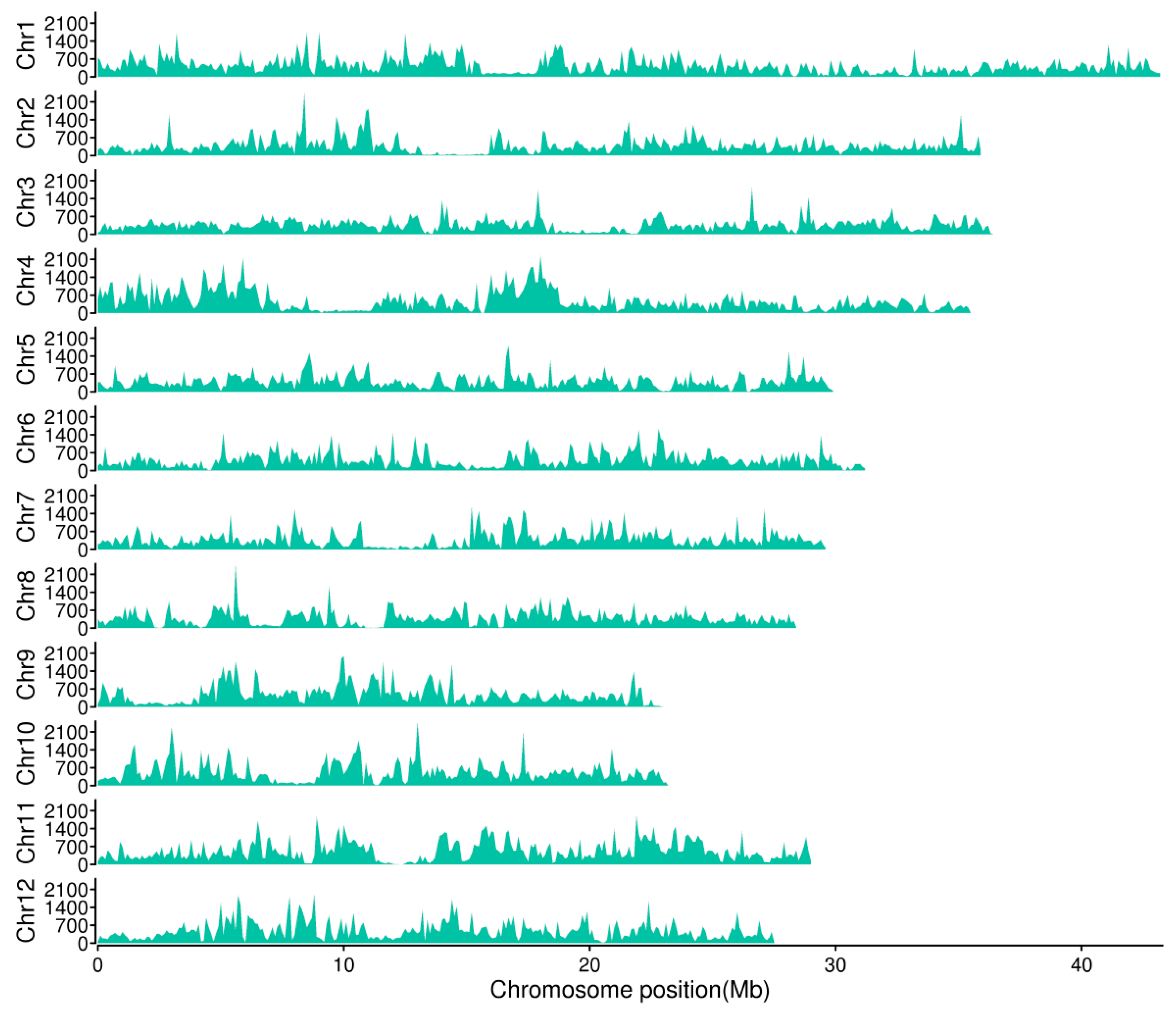
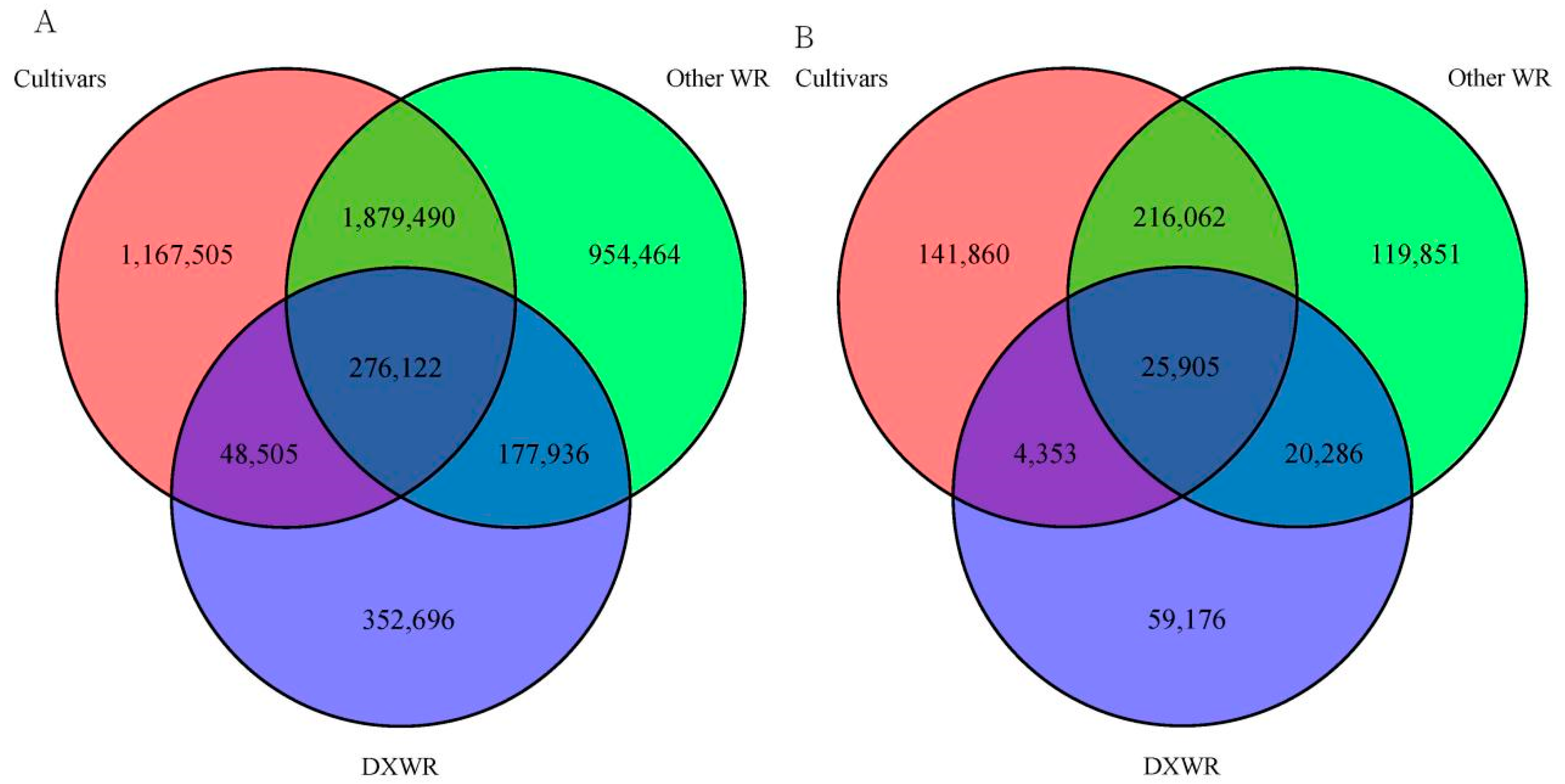
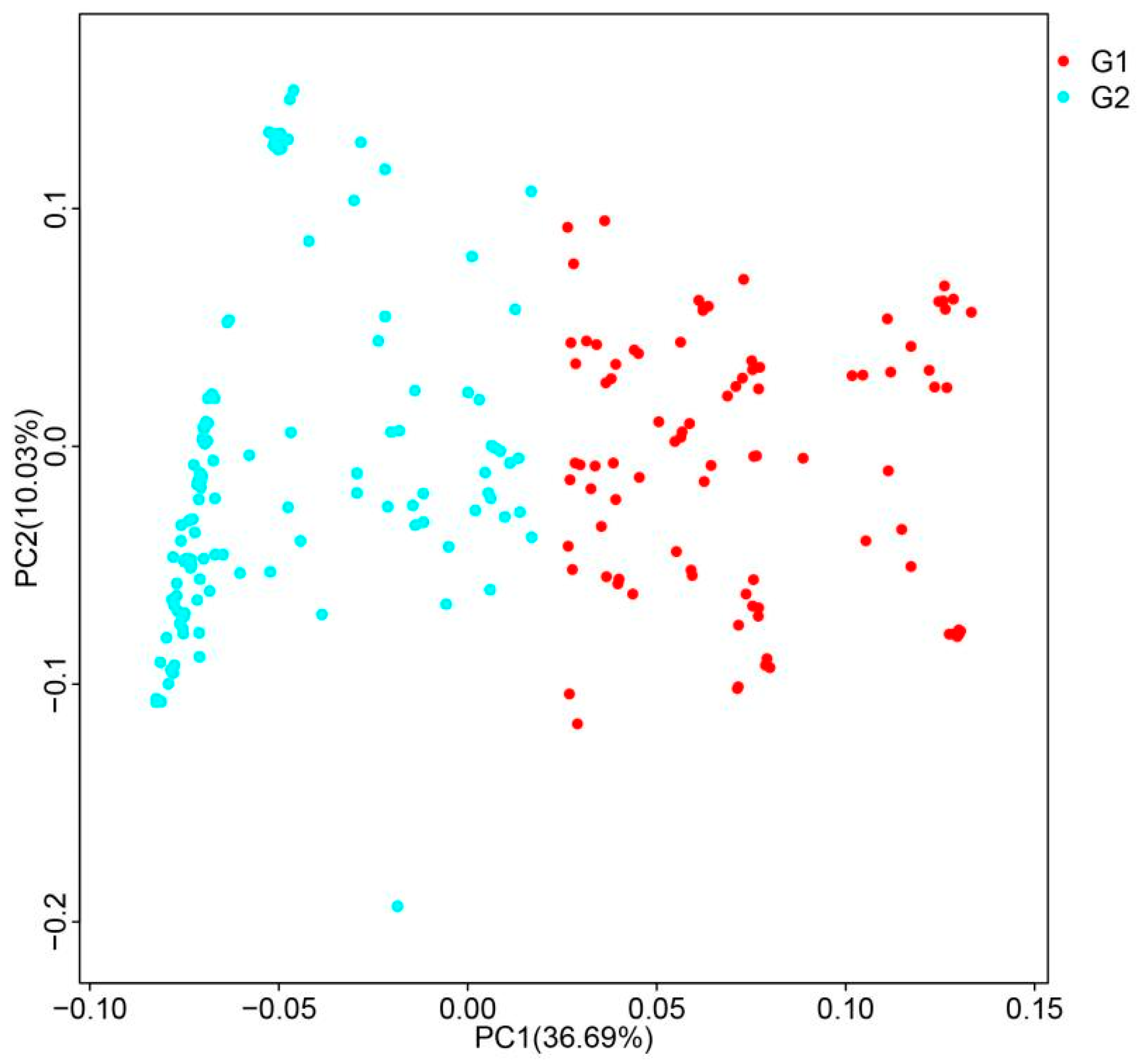
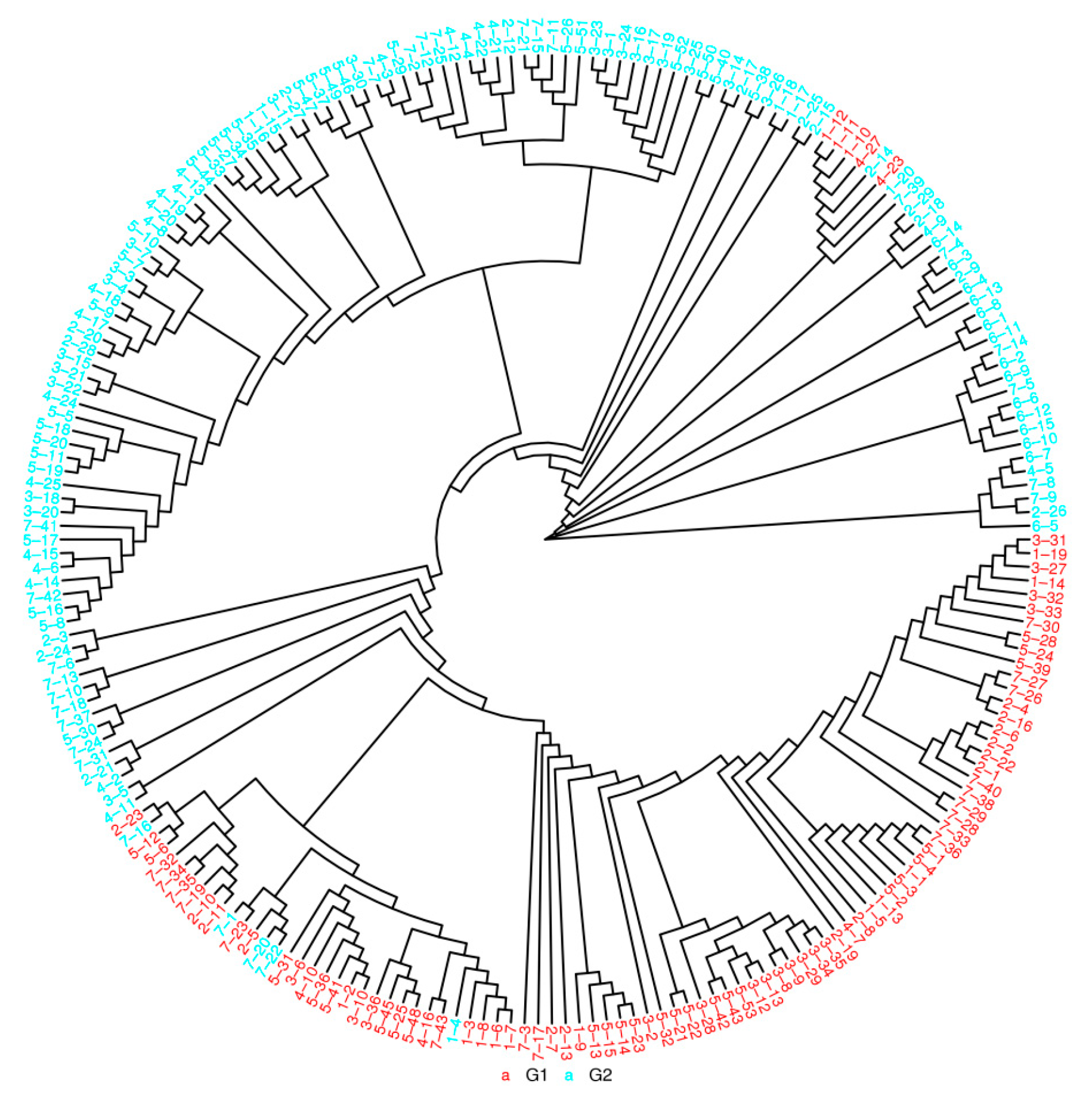
| Chr. ID | Length (bp) | SNPs | Indels | ||
|---|---|---|---|---|---|
| Number | Density (/Mb) | Number | Density (/Mb) Density (/Mb) | ||
| Chr1 | 43,270,923 | 163,739 | 3784.04 | 26,407 | 610.27 |
| Chr2 | 35,937,250 | 122,702 | 3414.34 | 19,631 | 546.26 |
| Chr3 | 36,413,819 | 116,424 | 3197.25 | 19,296 | 529.91 |
| Chr4 | 35,502,694 | 152,174 | 4286.27 | 22,104 | 622.6 |
| Chr5 | 29,958,434 | 109,666 | 3660.61 | 15,839 | 528.7 |
| Chr6 | 31,248,787 | 115,604 | 3699.47 | 18,518 | 592.6 |
| Chr7 | 29,697,621 | 104,163 | 3507.45 | 16,614 | 559.44 |
| Chr8 | 28,443,022 | 101,155 | 3556.41 | 14,915 | 524.38 |
| Chr9 | 23,012,720 | 102,698 | 4462.66 | 14,273 | 620.22 |
| Chr10 | 23,207,287 | 109,749 | 4729.07 | 16,041 | 691.21 |
| Chr11 | 29,021,106 | 136,145 | 4691.24 | 20,193 | 695.8 |
| Chr12 | 27,531,856 | 120,186 | 4365.34 | 17,842 | 648.05 |
| Total | 373,245,519 | 1,454,405 | 3896.64 | 221,673 | 593.91 |
| Category | SNP | InDel | ||
|---|---|---|---|---|
| Number | Ratio (%) | Number | Ratio (%) | |
| intergenic | 571,390 | 39.29 | 90,930 | 41.03 |
| upstream/downstream | 402,626 | 27.68 | 68,207 | 30.77 |
| upstream | 207,582 | 14.27 | 35,202 | 15.88 |
| downstream | 168,030 | 11.55 | 28,060 | 12.66 |
| upstream and downstream | 27,014 | 1.86 | 4945 | 2.23 |
| genic | 480,305 | 33.03 | 62,508 | 28.20 |
| intronic | 227,355 | 15.63 | 37,967 | 17.13 |
| exonic | 208,061 | 14.31 | 12,423 | 5.60 |
| synonymous/nonframeshift | 81,048 | 5.57 | 4410 | 1.99 |
| nonsynonymous/frameshift | 121,427 | 8.35 | 7748 | 3.50 |
| stopgain | 5067 | 0.35 | 246 | 0.11 |
| stoploss | 519 | 0.04 | 19 | 0.01 |
| 5′UTR | 15,372 | 1.06 | 5067 | 2.29 |
| 3′UTR | 28,205 | 1.94 | 6887 | 3.11 |
| splicing | 1312 | 0.09 | 164 | 0.07 |
| Subgroup | ZTC | STS | KXL | ZT | AJS | DTS | DT | DTX | DTXC | Number |
|---|---|---|---|---|---|---|---|---|---|---|
| G1 | 11 | 5 | 13 | 14 | 4 | 14 | 15 | 13 | 0 | 89 |
| G2 | 12 | 22 | 8 | 15 | 18 | 6 | 18 | 17 | 15 | 131 |
| Number | 23 | 27 | 21 | 29 | 22 | 20 | 33 | 30 | 15 | 220 |
| G1(%) | 48 | 19 | 62 | 48 | 18 | 70 | 45 | 43 | 0 | 40.5 |
| G2(%) | 52 | 81 | 38 | 52 | 82 | 30 | 55 | 57 | 100 | 59.5 |
| Trait | G1 (n = 15) | G2 (n = 15) | F | ||||||
|---|---|---|---|---|---|---|---|---|---|
| M ± SD | Min | Max | CV (%) | M ± SD | Min | Max | CV (%) | ||
| Plant height | 121.67 ± 22.19 | 96.33 | 137.67 | 18.00 | 118.08 ± 21.32 | 91.33 | 129.67 | 18.00 | 0.76 |
| Tiller | 32.00 ± 7.36 | 24.33 | 39.00 | 23.00 | 38.17 ± 12.67 | 28.67 | 54.00 | 33.00 | 0.34 |
| DR | 15.33 ± 3.28 | 11.68 | 18.00 | 21.00 | 28.92 ± 5.13 | 23.67 | 36.33 | 18.00 | 0.01 ** |
| SR | 64.67 ± 8.50 | 56.00 | 73.00 | 13.00 | 73.75 ± 9.92 | 58.33 | 85.67 | 13.00 | 0.54 |
| TR | 80.00 ± 8.19 | 74.00 | 89.73 | 10.00 | 102.67 ± 12.36 | 87.67 | 112.33 | 12.00 | 0.08 |
| RDR(%) | 19.28 ± 4.63 | 15.22 | 24.32 | 24.00 | 28.36 ± 5.51 | 21.65 | 42.46 | 19.00 | 0.03 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nie, Y.; Hou, G.; Xia, H.; Wang, L.; Lei, J.; Chen, H.; Chen, L.; Luo, L. Genetic Diversity and Population Differentiation of Dongxiang Wild Rice (Oryza rufipogon Griff.) Based on SNP Markers. Agronomy 2022, 12, 3056. https://doi.org/10.3390/agronomy12123056
Nie Y, Hou G, Xia H, Wang L, Lei J, Chen H, Chen L, Luo L. Genetic Diversity and Population Differentiation of Dongxiang Wild Rice (Oryza rufipogon Griff.) Based on SNP Markers. Agronomy. 2022; 12(12):3056. https://doi.org/10.3390/agronomy12123056
Chicago/Turabian StyleNie, Yuanyuan, Guihua Hou, Hui Xia, Lei Wang, Jianguo Lei, Hong Chen, Liang Chen, and Lijun Luo. 2022. "Genetic Diversity and Population Differentiation of Dongxiang Wild Rice (Oryza rufipogon Griff.) Based on SNP Markers" Agronomy 12, no. 12: 3056. https://doi.org/10.3390/agronomy12123056
APA StyleNie, Y., Hou, G., Xia, H., Wang, L., Lei, J., Chen, H., Chen, L., & Luo, L. (2022). Genetic Diversity and Population Differentiation of Dongxiang Wild Rice (Oryza rufipogon Griff.) Based on SNP Markers. Agronomy, 12(12), 3056. https://doi.org/10.3390/agronomy12123056





