The Roles of MADS-Box Genes from Root Growth to Maturity in Arabidopsis and Rice
Abstract
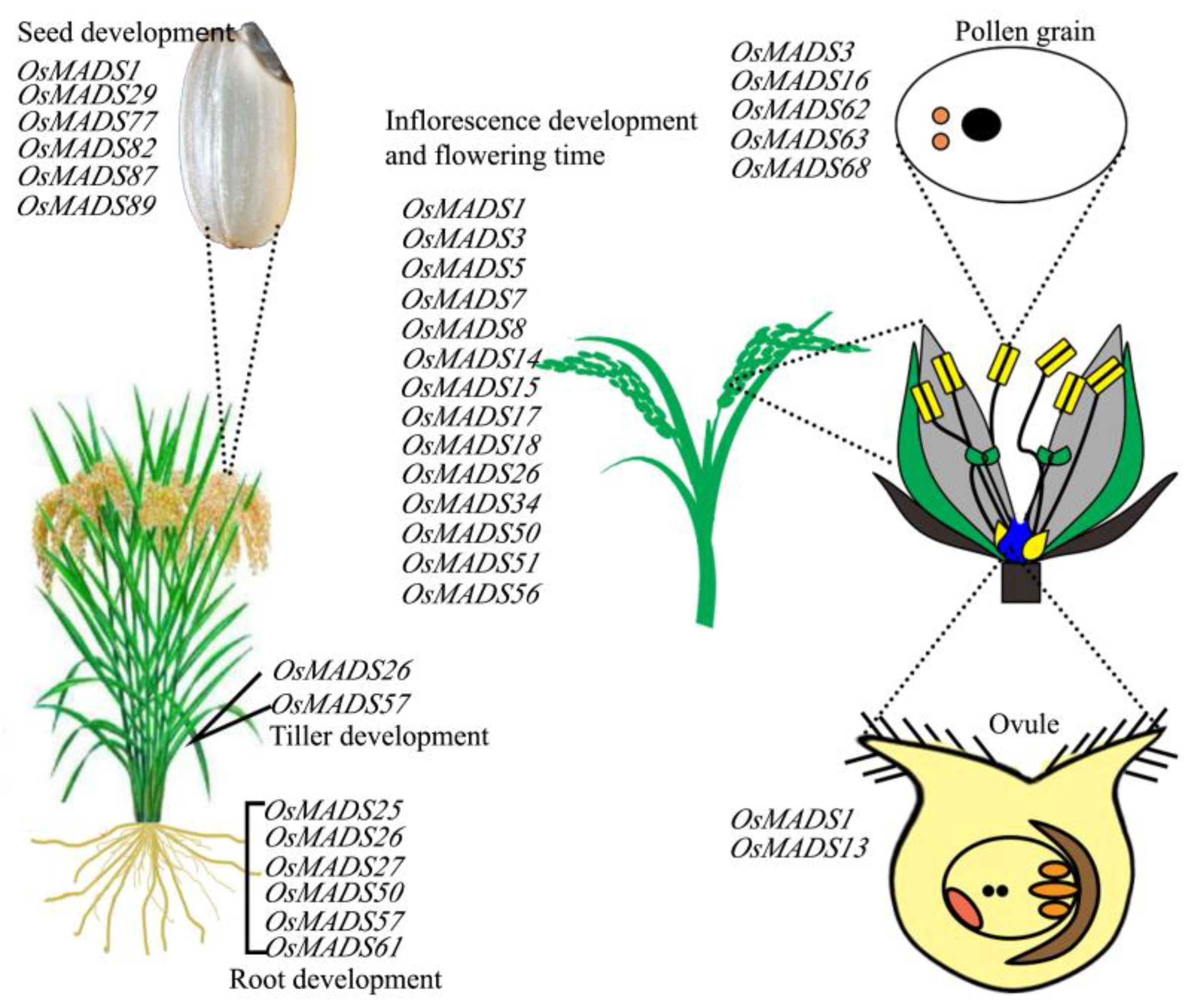
:1. Introduction
2. MADS-Box Gene Expression Profiles during Root Development
3. MADS-Box Gene Expression Profiles during Leaf Development in Rice
4. MADS-Box Genes Are Responsible for Inflorescence Branching
| Gene | Locus ID | Function | Reference |
|---|---|---|---|
| STK/AGL11 | At4g09960 | Ovule development | [19,30] |
| XAL1/AGL12 | At1g71692 | Root development; transition to flowering | [73] |
| AGL15 | AT5g13790 | Embryogenesis; sepal and petal longevity; flowering repressor with AGL18; fruit maturation | [74,75,76] |
| AGL18 | At3g57390 | Flowering repressor with AGL15 | [77] |
| AGL16 | At3g57230 | Number and distribution of stomata | [78] |
| AGL17 | At2g22630 | Root formation; Flowering activator | [31,32,79] |
| AGL6 | At2g45650 | Flowering activator; lateral organ development | [80] |
| FLC/AGL25 | At5g10140 | Juvenile-to-adult transition; flowering repressor; flowering initiation, flower organ development | [81,82] |
| MAF2/AGL31 | At5g65050 | Flowering repressor | [83] |
| AGL19 | At4g22950 | Flowering activator | [84] |
| FUL/AGL8 | At5g60910 | Meristem identity specification; annual life cycle regulator with SOC1; fruit development | [62,85] |
| AP1/AGL7 | At1g69120 | Homeotic A-class gene; meristem identity specification | [62] |
| AGL24 | At4g24540 | Flowering activator | [86] |
| AGL23 | At1g65360 | Embryo sac development | [87] |
| AGL28 | At1g01530 | Flowering activator | [88] |
| AGL42 | At5g62165 | A marker for quiescent center identity cells | [42] |
| AGL62 | At5g60440 | Central cell development | [89] |
5. Communicating Role from Vegetative to Flowering Transition
6. Molecular Events at the Shoot Apical Meristem in Response to Photoperiodic Induction
7. Floret Pattern Initiation and Development
| Gene | Genomic Identity | Function | Reference |
|---|---|---|---|
| OsMADS1 | LOC_Os03g11614 | Involved in the early stages of rice floret development | [161] |
| OsMADS3 | LOC_Os01g10504 | Meristem function in early and late floral development, involved in the formation of stamens and ovules | [147] |
| OsMADS5 | LOC_Os06g06750 | Expressed strongly across a broad range of reproductive stages and tissues | [162] |
| OsMADS7 | LOC_Os08g41950 | Improves the stability of rice amylose content at high temperature | [163] |
| OsMADS8 | LOC_Os09g32948 | Inflorescence branch meristems | [152] |
| OsMADS13 | LOC_Os12g10540 | Ovule identity | [148] |
| OsMADS14 | LOC_Os03g54160 | Flowering activator | [112] |
| OsMADS15 | LOC_Os07g01820 | Flowering activator | [112] |
| OsMADS16 | LOC_Os06g49840 | Regulation of floral organ development and pollen formation | [164] |
| OsMADS17 | LOC_Os04g49150 | Regulates hormone signaling and floral identity | [103] |
| OsMADS18 | LOC_Os07g41370 | Flowering activator | [112] |
| OsMADS25 | LOC_Os04g23910 | OsMADS25 overexpression results in more lateral roots | [29] |
| OsMADS26 | LOC_Os08g02070 | Expressed in rice leaves and inflorescences | [165] |
| OsMADS27 | LOC_Os02g36924 | Produced more lateral roots | [50] |
| OsMADS29 | LOC_Os02g07430 | Involved in programmed cell death (PCD) in the developing embryonic cell nuclear region | [166,167] |
| OsMADS34 | LOC_Os03g54170 | Involved in development of the inflorescence | [152] |
| OsMADS50 | LOC_Os03g03100 | Flowering activator | [107] |
| OsMADS51 | LOC_Os01g69850 | Flowering activator | [113] |
| OsMADS56 | LOC_Os10g39130 | Flowering suppressor | [107] |
| OsMADS57 | LOC_Os02g49840 | Expressed in root vasculature | [35,44] |
| OsMADS61 | LOC_Os04g38770 | Expressed in root vasculature | [35,44] |
| OsMADS62 | LOC_Os08g38590 | Rice anther development | [168] |
| OsMADS63 | LOC_Os06g11970 | Rice anther development | [168] |
| OsMADS68 | LOC_Os11g43740 | Pollen development | [153] |
| OsMADS77 | LOC_Os09g02780 | Endosperm development | [169] |
| OsMADS87 | LOC_Os03g38610 | Endosperm development | [170] |
| OsMADS89 | LOC_Os01g18440 | Endosperm development | [170] |
8. MADS-Box Genes Play an Important Role in Seed Setting and Development
9. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schilling, S.; Pan, S.; Kennedy, A.; Melzer, R. MADS-box genes and crop domestication: The jack of all traits. J. Exp. Bot. 2018, 69, 1447–1469. [Google Scholar] [CrossRef]
- Ma, J.; Yang, Y.; Luo, W.; Yang, C.; Ding, P.; Liu, Y.; Qiao, L.; Chang, Z.; Geng, H.; Wang, P. Genome-wide identification and analysis of the MADS-box gene family in bread wheat (Triticum aestivum L.). PLoS ONE 2017, 12, e0181443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Bodt, S.; Raes, J.; Van de Peer, Y.; Theißen, G. And then there were many: MADS goes genomic. Trends Plant Sci. 2003, 8, 475–483. [Google Scholar] [CrossRef]
- Theißen, G.; Kim, J.T.; Saedler, H. Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 1996, 43, 484–516. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Khurana, P. Overexpression of TaMADS from wheat promotes flowering by upregulating expression of floral promoters and provides protection against thermal stress. Plant Gene 2019, 17, 100168. [Google Scholar] [CrossRef]
- Masiero, S.; Colombo, L.; Grini, P.E.; Schnittger, A.; Kater, M.M. The emerging importance of type I MADS box transcription factors for plant reproduction. Plant Cell 2011, 23, 865–872. [Google Scholar] [CrossRef] [Green Version]
- Kaufmann, K.; Melzer, R.; Theißen, G. MIKC-type MADS-domain proteins: Structural modularity, protein interactions and network evolution in land plants. Gene 2005, 347, 183–198. [Google Scholar] [CrossRef]
- Masiero, S.; Imbriano, C.; Ravasio, F.; Favaro, R.; Pelucchi, N.; Gorla, M.S.; Mantovani, R.; Colombo, L.; Kater, M.M. Ternary complex formation between MADS-box transcription factors and the histone fold protein NF-YB. J. Biol. Chem. 2002, 277, 26429–26435. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Fanning, L.; Jack, T. The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. Plant J. 2003, 33, 47–59. [Google Scholar] [CrossRef]
- Van Dijk, A.D.; Morabito, G.; Fiers, M.; van Ham, R.C.; Angenent, G.C.; Immink, R.G. Sequence motifs in MADS transcription factors responsible for specificity and diversification of protein-protein interaction. PLoS Comput. Biol. 2010, 6, e1001017. [Google Scholar] [CrossRef] [Green Version]
- Callens, C.; Tucker, M.R.; Zhang, D.; Wilson, Z.A. Dissecting the role of MADS-box genes in monocot floral development and diversity. J. Exp. Bot. 2018, 69, 2435–2459. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Kitagawa, M.; Ihashi, N.; Yabe, K.; Kimbara, J.; Yasuda, J.; Ito, H.; Inakuma, T.; Hiroi, S.; Kasumi, T. DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J. 2008, 55, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Favaro, R.; Pinyopich, A.; Battaglia, R.; Kooiker, M.; Borghi, L.; Ditta, G.; Yanofsky, M.F.; Kater, M.M.; Colombo, L. MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 2003, 15, 2603–2611. [Google Scholar] [CrossRef] [Green Version]
- Fang, S.-C.; Fernandez, D.E. Effect of regulated overexpression of the MADS domain factor AGL15 on flower senescence and fruit maturation. Plant Physiol. 2002, 130, 78–89. [Google Scholar] [CrossRef] [Green Version]
- Benedito, V.; Angenent, G.; Van Tuyl, J.; Krens, F. Lilium longiflorum and molecular floral development: The ABCDE model. In Proceedings of the XXI International Eucarpia Symposium on Classical versus Molecular Breeding of Ornamentals-Part II 651, Munich, Germany, 25–29 August 2003; pp. 83–89. [Google Scholar]
- Kater, M.M.; Dreni, L.; Colombo, L. Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J. Exp. Bot. 2006, 57, 3433–3444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelaz, S.; Ditta, G.S.; Baumann, E.; Wisman, E.; Yanofsky, M.F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 2000, 405, 200–203. [Google Scholar] [CrossRef]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef]
- Pinyopich, A.; Ditta, G.S.; Savidge, B.; Liljegren, S.J.; Baumann, E.; Wisman, E.; Yanofsky, M.F. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 2003, 424, 85–88. [Google Scholar] [CrossRef]
- Thei, G.; Saedler, H. Plant biology: Floral quartets. Nature 2001, 409, 469–472. [Google Scholar]
- Ditta, G.; Pinyopich, A.; Robles, P.; Pelaz, S.; Yanofsky, M.F. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 2004, 14, 1935–1940. [Google Scholar] [CrossRef] [Green Version]
- Smaczniak, C.; Immink, R.G.; Angenent, G.C.; Kaufmann, K. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 2012, 139, 3081–3098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, W.; Chen, D.; Kaufmann, K. Molecular mechanisms of floral organ specification by MADS domain proteins. Curr. Opin. Plant Biol. 2016, 29, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, M.E. Changing MADS-box transcription factor protein–protein interactions as a mechanism for generating floral morphological diversity. Integr. Comp. Biol. 2017, 57, 1312–1321. [Google Scholar] [CrossRef] [PubMed]
- Bloomer, R.; Dean, C. Fine-tuning timing: Natural variation informs the mechanistic basis of the switch to flowering in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 5439–5452. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, C.; Dean, C. The FLC locus: A platform for discoveries in epigenetics and adaptation. Annu. Rev. Cell Dev. Biol. 2017, 33, 555–575. [Google Scholar] [CrossRef]
- Theißen, G.; Rümpler, F.; Gramzow, L. Array of MADS-box genes: Facilitator for rapid adaptation? Trends Plant Sci. 2018, 23, 563–576. [Google Scholar] [CrossRef]
- Theißen, G.; Melzer, R.; Rümpler, F. MADS-domain transcription factors and the floral quartet model of flower development: Linking plant development and evolution. Development 2016, 143, 3259–3271. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.; Liu, Y.; Zhang, A.; Su, S.; Yan, A.; Huang, L.; Ali, I.; Liu, Y.; Forde, B.G.; Gan, Y. MADS-box transcription factor OsMADS25 regulates root development through affection of nitrate accumulation in rice. PLoS ONE 2015, 10, e0135196. [Google Scholar] [CrossRef]
- Rounsley, S.D.; Ditta, G.S.; Yanofsky, M.F. Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 1995, 7, 1259–1269. [Google Scholar]
- Becker, A.; Theißen, G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 2003, 29, 464–489. [Google Scholar] [CrossRef]
- Alvarez-Buylla, E.R.; García-Ponce, B.; Sánchez, M.d.l.P.; Espinosa-Soto, C.; García-Gómez, M.L.; Piñeyro-Nelson, A.; Garay-Arroyo, A. MADS-box genes underground becoming mainstream: Plant root developmental mechanisms. New Phytol. 2019, 223, 1143–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Forde, B.G. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 1998, 279, 407–409. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Buylla, E.R.; Liljegren, S.J.; Pelaz, S.; Gold, S.E.; Burgeff, C.; Ditta, G.S.; Vergara-Silva, F.; Yanofsky, M.F. MADS-box gene evolution beyond flowers: Expression in pollen, endosperm, guard cells, roots and trichomes. Plant J. 2000, 24, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Su, S.; Xu, Y.; Zhao, Y.; Yan, A.; Huang, L.; Ali, I.; Gan, Y. The effects of fluctuations in the nutrient supply on the expression of five members of the AGL17 clade of MADS-box genes in rice. PLoS ONE 2014, 9, e105597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gan, Y.; Filleur, S.; Rahman, A.; Gotensparre, S.; Forde, B.G. Nutritional regulation of ANR1 and other root-expressed MADS-box genes in Arabidopsis thaliana. Planta 2005, 222, 730–742. [Google Scholar] [CrossRef] [PubMed]
- Gan, Y.; Bernreiter, A.; Filleur, S.; Abram, B.; Forde, B.G. Overexpressing the ANR1 MADS-box gene in transgenic plants provides new insights into its role in the nitrate regulation of root development. Plant Cell Physiol. 2012, 53, 1003–1016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, L.-H.; Miao, Z.-Q.; Qi, G.-F.; Wu, J.; Cai, X.-T.; Mao, J.-L.; Xiang, C.-B. MADS-box transcription factor AGL21 regulates lateral root development and responds to multiple external and physiological signals. Mol. Plant 2014, 7, 1653–1669. [Google Scholar] [CrossRef] [Green Version]
- Remans, T.; Nacry, P.; Pervent, M.; Filleur, S.; Diatloff, E.; Mounier, E.; Tillard, P.; Forde, B.G.; Gojon, A. The Arabidopsis NRT1. 1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Natl. Acad. Sci. USA 2006, 103, 19206–19211. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; Wang, H.; Hamera, S.; Chen, X.; Fang, R. miR444a has multiple functions in the rice nitrate-signaling pathway. Plant J. 2014, 78, 44–55. [Google Scholar] [CrossRef]
- Zhao, P.X.; Zhang, J.; Chen, S.Y.; Wu, J.; Xia, J.Q.; Sun, L.Q.; Ma, S.S.; Xiang, C.B. Arabidopsis MADS-box factor AGL16 is a negative regulator of plant response to salt stress by downregulating salt-responsive genes. New Phytol. 2021, 232, 2418–2439. [Google Scholar] [CrossRef]
- Nawy, T.; Lee, J.-Y.; Colinas, J.; Wang, J.Y.; Thongrod, S.C.; Malamy, J.E.; Birnbaum, K.; Benfey, P.N. Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 2005, 17, 1908–1925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hacham, Y.; Holland, N.; Butterfield, C.; Ubeda-Tomas, S.; Bennett, M.J.; Chory, J.; Savaldi-Goldstein, S. Brassinosteroid perception in the epidermis controls root meristem size. Development 2011, 138, 839–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puig, J.; Meynard, D.; Khong, G.N.; Pauluzzi, G.; Guiderdoni, E.; Gantet, P. Analysis of the expression of the AGL17-like clade of MADS-box transcription factors in rice. Gene Expr. Patterns 2013, 13, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Xu, N.; Chen, H.; Wang, G.; Huang, J. OsMADS25 regulates root system development via auxin signalling in rice. Plant J. 2018, 95, 1004–1022. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Liang, Z.; Chen, S.; Sun, H.; Fan, X.; Wang, C.; Xu, G.; Zhang, Y. A transcription factor, OsMADS57, regulates long-distance nitrate transport and root elongation. Plant Physiol. 2019, 180, 882–895. [Google Scholar] [CrossRef]
- Jia, Z.; von Wirén, N. Signaling pathways underlying nitrogen-dependent changes in root system architecture: From model to crop species. J. Exp. Bot. 2020, 71, 4393–4404. [Google Scholar] [CrossRef]
- Wu, J.; Yu, C.; Huang, L.; Gan, Y. A rice transcription factor, OsMADS57, positively regulates high salinity tolerance in transgenic Arabidopsis thaliana and Oryza sativa plants. Physiol. Plant. 2021, 173, 1120–1135. [Google Scholar] [CrossRef]
- Shao, Y.; Zhou, H.-Z.; Wu, Y.; Zhang, H.; Lin, J.; Jiang, X.; He, Q.; Zhu, J.; Li, Y.; Yu, H. OsSPL3, an SBP-domain protein, regulates crown root development in rice. Plant Cell 2019, 31, 1257–1275. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Xu, N.; Wu, Q.; Yu, B.; Chu, Y.; Li, X.; Huang, J.; Jin, L. OsMADS27 regulates the root development in a NO3−—Dependent manner and modulates the salt tolerance in rice (Oryza sativa L.). Plant Sci. 2018, 277, 20–32. [Google Scholar] [CrossRef]
- Duan, K.; Li, L.; Hu, P.; Xu, S.P.; Xu, Z.H.; Xue, H.W. A brassinolide-suppressed rice MADS-box transcription factor, OsMDP1, has a negative regulatory role in BR signaling. Plant J. 2006, 47, 519–531. [Google Scholar] [CrossRef]
- Lee, S.; Woo, Y.-M.; Ryu, S.-I.; Shin, Y.-D.; Kim, W.T.; Park, K.Y.; Lee, I.-J.; An, G. Further characterization of a rice AGL12 group MADS-box gene, OsMADS26. Plant Physiol. 2008, 147, 156–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, F.; Wang, D.; Zhao, X.; Zhang, T.; Sun, H.; Zhu, L.; Zhang, F.; Li, L.; Li, Q.; Tao, D. Identification of rhizome-specific genes by genome-wide differential expression analysis in Oryza longistaminata. BMC Plant Biol. 2011, 11, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krugman, T.; Peleg, Z.; Quansah, L.; Chagué, V.; Korol, A.B.; Nevo, E.; Saranga, Y.; Fait, A.; Chalhoub, B.; Fahima, T. Alteration in expression of hormone-related genes in wild emmer wheat roots associated with drought adaptation mechanisms. Funct. Integr. Genom. 2011, 11, 565–583. [Google Scholar] [CrossRef]
- Lee, S.; Choi, S.C.; An, G. Rice SVP-group MADS-box proteins, OsMADS22 and OsMADS55, are negative regulators of brassinosteroid responses. Plant J. 2008, 54, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Ma, T.; Ma, L.; Lin, X. Ectopic expression of PvSOC1, a homolog of SOC1 from Phyllostachys violascens, promotes flowering in Arabidopsis and rice. Acta Physiol. Plant. 2016, 38, 1–9. [Google Scholar] [CrossRef]
- Aceto, S.; Gaudio, L. The MADS and the beauty: Genes involved in the development of orchid flowers. Curr. Genom. 2011, 12, 342–356. [Google Scholar] [CrossRef] [Green Version]
- Claßen-Bockhoff, R.; Bull-Hereñu, K. Towards an ontogenetic understanding of inflorescence diversity. Ann. Bot. 2013, 112, 1523–1542. [Google Scholar] [CrossRef] [Green Version]
- Kaufmann, K.; Wellmer, F.; Muiño, J.M.; Ferrier, T.; Wuest, S.E.; Kumar, V.; Serrano-Mislata, A.; Madueno, F.; Krajewski, P.; Meyerowitz, E.M. Orchestration of floral initiation by APETALA1. Science 2010, 328, 85–89. [Google Scholar] [CrossRef] [Green Version]
- Moyroud, E.; Minguet, E.G.; Ott, F.; Yant, L.; Posé, D.; Monniaux, M.; Blanchet, S.; Bastien, O.; Thévenon, E.; Weigel, D. Prediction of regulatory interactions from genome sequences using a biophysical model for the Arabidopsis LEAFY transcription factor. Plant Cell 2011, 23, 1293–1306. [Google Scholar] [CrossRef] [Green Version]
- Winter, C.M.; Austin, R.S.; Blanvillain-Baufumé, S.; Reback, M.A.; Monniaux, M.; Wu, M.-F.; Sang, Y.; Yamaguchi, A.; Yamaguchi, N.; Parker, J.E. LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev. Cell 2011, 20, 430–443. [Google Scholar] [CrossRef]
- Ferrándiz, C.; Gu, Q.; Martienssen, R.; Yanofsky, M.F. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 2000, 127, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Teo, Z.W.N.; Bi, Y.; Song, S.; Xi, W.; Yang, X.; Yin, Z.; Yu, H. A conserved genetic pathway determines inflorescence architecture in Arabidopsis and rice. Dev. Cell 2013, 24, 612–622. [Google Scholar] [CrossRef] [Green Version]
- Goslin, K.; Zheng, B.; Serrano-Mislata, A.; Rae, L.; Ryan, P.T.; Kwaśniewska, K.; Thomson, B.; Ó’Maoiléidigh, D.S.; Madueño, F.; Wellmer, F.; et al. Transcription factor interplay between LEAFY and APETALA1/CAULIFLOWER during floral initiation. Physiol. Plant. 2017, 174, 1097–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pajoro, A.; Madrigal, P.; Muiño, J.M.; Matus, J.T.; Jin, J.; Mecchia, M.A.; Debernardi, J.M.; Palatnik, J.F.; Balazadeh, S.; Arif, M. Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 2014, 15, 1–19. [Google Scholar] [CrossRef]
- Winter, C.M.; Yamaguchi, N.; Wu, M.F.; Wagner, D. Transcriptional programs regulated by both LEAFY and APETALA1 at the time of flower formation. Physiol. Plant. 2015, 155, 55–73. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, A.; Wu, M.F.; Yang, L.; Wu, G.; Poethig, R.S.; Wagner, D. The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. cell 2009, 17, 268–278. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, M.; Shimamoto, K.; Kyozuka, J. Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 2002, 29, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Carmona, M.J.; Calonje, M.; Martínez-Zapater, J.M. The FT/TFL1 gene family in grapevine. Plant Mol. Biol. 2007, 63, 637–650. [Google Scholar] [CrossRef] [PubMed]
- Danilevskaya, O.N.; Meng, X.; Ananiev, E.V. Concerted modification of flowering time and inflorescence architecture by ectopic expression of TFL1-like genes in maize. Plant Physiol. 2010, 153, 238–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Hu, W.; Wang, L.; Lin, C.; Cong, B.; Sun, C.; Luo, D. TFL1/CEN-like genes control intercalary meristem activity and phase transition in rice. Plant Sci. 2005, 168, 1393–1408. [Google Scholar] [CrossRef]
- Meng, Q.; Li, X.; Zhu, W.; Yang, L.; Liang, W.; Dreni, L.; Zhang, D. Regulatory network and genetic interactions established by OsMADS34 in rice inflorescence and spikelet morphogenesis. J. Integr. Plant Biol. 2017, 59, 693–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tapia-López, R.; García-Ponce, B.; Dubrovsky, J.G.; Garay-Arroyo, A.; Pérez-Ruíz, R.V.; Kim, S.-H.; Acevedo, F.; Pelaz, S.; Alvarez-Buylla, E.R. An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol. 2008, 146, 1182–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez, D.E.; Heck, G.R.; Perry, S.E.; Patterson, S.E.; Bleecker, A.B.; Fang, S.-C. The embryo MADS domain factor AGL15 acts postembryonically: Inhibition of perianth senescence and abscission via constitutive expression. Plant Cell 2000, 12, 183–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heck, G.R.; Perry, S.E.; Nichols, K.W.; Fernandez, D.E. AGL15, a MADS domain protein expressed in developing embryos. Plant Cell 1995, 7, 1271–1282. [Google Scholar] [PubMed] [Green Version]
- Harding, E.W.; Tang, W.; Nichols, K.W.; Fernandez, D.E.; Perry, S.E. Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-Like 15. Plant Physiol. 2003, 133, 653–663. [Google Scholar] [CrossRef] [Green Version]
- Adamczyk, B.J.; Lehti-Shiu, M.D.; Fernandez, D.E. The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J. 2007, 50, 1007–1019. [Google Scholar] [CrossRef]
- Kutter, C.; Schoöb, H.; Stadler, M.; Meins, F., Jr.; Si-Ammour, A. MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell 2007, 19, 2417–2429. [Google Scholar] [CrossRef] [Green Version]
- Han, P.; García-Ponce, B.; Fonseca-Salazar, G.; Alvarez-Buylla, E.R.; Yu, H. AGAMOUS-LIKE 17, a novel flowering promoter, acts in a FT-independent photoperiod pathway. Plant J. 2008, 55, 253–265. [Google Scholar] [CrossRef]
- Yoo, S.K.; Wu, X.; Lee, J.S.; Ahn, J.H. AGAMOUS-LIKE 6 is a floral promoter that negatively regulates the FLC/MAF clade genes and positively regulates FT in Arabidopsis. Plant J. 2011, 65, 62–76. [Google Scholar] [CrossRef]
- Chiang, G.C.; Barua, D.; Kramer, E.M.; Amasino, R.M.; Donohue, K. Major flowering time gene, FLOWERING LOCUS C, regulates seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2009, 106, 11661–11666. [Google Scholar] [CrossRef] [Green Version]
- Deng, W.; Ying, H.; Helliwell, C.A.; Taylor, J.M.; Peacock, W.J.; Dennis, E.S. FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 6680–6685. [Google Scholar] [CrossRef] [Green Version]
- Ratcliffe, O.J.; Kumimoto, R.W.; Wong, B.J.; Riechmann, J.L. Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 2003, 15, 1159–1169. [Google Scholar] [CrossRef] [Green Version]
- Schönrock, N.; Bouveret, R.; Leroy, O.; Borghi, L.; Köhler, C.; Gruissem, W.; Hennig, L. Polycomb-group proteins repressthe floral activator AGL19 in the FLC-independent vernalization pathway. Genes Dev. 2006, 20, 1667–1678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melzer, S.; Lens, F.; Gennen, J.; Vanneste, S.; Rohde, A.; Beeckman, T. Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat. Genet. 2008, 40, 1489–1492. [Google Scholar] [CrossRef] [Green Version]
- Michaels, S.D.; Ditta, G.; Gustafson-Brown, C.; Pelaz, S.; Yanofsky, M.; Amasino, R.M. AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J. 2003, 33, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Masiero, S.; Vanzulli, S.; Lardelli, P.; Kater, M.M.; Colombo, L. AGL23, a type I MADS-box gene that controls female gametophyte and embryo development in Arabidopsis. Plant J. 2008, 54, 1037–1048. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.K.; Lee, J.S.; Ahn, J.H. Overexpression of AGAMOUS-LIKE 28 (AGL28) promotes flowering by upregulating expression of floral promoters within the autonomous pathway. Biochem. Biophys. Res. Commun. 2006, 348, 929–936. [Google Scholar] [CrossRef]
- Kang, I.-H.; Steffen, J.G.; Portereiko, M.F.; Lloyd, A.; Drews, G.N. The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 2008, 20, 635–647. [Google Scholar] [CrossRef] [Green Version]
- Sridhar, V.; Shankar, V.G.; Sujatha, M.; Suman, A. Genetic control of floral transition in cereals. Indian J. Plant Genet. Resour. 2008, 21, 1–16. [Google Scholar]
- Weigel, D. Genetic hierarchy controlling flower development. Mol. Basis Morphog. 1993, 93–107. [Google Scholar]
- Schmid, M.; Uhlenhaut, N.H.; Godard, F.; Demar, M.; Bressan, R.; Weigel, D.; Lohmann, J.U. Dissection of floral induction pathways using global expression analysis. Development 2003, 130, 6001–6012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lal, S.; Pacis, L.B.; Smith, H.M. Regulation of the SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE genes/microRNA156 module by the homeodomain proteins PENNYWISE and POUND-FOOLISH in Arabidopsis. Mol. Plant 2011, 4, 1123–1132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, J.H.; Ju, Y.; Seo, P.J.; Lee, J.H.; Park, C.M. The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J. 2012, 69, 577–588. [Google Scholar] [CrossRef]
- Torti, S.; Fornara, F.; Vincent, C.; Andrés, F.; Nordström, K.; Göbel, U.; Knoll, D.; Schoof, H.; Coupland, G. Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell 2012, 24, 444–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, X.; Wang, Y.; He, Y. Photoperiodic regulation of flowering time through periodic histone deacetylation of the florigen gene FT. PLoS Biol. 2013, 11, e1001649. [Google Scholar] [CrossRef]
- Fernandez, D.E.; Wang, C.-T.; Zheng, Y.; Adamczyk, B.J.; Singhal, R.; Hall, P.K.; Perry, S.E. The MADS-domain factors AGAMOUS-LIKE15 and AGAMOUS-LIKE18, along with SHORT VEGETATIVE PHASE and AGAMOUS-LIKE24, are necessary to block floral gene expression during the vegetative phase. Plant Physiol. 2014, 165, 1591–1603. [Google Scholar] [CrossRef] [PubMed]
- Serivichyaswat, P.; Ryu, H.-S.; Kim, W.; Kim, S.; Chung, K.S.; Kim, J.J.; Ahn, J.H. Expression of the floral repressor miRNA156 is positively regulated by the AGAMOUS-like proteins AGL15 and AGL18. Mol. Cells 2015, 38, 259. [Google Scholar] [CrossRef]
- Amasino, R. Seasonal and developmental timing of flowering. Plant J. 2010, 61, 1001–1013. [Google Scholar] [CrossRef]
- Lee, J.; Lee, I. Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 2010, 61, 2247–2254. [Google Scholar] [CrossRef] [Green Version]
- Srikanth, A.; Schmid, M. Regulation of flowering time: All roads lead to Rome. Cell. Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar] [CrossRef]
- Yu, S.; Galvão, V.C.; Zhang, Y.-C.; Horrer, D.; Zhang, T.-Q.; Hao, Y.-H.; Feng, Y.-Q.; Wang, S.; Schmid, M.; Wang, J.-W. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA PROMOTER BINDING–LIKE transcription factors. Plant Cell 2012, 24, 3320–3332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arora, R.; Agarwal, P.; Ray, S.; Singh, A.K.; Singh, V.P.; Tyagi, A.K.; Kapoor, S. MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genom. 2007, 8, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tadege, M.; Sheldon, C.C.; Helliwell, C.A.; Upadhyaya, N.M.; Dennis, E.S.; Peacock, W.J. Reciprocal control of flowering time by OsSOC1 in transgenic Arabidopsis and by FLC in transgenic rice. Plant Biotechnol. J. 2003, 1, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, J.; Han, J.J.; Han, M.J.; An, G. Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J. 2004, 38, 754–764. [Google Scholar] [CrossRef] [PubMed]
- Nam, J.; Kaufmann, K.; Theißen, G.; Nei, M. A simple method for predicting the functional differentiation of duplicate genes and its application to MIKC-type MADS-box genes. Nucleic Acids Res. 2005, 33, e12. [Google Scholar] [CrossRef] [Green Version]
- Ryu, C.H.; Lee, S.; Cho, L.H.; Kim, S.L.; Lee, Y.S.; Choi, S.C.; Jeong, H.J.; Yi, J.; Park, S.J.; Han, C.D. OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ. 2009, 32, 1412–1427. [Google Scholar] [CrossRef]
- Kang, H.-G.; An, G. Isolation and characterization of a rice MADS box gene belonging to the AGL2 gene family. Mol. Cells 1997, 7, 45–51. [Google Scholar]
- Kang, H.-G.; Jang, S.; Chung, J.-E.; Cho, Y.-G.; An, G. Characterization of two rice MADS box genes that control flowering time. Mol. Cells 1997, 7, 559–566. [Google Scholar]
- Zhao, X.Y.; Cheng, Z.J.; Zhang, X.S. Overexpression of TaMADS1, a SEPALLATA-like gene in wheat, causes early flowering and the abnormal development of floral organs in Arabidopsis. Planta 2006, 223, 698–707. [Google Scholar] [CrossRef]
- Su, Y.; Liu, J.; Liang, W.; Dou, Y.; Fu, R.; Li, W.; Feng, C.; Gao, C.; Zhang, D.; Kang, Z. Wheat AGAMOUS LIKE 6 transcription factors function in stamen development by regulating the expression of Ta APETALA3. Development 2019, 146, dev177527. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, K.; Yasuno, N.; Sato, Y.; Yoda, M.; Yamazaki, R.; Kimizu, M.; Yoshida, H.; Nagamura, Y.; Kyozuka, J. Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 2012, 24, 1848–1859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.L.; Lee, S.; Kim, H.J.; Nam, H.G.; An, G. OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol. 2007, 145, 1484–1494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wigge, P.A.; Kim, M.C.; Jaeger, K.E.; Busch, W.; Schmid, M.; Lohmann, J.U.; Weigel, D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 2005, 309, 1056–1059. [Google Scholar] [CrossRef] [PubMed]
- Andrés, F.; Coupland, G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012, 13, 627–639. [Google Scholar] [CrossRef]
- Samach, A.; Onouchi, H.; Gold, S.E.; Ditta, G.S.; Schwarz-Sommer, Z.; Yanofsky, M.F.; Coupland, G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 2000, 288, 1613–1616. [Google Scholar] [CrossRef] [Green Version]
- Abe, M.; Kobayashi, Y.; Yamamoto, S.; Daimon, Y.; Yamaguchi, A.; Ikeda, Y.; Ichinoki, H.; Notaguchi, M.; Goto, K.; Araki, T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 2005, 309, 1052–1056. [Google Scholar] [CrossRef]
- Jang, S.; Torti, S.; Coupland, G. Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J. 2009, 60, 614–625. [Google Scholar] [CrossRef]
- D’Aloia, M.; Bonhomme, D.; Bouché, F.; Tamseddak, K.; Ormenese, S.; Torti, S.; Coupland, G.; Périlleux, C. Cytokinin promotes flowering of Arabidopsis via transcriptional activation of the FT paralogue TSF. Plant J. 2011, 65, 972–979. [Google Scholar] [CrossRef] [Green Version]
- Teper-Bamnolker, P.; Samach, A. The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 2005, 17, 2661–2675. [Google Scholar] [CrossRef] [Green Version]
- Lifschitz, E.; Eviatar, T.; Rozman, A.; Shalit, A.; Goldshmidt, A.; Amsellem, Z.; Alvarez, J.P.; Eshed, Y. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 2006, 103, 6398–6403. [Google Scholar] [CrossRef] [Green Version]
- Giavalisco, P.; Kapitza, K.; Kolasa, A.; Buhtz, A.; Kehr, J. Towards the proteome of Brassica napus phloem sap. Proteomics 2006, 6, 896–909. [Google Scholar] [CrossRef] [PubMed]
- Michaels, S.D.; Amasino, R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 1999, 11, 949–956. [Google Scholar] [CrossRef] [Green Version]
- Sheldon, C.C.; Burn, J.E.; Perez, P.P.; Metzger, J.; Edwards, J.A.; Peacock, W.J.; Dennis, E.S. The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 1999, 11, 445–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartmann, U.; Höhmann, S.; Nettesheim, K.; Wisman, E.; Saedler, H.; Huijser, P. Molecular cloning of SVP: A negative regulator of the floral transition in Arabidopsis. Plant J. 2000, 21, 351–360. [Google Scholar] [CrossRef]
- Fujiwara, S.; Oda, A.; Yoshida, R.; Niinuma, K.; Miyata, K.; Tomozoe, Y.; Tajima, T.; Nakagawa, M.; Hayashi, K.; Coupland, G. Circadian clock proteins LHY and CCA1 regulate SVP protein accumulation to control flowering in Arabidopsis. Plant Cell 2008, 20, 2960–2971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Liu, C.; Shen, L.; Wu, Y.; Chen, H.; Robertson, M.; Helliwell, C.A.; Ito, T.; Meyerowitz, E.; Yu, H. A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 2008, 15, 110–120. [Google Scholar] [CrossRef] [Green Version]
- Searle, I.; He, Y.; Turck, F.; Vincent, C.; Fornara, F.; Kröber, S.; Amasino, R.A.; Coupland, G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006, 20, 898–912. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Yoo, S.J.; Park, S.H.; Hwang, I.; Lee, J.S.; Ahn, J.H. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 2007, 21, 397–402. [Google Scholar] [CrossRef] [Green Version]
- Taoka, K.-i.; Ohki, I.; Tsuji, H.; Furuita, K.; Hayashi, K.; Yanase, T.; Yamaguchi, M.; Nakashima, C.; Purwestri, Y.A.; Tamaki, S. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 2011, 476, 332–335. [Google Scholar] [CrossRef]
- Fornara, F.; Panigrahi, K.C.; Gissot, L.; Sauerbrunn, N.; Rühl, M.; Jarillo, J.A.; Coupland, G. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 2009, 17, 75–86. [Google Scholar] [CrossRef] [Green Version]
- Jaeger, K.E.; Pullen, N.; Lamzin, S.; Morris, R.J.; Wigge, P.A. Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell 2013, 25, 820–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pnueli, L.; Hareven, D.; Broday, L.; Hurwitz, C.; Lifschitz, E. The TM5 MADS box gene mediates organ differentiation in the three inner whorls of tomato flowers. Plant Cell 1994, 6, 175–186. [Google Scholar] [CrossRef]
- Savidge, B.; Rounsley, S.D.; Yanofsky, M.F. Temporal relationship between the transcription of two Arabidopsis MADS box genes and the floral organ identity genes. Plant Cell 1995, 7, 721–733. [Google Scholar] [PubMed] [Green Version]
- Yu, H.; Xu, Y.; Tan, E.L.; Kumar, P.P. AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proc. Natl. Acad. Sci. USA 2002, 99, 16336–16341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burgeff, C.; Liljegren, S.J.; Tapia-López, R.; Yanofsky, M.F.; Alvarez-Buylla, E.R. MADS-box gene expression in lateral primordia, meristems and differentiated tissues of Arabidopsis thaliana roots. Planta 2002, 214, 365–372. [Google Scholar] [CrossRef]
- Adamczyk, B.J.; Fernandez, D.E. MIKC* MADS domain heterodimers are required for pollen maturation and tube growth in Arabidopsis. Plant Physiol. 2009, 149, 1713–1723. [Google Scholar] [CrossRef] [Green Version]
- Portereiko, M.F.; Lloyd, A.; Steffen, J.G.; Punwani, J.A.; Otsuga, D.; Drews, G.N. AGL80 is required for central cell and endosperm development in Arabidopsis. Plant Cell 2006, 18, 1862–1872. [Google Scholar] [CrossRef] [Green Version]
- Crane, P.R.; Friis, E.M.; Pedersen, K.R. The origin and early diversification of angiosperms. Nature 1995, 374, 27–33. [Google Scholar] [CrossRef]
- Wu, F.; Shi, X.; Lin, X.; Liu, Y.; Chong, K.; Theißen, G.; Meng, Z. The ABC s of flower development: Mutational analysis of AP 1/FUL-like genes in rice provides evidence for a homeotic (A)-function in grasses. Plant J. 2017, 89, 310–324. [Google Scholar] [CrossRef]
- Xu, G.; Kong, H. Duplication and divergence of floral MADS-box genes in grasses: Evidence for the generation and modification of novel regulators. J. Integr. Plant Biol. 2007, 49, 927–939. [Google Scholar] [CrossRef]
- Wang, K.; Tang, D.; Hong, L.; Xu, W.; Huang, J.; Li, M.; Gu, M.; Xue, Y.; Cheng, Z. DEP and AFO regulate reproductive habit in rice. PLoS Genet. 2010, 6, e1000818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, S.R.; Prasad, K.; Vijayraghavan, U. Divergent regulatory OsMADS2 functions control size, shape and differentiation of the highly derived rice floret second-whorl organ. Genetics 2007, 176, 283–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, H.; Itoh, J.I.; Ohmori, S.; Miyoshi, K.; Horigome, A.; Uchida, E.; Kimizu, M.; Matsumura, Y.; Kusaba, M.; Satoh, H. Superwoman1-cleistogamy, a hopeful allele for gene containment in GM rice. Plant Biotechnol. J. 2007, 5, 835–846. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.-G.; Ohmori, S.; Kimizu, M.; Yoshida, H. Unequal genetic redundancy of rice PISTILLATA orthologs, OsMADS2 and OsMADS4, in lodicule and stamen development. Plant Cell Physiol. 2008, 49, 853–857. [Google Scholar] [CrossRef] [Green Version]
- Hama, E.; Takumi, S.; Ogihara, Y.; Murai, K. Pistillody is caused by alterations to the class-B MADS-box gene expression pattern in alloplasmic wheats. Planta 2004, 218, 712–720. [Google Scholar]
- Yasui, Y.; Tanaka, W.; Sakamoto, T.; Kurata, T.; Hirano, H.-Y. Genetic enhancer analysis reveals that FLORAL ORGAN NUMBER2 and OsMADS3 co-operatively regulate maintenance and determinacy of the flower meristem in rice. Plant Cell Physiol. 2017, 58, 893–903. [Google Scholar] [CrossRef] [Green Version]
- Dreni, L.; Jacchia, S.; Fornara, F.; Fornari, M.; Ouwerkerk, P.B.; An, G.; Colombo, L.; Kater, M.M. The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J. 2007, 52, 690–699. [Google Scholar] [CrossRef]
- Trevaskis, B.; Tadege, M.; Hemming, M.N.; Peacock, W.J.; Dennis, E.S.; Sheldon, C. Short vegetative phase-like MADS-box genes inhibit floral meristem identity in barley. Plant Physiol. 2007, 143, 225–235. [Google Scholar] [CrossRef] [Green Version]
- Sentoku, N.; Kato, H.; Kitano, H.; Imai, R. OsMADS22, an STMADS11-like MADS-box gene of rice, is expressed in non-vegetative tissues and its ectopic expression induces spikelet meristem indeterminacy. Mol. Genet. Genom. 2005, 273, 1–9. [Google Scholar] [CrossRef]
- Hu, Y.; Liang, W.; Yin, C.; Yang, X.; Ping, B.; Li, A.; Jia, R.; Chen, M.; Luo, Z.; Cai, Q. Interactions of OsMADS1 with floral homeotic genes in rice flower development. Mol. Plant 2015, 8, 1366–1384. [Google Scholar] [CrossRef]
- Cui, R.; Han, J.; Zhao, S.; Su, K.; Wu, F.; Du, X.; Xu, Q.; Chong, K.; Theißen, G.; Meng, Z. Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J. 2010, 61, 767–781. [Google Scholar] [CrossRef]
- Liu, Y.; Cui, S.; Wu, F.; Yan, S.; Lin, X.; Du, X.; Chong, K.; Schilling, S.; Theißen, G.; Meng, Z. Functional conservation of MIKC*-Type MADS box genes in Arabidopsis and rice pollen maturation. Plant Cell 2013, 25, 1288–1303. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Wilson, Z.A. Stamen specification and anther development in rice. Chin. Sci. Bull. 2009, 54, 2342–2353. [Google Scholar] [CrossRef]
- Kim, E.-J.; Hong, W.-J.; Kim, Y.-J.; Jung, K.-H. Transcriptome analysis of triple mutant for OsMADS62, OsMADS63, and OsMADS68 reveals the downstream regulatory mechanism for pollen germination in rice (Oryza sativa). Int. J. Mol. Sci. 2022, 23, 239. [Google Scholar] [CrossRef] [PubMed]
- Han, M.-J.; Jung, K.-H.; Yi, G.; Lee, D.-Y.; An, G. Rice Immature Pollen 1 (RIP1) is a regulator of late pollen development. Plant Cell Physiol. 2006, 47, 1457–1472. [Google Scholar] [CrossRef] [Green Version]
- Angenent, G.C.; Franken, J.; Busscher, M.; Weiss, D.; Van Tunen, A.J. Co-suppression of the petunia homeotic gene Fbp2 affects the identity of the generative meristem. Plant J. 1994, 5, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Pnueli, L.; Abu-Abeid, M.; Zamir, D.; Nacken, W.; Schwarz-Sommer, Z.; Lifschitz, E. The MADS box gene family in tomato: Temporal expression during floral development, conserved secondary structures and homology with homeotic genes from Antirrhinum and Arabidopsis. Plant J. 1991, 1, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Yanofsky, M.F.; Meyerowitz, E.M. AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 1991, 5, 484–495. [Google Scholar] [CrossRef] [Green Version]
- Liljegren, S.J.; Ditta, G.S.; Eshed, Y.; Savidge, B.; Bowman, J.L.; Yanofsky, M.F. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 2000, 404, 766–770. [Google Scholar] [CrossRef]
- Jeon, J.-S.; Jang, S.; Lee, S.; Nam, J.; Kim, C.; Lee, S.-H.; Chung, Y.-Y.; Kim, S.-R.; Lee, Y.H.; Cho, Y.-G. Leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 2000, 12, 871–884. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Liang, W.; Zhu, W.; Chen, M.; Ferrándiz, C.; Burton, R.A.; Dreni, L.; Zhang, D. Loss of LOFSEP transcription factor function converts spikelet to leaf-like structures in rice. Plant Physiol. 2018, 176, 1646–1664. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xu, H.; Feng, M.; Zhu, Y. Suppression of OsMADS7 in rice endosperm stabilizes amylose content under high temperature stress. Plant Biotechnol. J. 2018, 16, 18–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, L.; Duan, Y.; Ye, Y.; Cai, Z.; Wang, F.; Qu, X.; Qiu, R.; Wu, C.; Wu, W. Screening and analysis of proteins interacting with OsMADS16 in rice (Oryza sativa L.). PLoS ONE 2019, 14, e0221473. [Google Scholar] [CrossRef] [PubMed]
- Pelucchi, N.; Fornara, F.; Favalli, C.; Masiero, S.; Lago, C.; Pè, E.M.; Colombo, L.; Kater, M.M. Comparative analysis of rice MADS-box genes expressed during flower development. Sex. Plant Reprod. 2002, 15, 113–122. [Google Scholar] [CrossRef]
- Yang, X.; Wu, F.; Lin, X.; Du, X.; Chong, K.; Gramzow, L.; Schilling, S.; Becker, A.; Theißen, G.; Meng, Z. Live and let die-the Bsister MADS-box gene OsMADS29 controls the degeneration of cells in maternal tissues during seed development of rice (Oryza sativa). PLoS ONE 2012, 7, e51435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, L.-L.; Xue, H.-W. The MADS29 transcription factor regulates the degradation of the nucellus and the nucellar projection during rice seed development. Plant Cell 2012, 24, 1049–1065. [Google Scholar] [CrossRef] [Green Version]
- Wen, K.; Chen, Y.; Zhou, X.; Chang, S.; Feng, H.; Zhang, J.; Chu, Z.; Han, X.; Li, J.; Liu, J. OsCPK21 is required for pollen late-stage development in rice. J. Plant Physiol. 2019, 240, 153000. [Google Scholar] [CrossRef]
- Cheng, X.; Pan, M.; Zhiguo, E.; Zhou, Y.; Niu, B.; Chen, C. The maternally expressed polycomb group gene OsEMF2a is essential for endosperm cellularization and imprinting in rice. Plant Commun. 2021, 2, 100092. [Google Scholar] [CrossRef]
- Ishikawa, R.; Ohnishi, T.; Kinoshita, Y.; Eiguchi, M.; Kurata, N.; Kinoshita, T. Rice interspecies hybrids show precocious or delayed developmental transitions in the endosperm without change to the rate of syncytial nuclear division. Plant J. 2011, 65, 798–806. [Google Scholar] [CrossRef]
- Theissen, G.; Becker, A.; Di Rosa, A.; Kanno, A.; Kim, J.T.; Münster, T.; Winter, K.-U.; Saedler, H. A short history of MADS-box genes in plants. Plant Mol. Biol. 2000, 42, 115–149. [Google Scholar] [CrossRef]
- Yu, L.-H.; Wu, J.; Zhang, Z.-S.; Miao, Z.-Q.; Zhao, P.-X.; Wang, Z.; Xiang, C.-B. Arabidopsis MADS-box transcription factor AGL21 acts as environmental surveillance of seed germination by regulating ABI5 expression. Mol. Plant 2017, 10, 834–845. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Liang, W.; Hu, Y.; Zhu, L.; Yin, C.; Xu, J.; Dreni, L.; Kater, M.M.; Zhang, D. Rice MADS6 interacts with the floral homeotic genes SUPERWOMAN1, MADS3, MADS58, MADS13, and DROOPING LEAF in specifying floral organ identities and meristem fate. Plant Cell 2011, 23, 2536–2552. [Google Scholar] [CrossRef] [Green Version]
- Nayar, S.; Kapoor, M.; Kapoor, S. Post-translational regulation of rice MADS29 function: Homodimerization or binary interactions with other seed-expressed MADS proteins modulate its translocation into the nucleus. J. Exp. Bot. 2014, 65, 5339–5350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Begcy, K.; Liu, K.; Folsom, J.J.; Wang, Z.; Zhang, C.; Walia, H. Heat stress yields a unique MADS box transcription factor in determining seed size and thermal sensitivity. Plant Physiol. 2016, 171, 606–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folsom, J.J.; Begcy, K.; Hao, X.; Wang, D.; Walia, H. Rice Fertilization-Independent Endosperm1 regulates seed size under heat stress by controlling early endosperm development. Plant Physiol. 2014, 165, 238–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernardi, J.; Roig-Villanova, I.; Marocco, A.; Battaglia, R. Communicating across generations: The Bsister language. Plant Biosyst. Int. J. Deal. All Asp. Plant Biol. 2014, 148, 150–156. [Google Scholar]
- Kofuji, R.; Sumikawa, N.; Yamasaki, M.; Kondo, K.; Ueda, K.; Ito, M.; Hasebe, M. Evolution and divergence of the MADS-box gene family based on genome-wide expression analyses. Mol. Biol. Evol. 2003, 20, 1963–1977. [Google Scholar] [CrossRef]
- Yang, F.; Xu, F.; Wang, X.; Liao, Y.; Chen, Q.; Meng, X. Characterization and functional analysis of a MADS-box transcription factor gene (GbMADS9) from Ginkgo biloba. Sci. Hortic. 2016, 212, 104–114. [Google Scholar] [CrossRef]
- Nesi, N.; Debeaujon, I.; Jond, C.; Stewart, A.J.; Jenkins, G.I.; Caboche, M.; Lepiniec, L. The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell 2002, 14, 2463–2479. [Google Scholar] [CrossRef] [Green Version]
- De Folter, S.; Shchennikova, A.V.; Franken, J.; Busscher, M.; Baskar, R.; Grossniklaus, U.; Angenent, G.C.; Immink, R.G. A Bsister MADS-box gene involved in ovule and seed development in petunia and Arabidopsis. Plant J. 2006, 47, 934–946. [Google Scholar] [CrossRef]
- Deng, W.; Chen, G.; Peng, F.; Truksa, M.; Snyder, C.L.; Weselake, R.J. Transparent testa16 plays multiple roles in plant development and is involved in lipid synthesis and embryo development in canola. Plant Physiol. 2012, 160, 978–989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizzotti, C.; Mendes, M.A.; Caporali, E.; Schnittger, A.; Kater, M.M.; Battaglia, R.; Colombo, L. The MADS box genes SEEDSTICK and ARABIDOPSIS Bsister play a maternal role in fertilization and seed development. Plant J. 2012, 70, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, R.; Gramzow, L.; Melzer, R.; Theißen, G.; Becker, A. GORDITA (AGL63) is a young paralog of the Arabidopsis thaliana Bsister MADS box gene ABS (TT16) that has undergone neofunctionalization. Plant J. 2010, 63, 914–924. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Ubaldo, H.; Lozano-Sotomayor, P.; Ezquer, I.; Di Marzo, M.; Chávez Montes, R.A.; Gómez-Felipe, A.; Pablo-Villa, J.; Diaz-Ramirez, D.; Ballester, P.; Ferrándiz, C. New roles of NO TRANSMITTING TRACT and SEEDSTICK during medial domain development in Arabidopsis fruits. Development 2019, 146, dev172395. [Google Scholar] [CrossRef] [Green Version]
- Di Marzo, M.; Roig-Villanova, I.; Zanchetti, E.; Caselli, F.; Gregis, V.; Bardetti, P.; Chiara, M.; Guazzotti, A.; Caporali, E.; Mendes, M.A. MADS-box and bHLH transcription factors coordinate transmitting tract development in Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 526. [Google Scholar] [CrossRef]
- Balanzà, V.; Roig-Villanova, I.; Di Marzo, M.; Masiero, S.; Colombo, L. Seed abscission and fruit dehiscence required for seed dispersal rely on similar genetic networks. Development 2016, 143, 3372–3381. [Google Scholar] [CrossRef] [Green Version]
- Ezquer, I.; Mizzotti, C.; Nguema-Ona, E.; Gotté, M.; Beauzamy, L.; Viana, V.E.; Dubrulle, N.; Costa de Oliveira, A.; Caporali, E.; Koroney, A.-S. The developmental regulator SEEDSTICK controls structural and mechanical properties of the Arabidopsis seed coat. Plant Cell 2016, 28, 2478–2492. [Google Scholar] [CrossRef] [Green Version]
- Paolo, D.; Rotasperti, L.; Schnittger, A.; Masiero, S.; Colombo, L.; Mizzotti, C. The Arabidopsis MADS-domain transcription factor SEEDSTICK controls seed size via direct activation of E2Fa. Plants 2021, 10, 192. [Google Scholar] [CrossRef]
- Di Marzo, M.; Viana, V.E.; Banfi, C.; Cassina, V.; Corti, R.; Herrera-Ubaldo, H.; Babolin, N.; Guazzotti, A.; Kiegle, E.; Gregis, V. Cell wall modifications by α-XYLOSIDASE1 are required for the control of seed and fruit size. J. Exp. Bot. 2021. [Google Scholar] [CrossRef]
- Khong, G.N.; Pati, P.K.; Richaud, F.; Parizot, B.; Bidzinski, P.; Mai, C.D.; Bès, M.; Bourrié, I.; Meynard, D.; Beeckman, T. OsMADS26 negatively regulates resistance to pathogens and drought tolerance in rice. Plant Physiol. 2015, 169, 2935–2949. [Google Scholar] [CrossRef] [Green Version]
- Rensink, W.A.; Buell, C.R. Arabidopsis to rice. Applying knowledge from a weed to enhance our understanding of a crop species. Plant Physiol. 2004, 135, 622–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ram, H.; Soni, P.; Salvi, P.; Gandass, N.; Sharma, A.; Kaur, A.; Sharma, T.R. Insertional mutagenesis approaches and their use in rice for functional genomics. Plants 2019, 8, 310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, W.-J.; Jung, K.-H. Comparative analysis of flanking sequence tags of T-DNA/transposon insertional mutants and genetic variations of fast-neutron treated mutants in rice. J. Plant Biol. 2018, 61, 80–84. [Google Scholar] [CrossRef]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef]
- Montecillo, J.A.V.; Chu, L.L.; Bae, H. CRISPR-Cas9 system for plant genome editing: Current approaches and emerging developments. Agronomy 2020, 10, 1033. [Google Scholar] [CrossRef]
- Wu, J.-L.; Wu, C.; Lei, C.; Baraoidan, M.; Bordeos, A.; Madamba, M.R.S.; Ramos-Pamplona, M.; Mauleon, R.; Portugal, A.; Ulat, V.J. Chemical-and irradiation-induced mutants of indica rice IR64 for forward and reverse genetics. Plant Mol. Biol. 2005, 59, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Jankowicz-Cieslak, J.; Till, B.J. Chemical mutagenesis of seed and vegetatively propagated plants using EMS. Curr. Protoc. Plant Biol. 2016, 1, 617–635. [Google Scholar] [CrossRef]
- Aklilu, E. Review on forward and reverse genetics in plant breeding. All Life 2021, 14, 127–135. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shah, L.; Sohail, A.; Ahmad, R.; Cheng, S.; Cao, L.; Wu, W. The Roles of MADS-Box Genes from Root Growth to Maturity in Arabidopsis and Rice. Agronomy 2022, 12, 582. https://doi.org/10.3390/agronomy12030582
Shah L, Sohail A, Ahmad R, Cheng S, Cao L, Wu W. The Roles of MADS-Box Genes from Root Growth to Maturity in Arabidopsis and Rice. Agronomy. 2022; 12(3):582. https://doi.org/10.3390/agronomy12030582
Chicago/Turabian StyleShah, Liaqat, Amir Sohail, Rafiq Ahmad, Shihua Cheng, Liyong Cao, and Weixun Wu. 2022. "The Roles of MADS-Box Genes from Root Growth to Maturity in Arabidopsis and Rice" Agronomy 12, no. 3: 582. https://doi.org/10.3390/agronomy12030582
APA StyleShah, L., Sohail, A., Ahmad, R., Cheng, S., Cao, L., & Wu, W. (2022). The Roles of MADS-Box Genes from Root Growth to Maturity in Arabidopsis and Rice. Agronomy, 12(3), 582. https://doi.org/10.3390/agronomy12030582






