Transcriptome Analysis Reveals Genetic Factors Related to Callus Induction in Barley
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Condition
2.2. Sampling
2.3. Medium Preparation
2.4. RNA Extraction and Sequencing
2.5. Gene Validation and Expression Analysis
2.6. Statistical Analysis
3. Results
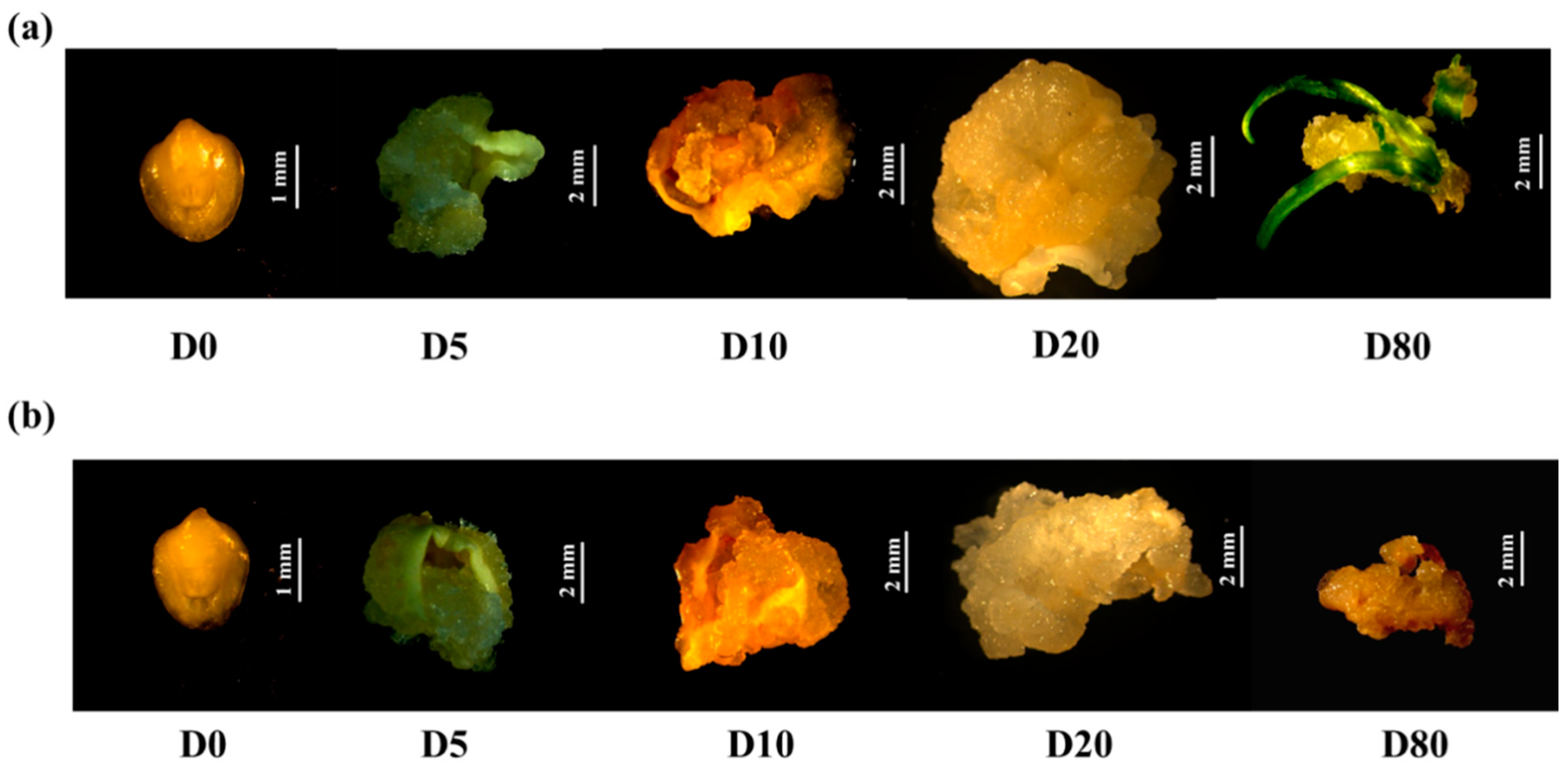
3.1. In Vitro Culture Performance of Immature Embryos
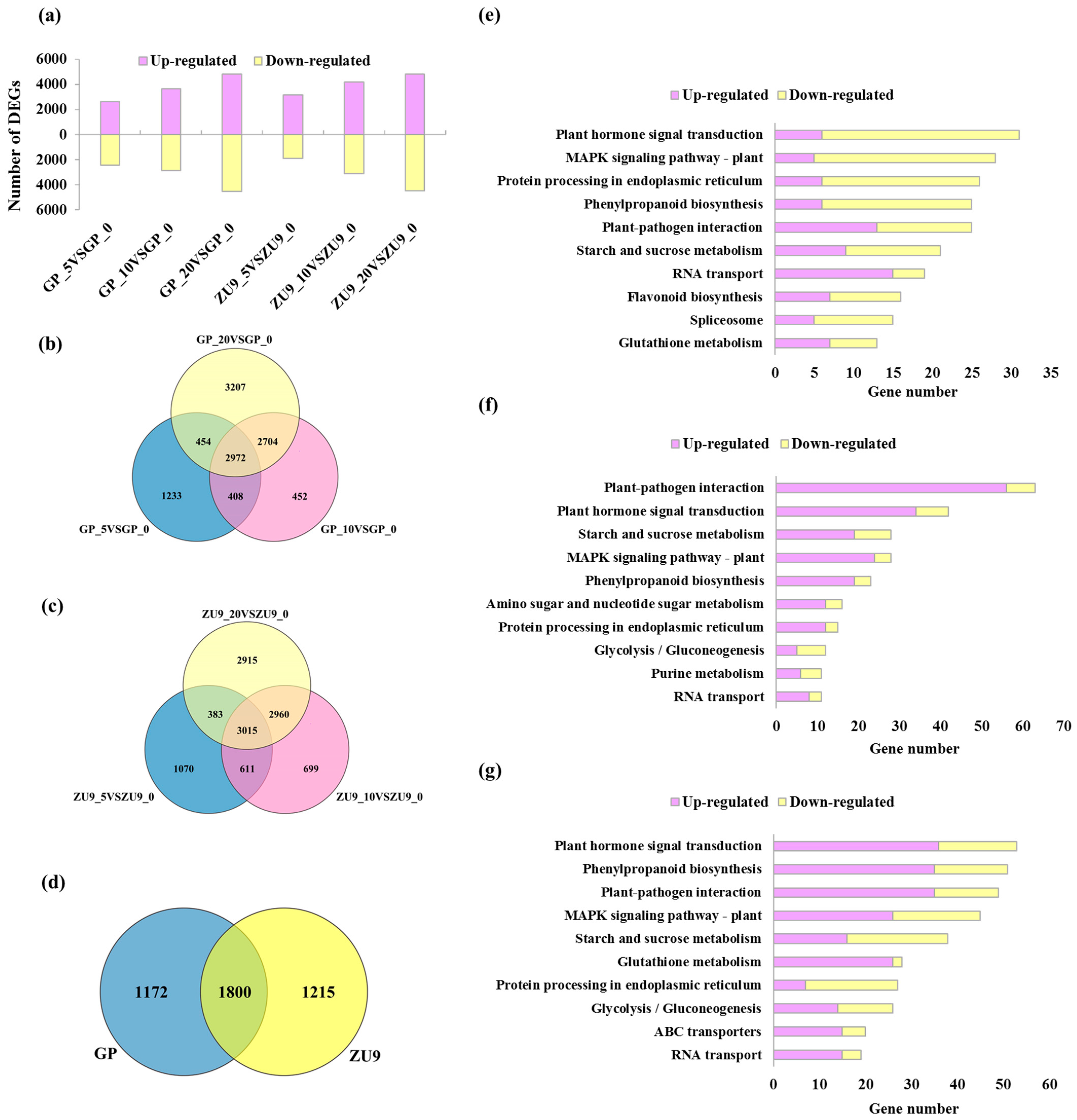
3.2. Global Analysis of Transcriptome Profiles in Barley Callus
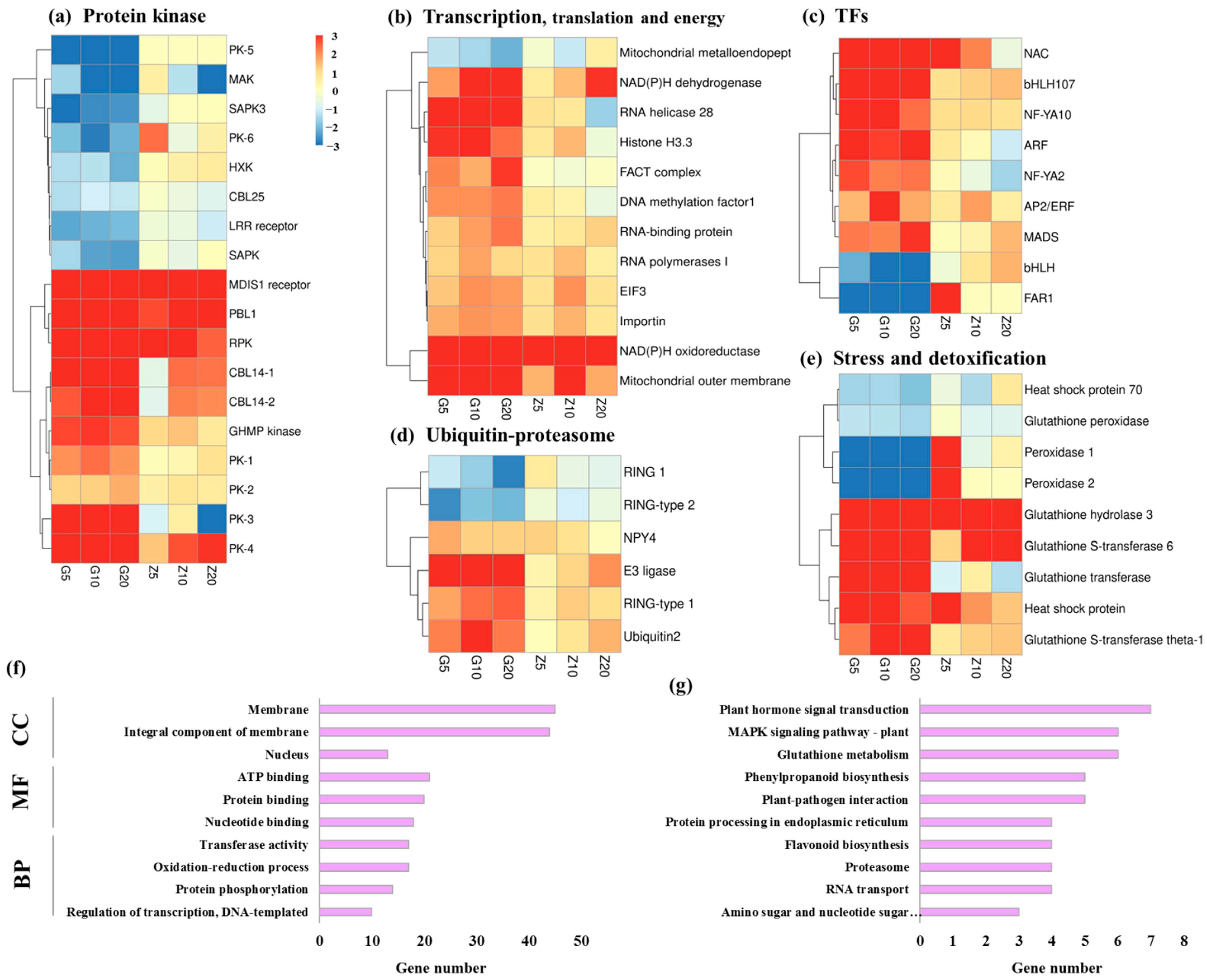
3.3. DEGs Involved in Biological Metabolism and Transcription Factors
3.4. Changes in the Expression of Phytohormone-Related Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Du, X.; Fang, T.; Liu, Y.; Huang, L.; Zang, M.; Wang, G.; Liu, J.; Fu, J. Transcriptome profiling predicts new genes to promote maize callus formation and transformation. Front. Plant Sci. 2019, 10, 1633. [Google Scholar] [CrossRef]
- Matres, J.M.; Hilscher, J.; Datta, A.; Armario-Najera, V.; Baysal, C.; He, W.; Huang, X.; Zhu, C.; Valizadeh-Kamran, R.; Trijatmiko, K.R.; et al. Genome editing in cereal crops: An overview. Transgenic Res. 2021, 30, 461–498. [Google Scholar] [CrossRef] [PubMed]
- Özgen, M.; Birsin, M.A.; Önde, S. The effect of hybrid vigor on callus induction and plant regeneration from mature embryo culture of barley (Hordeum vulgare). Plant Cell Tiss. Org. Cult. 2005, 82, 67–74. [Google Scholar] [CrossRef]
- Goetz, H. Genetic transformation of Triticeae cereals—Summary of almost three-decade’s development. Biotechnol. Adv. 2020, 40, 107484. [Google Scholar]
- Debernardi, J.M.; Tricoli, D.M.; Ercoli, M.F.; Hayta, S.; Ronald, P.; Palatnik, J.F.; Dubcovsky, J. A GRF–GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 2019, 38, 1274–1279. [Google Scholar] [CrossRef] [PubMed]
- Motte, H.; Vercauteren, A.; Depuydt, S.; Landschoot, S.; Geelen, D.; Werbrouck, S.; Goormachtig, S.; Vuylsteke, M.; Vereecke, D. Combining linkage and association mapping identifies RECEPTOR-LIKE PROTEIN KINASE1 as an essential Arabidopsis shoot regeneration gene. Proc. Natl. Acad. Sci. USA 2014, 111, 8305–8310. [Google Scholar] [CrossRef] [Green Version]
- Przetakiewicz, A.; Orczyk, W.; Nadolska-Orczyk, A. The effect of auxin on plant regeneration of wheat, barley and triticale. Plant Cell Tiss. Org. Cult. 2003, 73, 245–256. [Google Scholar] [CrossRef]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994, 6, 271–282. [Google Scholar] [CrossRef] [Green Version]
- Aldemita, R.R.; Hodges, T.K. Agrobacterium tumefaciens-mediated transformation of japonica and indica rice varieties. Planta 1996, 199, 612–617. [Google Scholar] [CrossRef]
- Hiei, Y.; Komari, T. Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat. Protoc. 2008, 3, 824–834. [Google Scholar] [CrossRef]
- Oliva, R.; Ji, C.; Atienza-Grande, G.; Huguet-Tapia, J.C.; Pérez-Quintero, A.; Li, T.; Eom, J.S.; Li, C.; Nguyen, H.; Liu, B.; et al. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 2019, 37, 1344–1350. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Liu, M.; Yan, Y.; Qing, C.; Zhang, X.; Zhang, Y.; Long, Y.; Wang, L.; Pan, L.; Zou, C.; et al. Genetic dissection of maize embryonic callus regenerative capacity using multi-locus genome-wide association studies. Front. Plant Sci. 2018, 9, 561. [Google Scholar] [CrossRef] [Green Version]
- Ishida, Y.; Saito, H.; Ohta, S.; Hiei, Y.; Komari, T.; Kumashiro, T. High efficiency transformation of maize (Zea mays L.) mediated by Agrobacterium tumefaciens. Nat. Biotechnol. 1996, 14, 745–750. [Google Scholar] [CrossRef] [PubMed]
- Frame, B.R.; Shou, H.; Chikwamba, R.K.; Zhang, Z.; Xiang, C.; Fonger, T.M.; Pegg, S.E.K.; Li, B.; Nettleton, D.S.; Pei, D.; et al. Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol. 2002, 129, 13–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mookkan, M.; Nelson-Vasilchik, K.; Hague, J.; Zhang, Z.; Kausch, A.P. Selectable marker independent transformation of recalcitrant maize inbred B73 and sorghum P898012 mediated by morphogenic regulators BABY BOOM and WUSCHEL2. Plant Cell Rep. 2017, 36, 1477–1491. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Ma, H.; Zhu, W.; Zhang, J.; Zhao, X.; Li, X. Seedling-derived leaf and root tip as alternative explants for callus induction and plant regeneration in maize. Physiol. Plant. 2021, 172, 1570–1580. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Jiang, Z.; Lu, S.; Lin, H.; Gao, S.; Peng, H.; Yuan, G.; Liu, L.; Zhang, Z.; Zhao, M.; et al. Combined small RNA and degradome sequencing reveals microRNA regulation during immature maize embryo dedifferentiation. Biochem. Biophys. Res. Commun. 2013, 441, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Ge, F.; Luo, X.; Huang, X.; Zhang, Y.; He, X.; Liu, M.; Lin, H.; Peng, H.; Li, L.; Zhang, Z.; et al. Genome-wide analysis of transcription factors involved in maize embryonic callus formation. Physiol. Plant. 2016, 158, 452–462. [Google Scholar] [CrossRef]
- Hiei, Y.; Ishida, Y.; Komari, T. Progress of cereal transformation technology mediated by Agrobacterium tumefaciens. Front. Plant Sci. 2014, 5, 628. [Google Scholar] [CrossRef] [Green Version]
- Hisano, H.; Matsuura, T.; Mod, I.C.; Yamane, M.; Sato, K. Endogenous hormone levels affect the regeneration ability of callus derived from different organs in barley. Plant Physiol. Biochem. 2016, 99, 66–72. [Google Scholar] [CrossRef]
- Fan, M.; Xu, C.; Xu, K.; Hu, Y. Lateral organ boundaries domain transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 2012, 22, 1169–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeuchi, M.; Favero, D.S.; Sakamoto, Y.; Iwase, A.; Coleman, D.; Rymen, B.; Sugimoto, K. Molecular mechanisms of plant regeneration. Annu. Rev. Plant Biol. 2019, 70, 377–406. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Kim, M.J.; Kim, N.Y.; Lee, S.H.; Kim, J. LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. Plant J. 2013, 73, 212–224. [Google Scholar] [CrossRef]
- Xu, C.; Cao, H.; Xu, E.; Zhang, S.; Hu, Y. Genome-wide identification of Arabidopsis LBD29 target genes reveals the molecular events behind auxin-induced cell reprogramming during callus formation. Plant Cell Physiol. 2018, 59, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Cao, H.; Zhang, Q.; Wang, H.; Xin, W.; Xu, E.; Zhang, S.; Yu, R.; Yu, D.; Hu, Y. Control of auxin-induced callus formation by bZIP59–LBD complex in Arabidopsis regeneration. Nat. Plants 2018, 4, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Ni, S.; Zhang, G. Transcriptome and metabolome analysis reveals regulatory networks and key genes controlling barley malting quality in responses to drought stress. Plant Physiol. Biochem. 2020, 152, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J.; et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef] [Green Version]
- Jayakodi, M.; Padmarasu, S.; Haberer, G.; Bonthala, V.S.; Gundlach, H.; Monatet, C.; Lux, T.; Kamal, N.; Lang, D.; Himmelbach, A.; et al. The barley pan-genome reveals the hidden legacy of mutation breeding. Nature 2020, 588, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Fu, L.; Su, T.; Ye, L.; Huang, L.; Kuang, L.; Wu, L.; Wu, D.; Chen, Z.; Zhang, G. Calmodulin HvCaM1 negatively regulates salt tolerance via modulation of HvHKT1s and HvCAMTA4. Plant Physiol. 2020, 183, 1650–1662. [Google Scholar] [CrossRef]
- Ganeshan, S.; Baga, M.; Harvey, B.L.; Rossnagel, B.G.; Scoles, G.J.; Chibbar, R.N. Production of multiple shoots from thidiazuron-treated mature embryos and leaf-base/apical meristems of barley (Hordeum vulgare). Plant Cell Tiss. Org. Cult. 2003, 73, 57–64. [Google Scholar] [CrossRef]
- Schreiber, M.; Mascher, M.; Wright, J.; Padmarasu, S.; Himmelbach, A.; Heavens, D.; Milne, L.; Clavijo, B.J.; Stein, N.; Waugh, R. Genome assembly of the barley ‘transformation reference’ cultivar golden promise. G3-Genes Genom. Genet. 2020, 10, 1823–1827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harwood, W.A. Advances and remaining challenges in the transformation of barley and wheat. J. Exp. Bot. 2012, 63, 1791–1798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harwood, W.A. A Protocol for High-Throughput Agrobacterium-Mediated Barley Transformation. In Cereal Genomics: Methods in Molecular Biology (Methods and Protocols); Humana Press: New York, NY, USA, 2014; Volume 1099, pp. 251–260. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, W.L.; Collins, H.M.; Singh, R.R.; Kibble, N.A.J.; Yap, K.; Taylor, J.; Fincher, G.B.; Burton, R.A. Method for hull-less barley transformation and manipulation of grain mixed-linkage beta-glucan. J. Integr. Plant Biol. 2018, 60, 382–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duclercq, J.; Sangwan-Norreel, B.; Catterou, M.; Sangwan, R.S. De novo shoot organogenesis: From art to science. Trends Plant Sci. 2011, 16, 597–606. [Google Scholar] [CrossRef]
- Kumlehn, J.; Serazetdinova, L.; Hensel, G.; Becker, D.; Loerz, H. Genetic transformation of barley (Hordeum vulgare L.) via infection of androgenetic pollen cultures with Agrobacterium tumefaciens. Plant Biotechnol. J. 2006, 4, 251–261. [Google Scholar] [CrossRef]
- Han, Y.; Broughton, S.; Liu, L.; Zhang, X.; Zeng, J.; He, X.; Li, C. Highly efficient and genotype-independent barley gene editing based on anther culture. Plant Commun. 2021, 2, 100082. [Google Scholar] [CrossRef]
- Han, Y.; Jin, X.; Wu, F.; Zhang, G. Genotypic differences in callus induction and plant regeneration from mature embryos of barley (Hordeum vulgare L.). J. Zhejiang Univ. Sci. B 2011, 12, 399–407. [Google Scholar] [CrossRef] [Green Version]
- Murray, F.; Brettell, R.; Matthews, P.; Bishop, D.; Jacobsen, J. Comparison of Agrobacterium-mediated transformation of four barley cultivars using the GFP and GUS reporter genes. Plant Cell Rep. 2004, 22, 397–402. [Google Scholar] [CrossRef]
- Zhao, J.; Li, H.; Fu, S.; Chen, B.; Sun, W.; Zhang, J.; Zhang, J. An iTRAQ-Based Proteomics Approach to Clarify the Molecular Physiology of Somatic Embryo Development in Prince Rupprecht’s Larch (Larix principis-rupprechtii Mayr). PLoS ONE 2015, 10, e0119987. [Google Scholar] [CrossRef] [Green Version]
- Botini, N.; Almeida, F.A.; Cruz, K.Z.C.M.; Reis, R.S.; Vale, E.M.; Garcia, A.B.; Santa-Catarina, C.; Silveira, V. Stage-specific protein regulation during somatic embryo development of Carica papaya L. ‘Golden’. BBA-Proteins Proteom. 2021, 1869, 140561. [Google Scholar] [CrossRef] [PubMed]
- Heringer, A.S.; Santa-Catarina, C.; Silveira, V. Insights from proteomic studies into plant somatic embryogenesis. Proteomics 2018, 18, e1700265. [Google Scholar] [CrossRef] [PubMed]
- Arnholdt-Schmitt, B.; Ragonezi, C.; Cardoso, H. Do mitochondria play a central role in stress-induced somatic embryogenesis? In In Vitro Embryogenesis in Higher Plants; Humana Press: New York, NY, USA, 2016; Volume 1359, pp. 87–100. [Google Scholar]
- Guerra, D.D.; Callis, J. Ubiquitin on the move: The ubiquitin modification system plays diverse roles in the regulation of endoplasmic reticulum- and plasma membrane-localized proteins. Plant Physiol. 2012, 160, 56–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauer, N.; Škiljaica, A.; Malenica, N.; Razdorov, G.; Klasić, M.; Juranić, M.; Močibob, M.; Sprunck, S.; Dresselhaus, T.; Levanić, D.L. The MATH-BTB protein TaMAB2 accumulates in ubiquitin-containing foci and interacts with the translation initiation machinery in Arabidopsis. Front. Plant Sci. 2019, 10, 1469. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Wang, B.; Wang, X.; Zhang, Y.; Dong, M.; Zhang, J. iTRAQ-based comparative proteomic analysis of embryogenic and non-embryogenic tissues of Prince Rupprecht’s larch (Larix principis-rupprechtii Mayr). Plant Cell Tiss. Org. Cult. 2015, 120, 655–669. [Google Scholar] [CrossRef]
- Fehér, A. Somatic embryogenesis—Stress-induced remodeling of plant cell fate. BBA-Gene Regul. Mech. 2015, 1849, 385–402. [Google Scholar] [CrossRef]
- Hu, C.; Lin, S.; Chi, W.; Charng, Y. Recent gene duplication and subfunctionalization produced a mitochondrial GrpE, the nucleotide exchange factor of the Hsp70 complex, specialized in thermotolerance to chronic heat stress in Arabidopsis. Plant Physiol. 2012, 158, 747–758. [Google Scholar] [CrossRef] [Green Version]
- Gulzar, B.; Mujib, A.; Rajam, M.V.; Frukh, A.; Zafar, N. Identification of somatic embryogenesis (SE) related proteins through label-free shotgun proteomic method and cellular role in Catharanthus roseus (L.) G. Don. Plant Cell Tiss. Org. Cult. 2019, 137, 225–237. [Google Scholar] [CrossRef]
- Zhang, J.; Ma, H.; Chen, S.; Ji, M.; Perl, A.; Kovacs, L.; Chen, S. Stress response proteins’ differential expression in embryogenic and non-embryogenic callus of Vitis vinifera L. cv. Cabernet Sauvignon—A proteomic approach. Plant Sci. 2009, 177, 103–113. [Google Scholar] [CrossRef]
- Lyngved, R.; Renaut, J.; Hausman, J.F.; Iversen, T.H.; Hvoslef-Eide, A.K. Embryo-specific proteins in Cyclamen persicum analyzed with 2-D DIGE. J. Plant Growth Regul. 2008, 27, 353. [Google Scholar] [CrossRef]
- Haq, S.; Khan, A.; Ali, M.; Gai, W.; Zhang, H.; Yu, Q.; Yang, S.; Wei, A.; Gong, Z. Knockdown of CaHSP60-6 confers enhanced sensitivity to heat stress in pepper (Capsicum annuum L.). Planta 2019, 250, 2127–2145. [Google Scholar] [CrossRef] [PubMed]
- Fraga, H.P.F.; Vieira, L.N.; Heringer, A.S.; Puttkammer, C.C.; Silveira, V.; Guerra, M.P. DNA methylation and proteome profiles of Araucaria angustifolia (Bertol.) Kuntze embryogenic cultures as affected by plant growth regulators supplementation. Plant Cell Tiss. Org. Cult. 2016, 125, 353–374. [Google Scholar] [CrossRef]
- Gong, H.; Jiao, Y.; Hu, W.; Pua, E.C. Expression of glutathione-S-transferase and its role in plant growth and development in vivo and shoot morphogenesis in vitro. Plant Mol. Biol. 2005, 57, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Vieira, L.D.; Santa-Catarina, C.; Fraga, H.P.D.; dos Santos, A.L.W.; Steinmacher, D.A.; Schlogl, P.S.; Silveira, V.; Steinera, N.; Floh, E.L.S.; Guerra, M.P. Glutathione improves early somatic embryogenesis in Araucaria angustifolia (Bert) O. Kuntze by alteration in nitric oxide emission. Plant Sci. 2012, 195, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Suo, J.; Zhou, C.; Zeng, Z.; Li, X.; Bian, H.; Wang, J.; Zhu, M.; Han, N. Identification of regulatory factors promoting embryogenic callus formation in barley through transcriptome analysis. BMC Plant Biol. 2021, 21, 145. [Google Scholar] [CrossRef]
- Stepanova, A.N.; Yun, J.; Robles, L.M.; Novak, O.; He, W.; Guo, H.; Ljung, K.; Alonso, J.M. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 2001, 23, 3961–3973. [Google Scholar] [CrossRef] [Green Version]
- Spartz, A.K.; Lee, S.H.; Wenger, J.P.; Gonzalez, N.; Itoh, H.; Inzé, D.; Peer, W.A.; Murphy, A.S.; Overvoorde, P.J.; Gray, W.M. The SAUR19 subfamily of small auxin-up RNA genes promote cell expansion. Plant J. 2012, 70, 978–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Guo, G.; Xu, H.; He, T.; Zong, Y.; Zhang, S.; Faheem, M.; Lu, R.; Zhou, L.; Liu, C. Comparative transcriptome analysis reveals compatible and recalcitrant genotypic response of barley microspore-derived embryogenic callus toward Agrobacterium infection. BMC Plant Biol. 2001, 21, 579. [Google Scholar] [CrossRef] [PubMed]
- Okushima, Y.; Fukaki, H.; Onoda, M.; Theologis, A.; Tasaka, M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 2007, 19, 118–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raspor, M.; Motyka, V.; Kaleri, A.R.; Ninković, S.; Tubić, L.; Cingel, A.; Ćosić, T. Integrating the roles for cytokinin and auxin in de novo shoot organogenesis: From hormone uptake to signaling outputs. Int. J. Mol. Sci. 2021, 22, 8554. [Google Scholar] [CrossRef]
- Su, Y.; Zhang, X. The hormonal control of regeneration in plants. Curr. Top. Dev. Biol. 2014, 108, 35–69. [Google Scholar] [PubMed]
- Perianez-Rodriguez, J.; Manzano, C.; Moreno-Risueno, M.A. Postembryonic organogenesis and plant regeneration from tissues: Two sides of the same coin? Front. Plant Sci. 2014, 5, 219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sang, Y.; Cheng, Z.; Zhang, X. Plant stem cells and de novo organogenesis. New Phytol. 2018, 218, 1334–1339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumoto-Kitano, M.; Kusumoto, T.; Tarkowski, P.; Kinoshita-Tsujimura, K.; Vaclavikova, K.; Miyawaki, K.; Kakimoto, T. Cytokinins are central regulators of cambial activity. Proc. Natl. Acad. Sci. USA 2008, 105, 20027–20031. [Google Scholar] [CrossRef] [Green Version]
- Tu, M.; Wang, W.; Yao, N.; Cai, C.; Liu, Y.; Lin, C.; Zuo, Z.; Zhu, Q. The transcriptional dynamics during de novo shoot organogenesis of Ma bamboo (Dendrocalamus latiflorus Munro): Implication of the contributions of the abiotic stress response in this process. Plant J. 2021, 107, 1513–1532. [Google Scholar] [CrossRef]
- Frank, M.; Cortleven, A.; Novak, O.; Schmulling, T. Root-derived trans-zeatin cytokinin protects Arabidopsis plants against photoperiod stress. Plant Cell Environ. 2020, 43, 2637–2649. [Google Scholar] [CrossRef] [PubMed]


| Gene id | Description | GP | ZU9 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| D5 a | D10 | D20 | Reg. b | D5 | D10 | D20 | Reg. | ||
| Auxin | |||||||||
| HORVU2Hr1G116980 | indole-3-pyruvate monooxygenase YUCCA5 | —— | —— | —— | Ns | −0.07 | −0.22 | −5.78 | Down |
| HORVU3Hr1G030390 | indole-3-pyruvate monooxygenase YUCCA6-like | −2.00 | −7.86 | −7.86 | Down | —— | —— | —— | Ns |
| HORVU5Hr1G028100 | indole-3-pyruvate monooxygenase YUCCA6-like | −8.25 | −8.25 | −1.45 | Down | —— | —— | —— | Ns |
| HORVU7Hr1G023880 | indole-3-pyruvate monooxygenase YUCCA10 | 7.90 | —— | 7.64 | Up | −6.34 | −4.39 | −11.28 | Down |
| HORVU1Hr1G072230 | tryptophan aminotransferase related 2 | 0.72 | −0.64 | −1.53 | Ns | 1.87 | 0.53 | 1.03 | Up |
| HORVU3Hr1G078620 | putative auxin efflux carrier component 5 | —— | —— | —— | Down | 6.92 | 6.07 | —— | Up |
| HORVU2Hr1G119400 | auxin-responsive protein IAA14-like | −0.18 | −0.47 | −3.61 | Down | 2.05 | 2.87 | 1.19 | Up |
| HORVU5Hr1G081180 | auxin-responsive protein IAA26-like | —— | —— | 7.29 | Up | −8.43 | −8.43 | −8.43 | Down |
| HORVU5Hr1G094240 | auxin-responsive protein IAA13-like | 5.82 | 2.28 | 4.26 | Up | 3.78 | 0.10 | −0.47 | Down |
| HORVU5Hr1G076740 | auxin-responsive protein SAUR50-like | 7.45 | —— | —— | Up | 2.09 | 0.66 | −1.11 | Down |
| HORVU5Hr1G085540 | auxin-responsive protein SAUR36 | 9.53 | —— | —— | Up | −6.46 | −6.46 | −6.46 | Down |
| Cytokinin | |||||||||
| HORVU3Hr1G063960 | adenylate isopentenyl transferase 1 | 11.67 | 9.67 | 10.55 | Up | 3.06 | −0.48 | −0.82 | Down |
| HORVU0Hr1G005240 | cytokinin dehydrogenase 8-like | 11.54 | —— | —— | Ns | —— | —— | 11.38 | Up |
| HORVU1Hr1G057860 | cytokinin dehydrogenase 2 | —— | 4.98 | 4.71 | Up | −0.91 | −1.50 | −2.27 | Down |
| HORVU2Hr1G090140 | cytokinin dehydrogenase 8-like | −8.29 | −8.29 | −3.40 | Down | —— | 5.48 | —— | Up |
| HORVU3Hr1G027460 | cytokinin oxidase/dehydrogenase | −8.58 | −8.58 | −8.58 | Down | —— | —— | —— | Ns |
| HORVU6Hr1G039680 | cytokinin oxidase/dehydrogenase | —— | —— | —— | Ns | −7.12 | −7.12 | −7.12 | Down |
| HORVU6Hr1G039690 | cytokinin oxidase/dehydrogenase | —— | —— | —— | Ns | −8.38 | −8.38 | −8.38 | Down |
| HORVU7Hr1G086710 | cytokinin dehydrogenase 10-like | —— | —— | 7.77 | Up | −6.64 | −6.64 | −6.64 | Down |
| HORVU2Hr1G064940 | histidine kinase 5 | −0.71 | −0.48 | −9.35 | Down | —— | —— | —— | Ns |
| HORVU3Hr1G069130 | histidine kinase 1 | 7.31 | 7.54 | 6.54 | Up | —— | —— | —— | Ns |
| HORVU5Hr1G012530 | histidine kinase 4 | −2.91 | 2.59 | 4.53 | Up | −2.10 | −1.86 | −1.50 | Down |
| HORVU4Hr1G001670 | histidine-containing phosphotransfer protein 2-like | —— | 10.32 | 11.00 | Up | —— | —— | —— | Ns |
| HORVU4Hr1G074350 | histidine-containing phosphotransfer protein 2-like | —— | 8.67 | 11.46 | Up | —— | —— | —— | Ns |
| HORVU7Hr1G049600 | histidine-containing phosphotransfer protein 2-like | 7.30 | 9.62 | —— | Up | —— | —— | —— | Ns |
| HORVU7Hr1G114450 | two-component response regulator ARR2 | —— | 6.79 | 6.40 | Up | —— | —— | —— | Ns |
| Gibberellin | |||||||||
| HORVU5Hr1G082380 | ent-kaur-16-ene synthase, chloroplastic isoform X1 | —— | —— | —— | Ns | 8.59 | 10.13 | 6.24 | Up |
| HORVU7Hr1G101720 | gibberellin 2-beta-dioxygenase 6-like | —— | —— | 7.25 | Up | −9.98 | −9.98 | −2.20 | Down |
| HORVU1Hr1G090800 | DELLA protein SLR1-like | —— | —— | 5.28 | Up | −6.81 | −6.81 | −0.50 | Down |
| Abscisic acid | |||||||||
| HORVU1Hr1G074200 | putative aldehyde oxidase-like protein | 10.37 | 9.08 | —— | Up | 1.30 | −1.71 | −7.62 | Down |
| HORVU3Hr1G062290 | aldehyde oxidase GLOX-like | —— | —— | —— | Ns | −6.48 | −6.48 | −6.48 | Down |
| HORVU4Hr1G019740 | aldehyde oxidase GLOX-like | 1.09 | 0.66 | −2.16 | Down | 11.44 | 9.54 | 8.83 | Up |
| HORVU0Hr1G032390 | abscisic acid receptor 8 | 11.57 | —— | 10.34 | Up | −0.64 | 0.23 | −10.94 | Down |
| HORVU3Hr1G040680 | abscisic acid receptor PYL2-like | 10.61 | —— | 8.75 | Up | 0.37 | −1.08 | −8.88 | Down |
| HORVU7Hr1G029040 | putative protein phosphatase 2C 6 | —— | 7.12 | 7.06 | Up | −0.75 | −0.29 | −1.30 | Down |
| HORVU0Hr1G032840 | serine/threonine-protein kinase SAPK4-like | —— | —— | 7.32 | Up | —— | —— | —— | Ns |
| Ethylene | |||||||||
| HORVU3Hr1G098750 | ACC synthase | —— | —— | —— | Ns | −3.93 | −4.65 | −0.97 | Down |
| HORVU0Hr1G001750 | ethylene receptor, partial | 7.19 | —— | —— | Up | −9.24 | −1.07 | −9.24 | Down |
| HORVU3Hr1G093140 | serine/threonine-protein kinase CTR1 | 0.79 | 0.42 | −3.29 | Ns | 2.67 | 2.12 | 2.07 | Up |
| HORVU2Hr1G098250 | ethylene-responsive transcription factor ERF105-like | 0.91 | 1.45 | 0.21 | Ns | 0.68 | 2.63 | 2.72 | Up |
| HORVU4Hr1G015350 | ethylene-responsive transcription factor ERF022-like | 1.74 | 2.19 | 0.55 | Up | 6.02 | 4.67 | 4.23 | Up |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Z.; Wang, F.; Tu, Y.; Xu, Y.; Shen, Q.; Zhang, G. Transcriptome Analysis Reveals Genetic Factors Related to Callus Induction in Barley. Agronomy 2022, 12, 749. https://doi.org/10.3390/agronomy12030749
Xu Z, Wang F, Tu Y, Xu Y, Shen Q, Zhang G. Transcriptome Analysis Reveals Genetic Factors Related to Callus Induction in Barley. Agronomy. 2022; 12(3):749. https://doi.org/10.3390/agronomy12030749
Chicago/Turabian StyleXu, Zhengyuan, Fengyue Wang, Yishan Tu, Yunfeng Xu, Qiufang Shen, and Guoping Zhang. 2022. "Transcriptome Analysis Reveals Genetic Factors Related to Callus Induction in Barley" Agronomy 12, no. 3: 749. https://doi.org/10.3390/agronomy12030749
APA StyleXu, Z., Wang, F., Tu, Y., Xu, Y., Shen, Q., & Zhang, G. (2022). Transcriptome Analysis Reveals Genetic Factors Related to Callus Induction in Barley. Agronomy, 12(3), 749. https://doi.org/10.3390/agronomy12030749








