Genetic Mapping and Candidate Gene Prediction of a QTL Related to Early Heading on Wild Emmer Chromosome 7BS in the Genetic Background of Common Wheat
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Genome and Transcriptome Sequencing for Checking the Chromosome Constitution
2.3. Experiment for Vernalization and Photoperiod Response
2.4. Observation of Wheat Young Spike Development
2.5. Field and Growth Room Experiment of the F2 Population from a Cross between CASL7BS and CS
2.6. Map Construction and QTL Mapping
3. Results
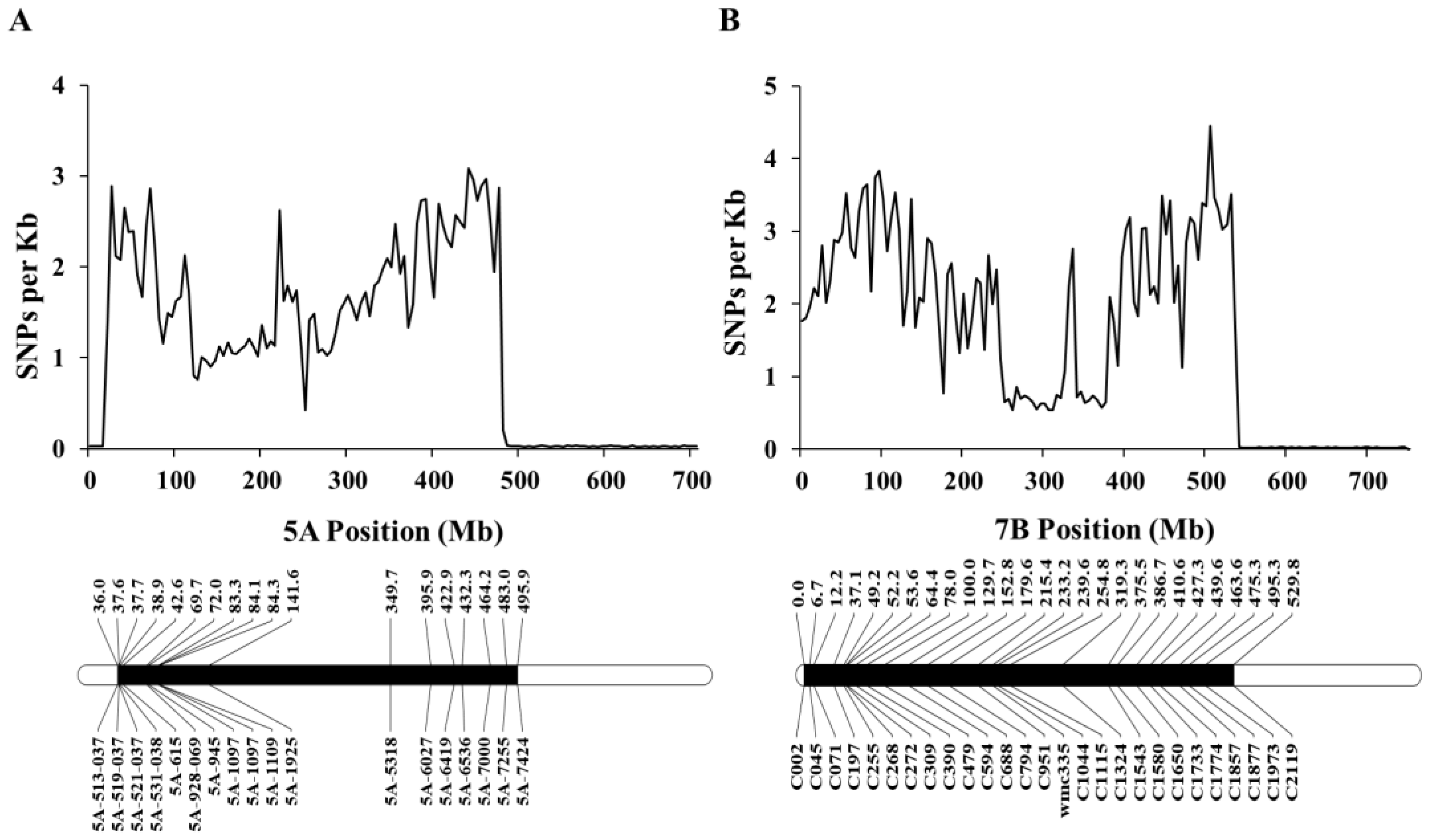
3.1. Chromosome Constitution of CASL7BS Identified by Transcriptome and Genome Sequencing
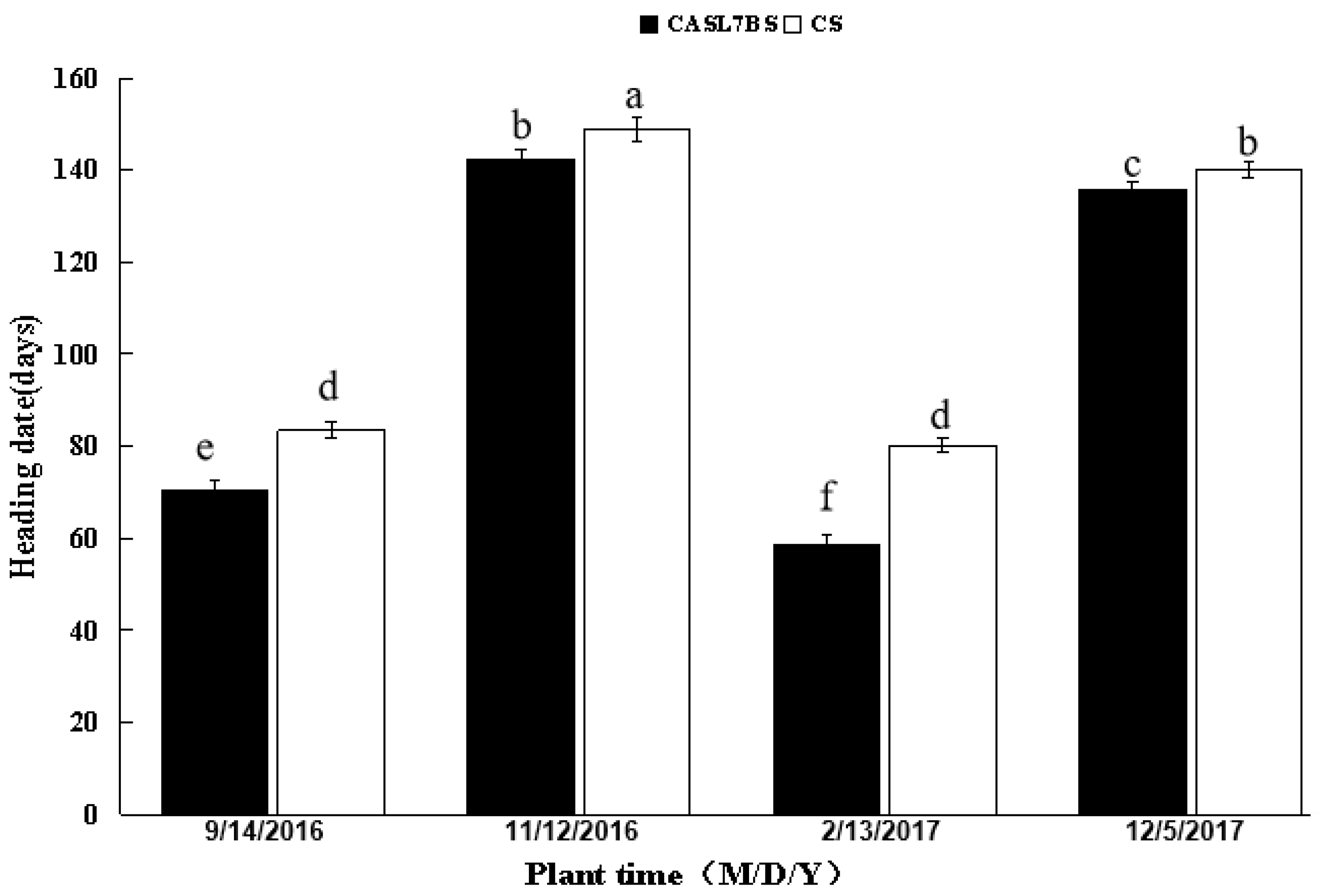
3.2. Consistent Early Heading of CASL7BS Compared to CS in Different Environments
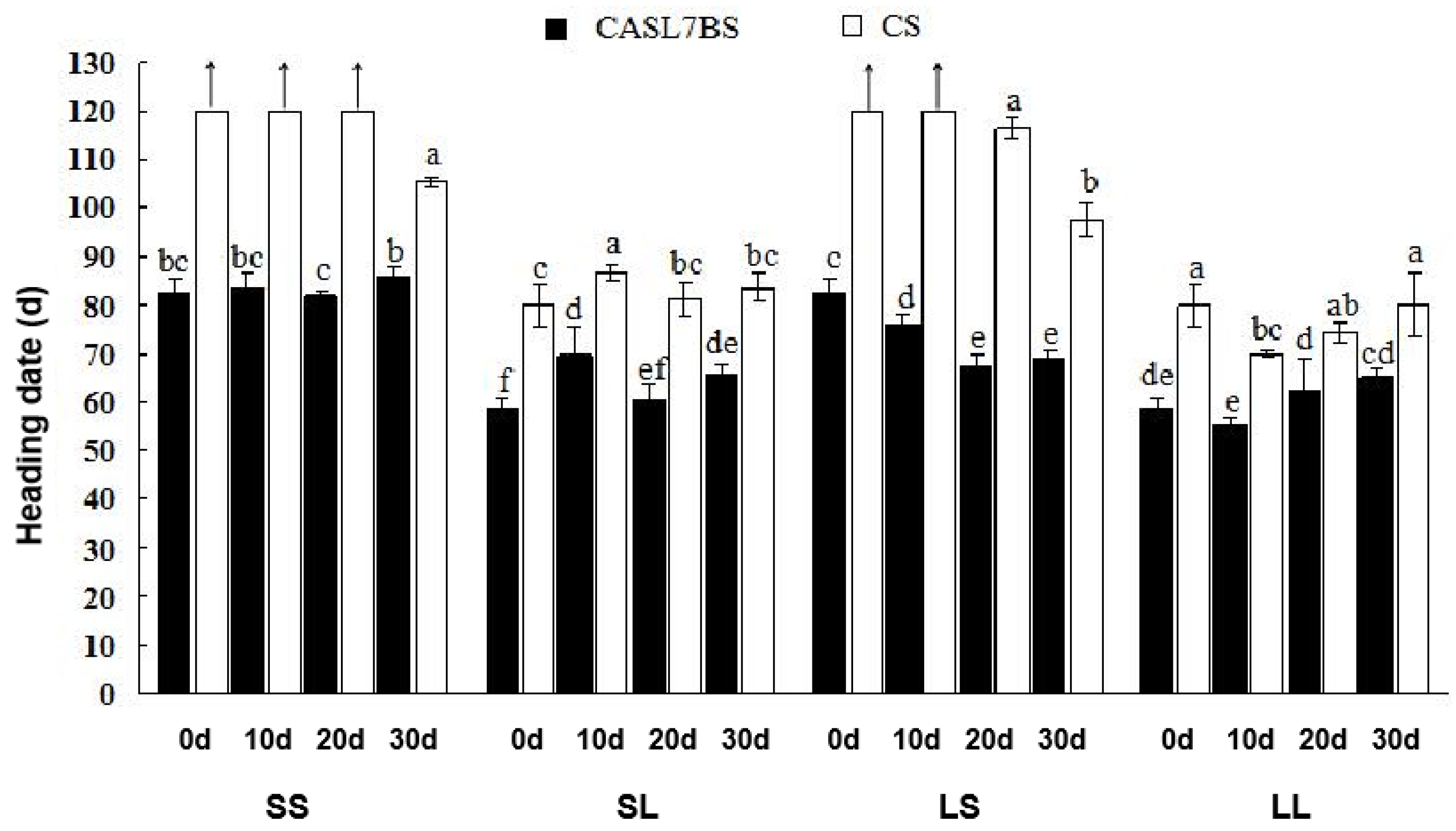
3.3. Different Responses of CS and CASL7BS to Vernalization and Photoperiod
3.4. Comparison of Young Spike Development between CS and CASL7BS
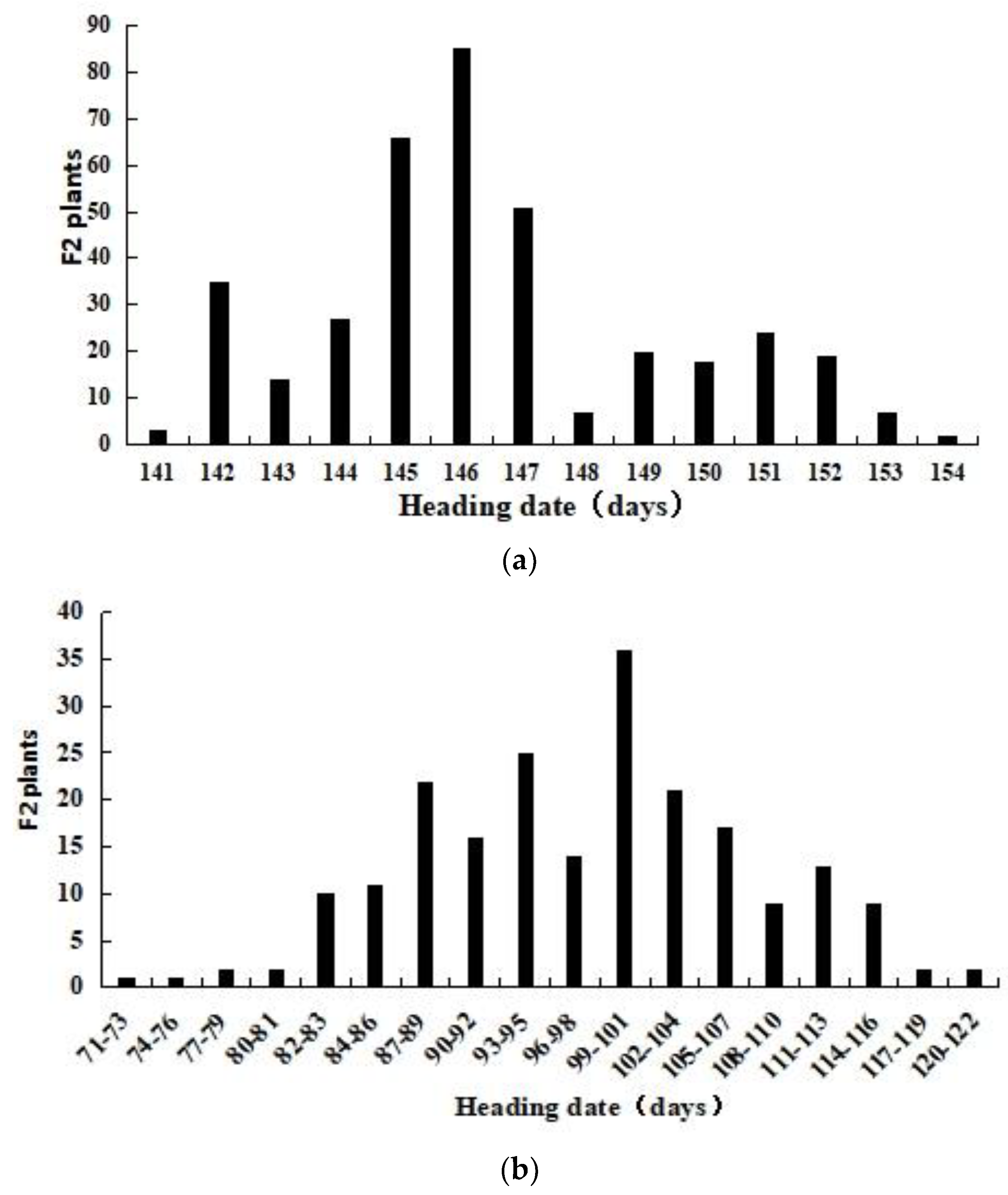
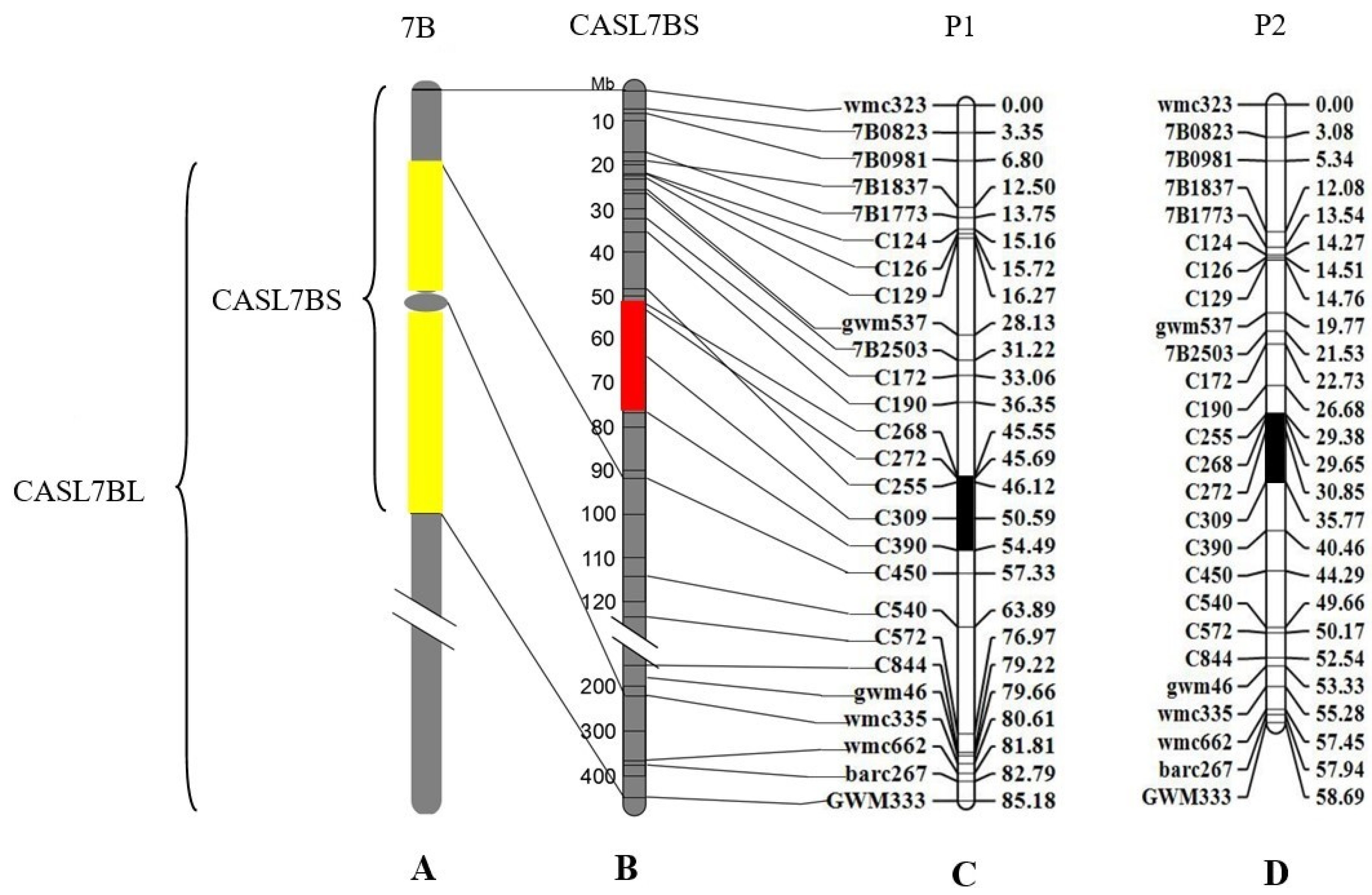
3.5. Genetic Mapping of the QTL Related to Early HD in CASL7BS Using F2 Population Derived from a Cross between CASL7BS and CS
3.6. Further Narrowing down the HD-Related QTL to about 3 Mb Region of Chromosome 7BS Using F3 and F4 Families
3.7. Candidate Gene Prediction
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lin, F.; Xue, S.L.; Tian, D.G.; Li, C.J.; Cao, Y.; Zhang, Z.Z.; Zhang, C.Q.; Ma, Z.Q. Mapping chromosomal regions affecting flowering time in a spring wheat RIL population. Euphytica 2008, 164, 769–777. [Google Scholar] [CrossRef]
- Kamran, A.; Iqbal, M.; Spaner, D. Flowering time in wheat (Triticum aestivum L.): A key factor for global adaptability. Euphytica 2014, 197, 1–26. [Google Scholar] [CrossRef]
- Shenoda, J.E.; Sanad, M.N.M.E.; Rizkalla, A.A.; El-Assal, S.; Ali, R.T.; Hussein, M.H. Effect of long-term heat stress on grain yield, pollen grain viability and germinability in bread wheat (Triticum aestivum L.) under field conditions. Heliyon 2021, 7, e07096. [Google Scholar] [CrossRef] [PubMed]
- Slafer, G.; Calderini, D.; Miralles, D. Yield Components and Compensation in Wheat: Opportunities for Further Increasing Yield Potential; CIMMYT: Mexico City, Mexico, 1996; pp. 101–133. [Google Scholar]
- Stelmakh, A.F. Genetic systems regulating flowering response in wheat. In Wheat: Prospects for Global Improvement, Proceedings of the 5th International Wheat Conference, Ankara, Turkey, 10–14 June 1996; Braun, H.J., Altay, F., Kronstad, W.E., Beniwal, S.P.S., McNab, A., Eds.; Springer: Dordrecht, The Netherlands, 1997; pp. 491–501. [Google Scholar]
- Wang, C.; Tian, Q.; Hou, Z.; Mucha, M.; Aukerman, M.; Olsen, O.A. The Arabidopsis thaliana AT PRP39-1 gene, encoding a tetratricopeptide repeat protein with similarity to the yeast pre-mRNA processing protein PRP39, affects flowering time. Plant Cell Rep. 2007, 26, 1357–1366. [Google Scholar] [CrossRef] [PubMed]
- Nishida, H.; Yoshida, T.; Kawakami, K.; Fujita, M.; Long, B.; Akashi, Y.; Laurie, D.A.; Kato, K. Structural variation in the 5′ upstream region of photoperiod-insensitive alleles Ppd-A1a and Ppd-B1a identified in hexaploid wheat (Triticum aestivum L.), and their effect on heading time. Mol. Breed. 2013, 31, 27–37. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, Z.; Cheng, M.; Chen, J.; Zhu, T.; Wang, R.; Liu, Y.; Qi, P.; Chen, G.; Jiang, Q.; et al. Uncovering the dispersion history, adaptive evolution and selection of wheat in China. Plant Biotechnol. J. 2018, 16, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Distelfeld, A.; Li, C.; Dubcovsky, J. Regulation of flowering in temperate cereals. Curr. Opin. Plant Biol. 2009, 12, 178–184. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Gao, M.; Zhang, J.; Zuo, A.; Shang, X.; Cui, D. Molecular characterization of vernalization and response genes in bread wheat from the Yellow and Huai Valley of China. BMC Plant Biol. 2013, 13, 199. [Google Scholar] [CrossRef] [Green Version]
- Yan, L.; Loukoianov, A.; Blechl, A.; Tranquilli, G.; Ramakrishna, W.; SanMiguel, P.; Bennetzen, J.L.; Echenique, V.; Dubcovsky, J. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 2004, 303, 1640–1644. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, T.; Nishida, H.; Zhu, J.; Nitcher, R.; Distelfeld, A.; Akashi, Y.; Kato, K.; Dubcovsky, J. Vrn-D4 is a vernalization gene located on the centromeric region of chromosome 5D in hexaploid wheat. Theor. Appl. Genet. 2010, 120, 543–552. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Dong, Q.; Deng, M.; Lin, D.; Xiao, J.; Cheng, P.; Xing, L.; Niu, Y.; Gao, C.; Zhang, W.; et al. The vernalization-induced long non-coding RNA VAS functions with the transcription factor TaRF2b to promote TaVRN1 expression for flowering in hexaploid wheat. Mol. Plant 2021, 14, 1525–1538. [Google Scholar] [CrossRef] [PubMed]
- Diallo, A.O.; Ali-Benali, M.A.; Badawi, M.; Houde, M.; Sarhan, F. Expression of vernalization responsive genes in wheat is associated with histone H3 trimethylation. Mol. Genet. Genom. 2012, 287, 575–590. [Google Scholar] [CrossRef] [PubMed]
- Adam, H.; Ouellet, F.; Kane, N.A.; Agharbaoui, Z.; Major, G.; Tominaga, Y.; Sarhan, F. Overexpression of TaVRN1 in Arabidopsis promotes early flowering and alters development. Plant Cell Physiol. 2007, 48, 1192–1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butterworth, K.; Reid, A.; Sayers, L.; Worland, A.J. A comparison of the pleiotropic effects of photoperiod insensitive genes Ppd-A1, Ppd-B1 and Ppd-D1 in a Mercia winter wheat background. Ann. Wheat News 2001, 47, 230–231. [Google Scholar]
- Shaw, L.M.; Turner, A.S.; Laurie, D.A. The impact of photoperiod insensitive Ppd-1a mutations on the photoperiod pathway across the three genomes of hexaploid wheat (Triticum aestivum). Plant J. 2012, 71, 71–84. [Google Scholar] [CrossRef]
- Zhou, W.; Wu, S.; Ding, M.; Li, J.; Shi, Z.; Wei, W.; Guo, J.; Zhang, H.; Jiang, Y.; Rong, J. Mapping of Ppd-B1, A major candidate gene for late heading on wild emmer chromosome arm 2bs and assessment of its interactions with early heading QTLs on 3AL. PLoS ONE 2016, 11, e0147377. [Google Scholar] [CrossRef] [Green Version]
- Scarth, R.; Law, C.N. The location of the photoperiod gene, Ppd2 and an additional genetic factor for ear-emergence time on chromosome 2B of wheat. Heredity 1983, 51, 607–619. [Google Scholar] [CrossRef]
- Worland, A.J.; Börner, A.; Korzun, V.; Li, W.M.; Petrovíc, S.; Sayers, E.J. The influence of photoperiod genes on the adaptability of European winter wheats. Euphytica 1998, 100, 385–394. [Google Scholar] [CrossRef]
- Royo, C.; Dreisigacker, S.; Alfaro, C.; Ammar, K.; Villegas, D. Effect of Ppd-1 genes on durum wheat flowering time and grain filling duration in a wide range of latitudes. J. Agric. Sci. 2016, 154, 612–631. [Google Scholar] [CrossRef] [Green Version]
- Beales, J.; Turner, A.; Griffiths, S.; Snape, J.W.; Laurie, D.A. A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor. Appl. Genet. 2007, 115, 721–733. [Google Scholar] [CrossRef]
- Khlestkina, E.K.; Giura, A.; Röder, M.S.; Börner, A. A new gene controlling the flowering response to photoperiod in wheat. Euphytica 2009, 165, 579–585. [Google Scholar] [CrossRef]
- Wang, B.; Jin, S.H.; Hu, H.Q.; Sun, Y.G.; Wang, Y.W.; Han, P.; Hou, B.K. UGT87A2, an Arabidopsis glycosyltransferase, regulates flowering time via FLOWERING LOCUS C. New Phytol. 2012, 194, 666–675. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Wada, T. Genetic analysis and selection experiment for narrow-sense earliness in wheat by using segregating hybrid progenies. Breed. Sci. 1999, 49, 233–238. [Google Scholar] [CrossRef]
- Gawroński, P.; Ariyadasa, R.; Himmelbach, A.; Poursarebani, N.; Kilian, B.; Stein, N.; Steuernagel, B.; Hensel, G.; Kumlehn, J.; Sehgal, S.K.; et al. A Distorted Circadian Clock Causes Early Flowering and Temperature-Dependent Variation in Spike Development in the Eps-3Am Mutant of Einkorn Wheat. Genetics 2014, 196, 1253–1261. [Google Scholar] [CrossRef] [Green Version]
- Zikhali, M.; Wingen, L.U.; Griffiths, S. Delimitation of the Earliness per se D1 (Eps-D1) flowering gene to a subtelomeric chromosomal deletion in bread wheat (Triticum aestivum). J. Exp. Bot. 2015, 67, 287–299. [Google Scholar] [CrossRef] [Green Version]
- Gawroński, P.; Schnurbusch, T. High-density mapping of the earliness per se-3Am (Eps-3Am) locus in diploid einkorn wheat and its relation to the syntenic regions in rice and Brachypodiumdistachyon L. Mol. Breed. 2012, 30, 1097–1108. [Google Scholar] [CrossRef]
- Bullrich, L.; Appendino, M.; Tranquilli, G.; Lewis, S.; Dubcovsky, J. Mapping of a thermo-sensitive earliness per se gene on Triticum monococcum chromosome 1Am. Theor. Appl. Genet. 2002, 105, 585–593. [Google Scholar] [CrossRef]
- van Beem, J.; Mohler, V.; Lukman, R.; van Ginkel, M.; William, M.; Crossa, J.; Worland, A.J. Analysis of genetic factors influencing the developmental rate of globally important CIMMYT wheat cultivars. Crop Sci. 2005, 45, 2113–2119. [Google Scholar] [CrossRef]
- Feldman, M.; Millet, E.; Abbo, S.; Balfourier, F.; Perretant, M. Exploitation of wild emmer wheat to increase yield and protein content in durum and common wheat. In Proceedings of the Genetic Resources Section Meeting of EUCARPIA, Clermont-Ferrand, France, 15–18 March 1994; pp. 151–161. [Google Scholar]
- Nevo, E. Genomic diversity in nature and domestication. In Plant Diversity Evolution: Genotypic Phenotypic Variation in Higher Plants; CABI ebook: Lismore, NSW, Australia, 2005; pp. 287–315. [Google Scholar]
- Xie, W.; Nevo, E. Wild emmer: Genetic resources, gene mapping and potential for wheat improvement. Euphytica 2008, 164, 603–614. [Google Scholar] [CrossRef]
- Ahmadi, H.; Nazarian, F. The inheritance and chromosomal location of morphological traits in wild wheat, Triticum turgidum L. ssp. dicoccoides. Euphytica 2007, 158, 103–108. [Google Scholar] [CrossRef]
- Millet, E.; Rong, J.K.; Qualset, C.O.; McGuire, P.E.; Bernard, M.; Sourdille, P.; Feldman, M. Production of chromosome-arm substitution lines of wild emmer in common wheat. Euphytica 2013, 190, 1–17. [Google Scholar] [CrossRef]
- Zhou, Y.; Cao, H.; Hou, M.; Nirasawa, S.; Tatsumi, E.; Foster, T.J.; Cheng, Y. Effect of konjac glucomannan on physical and sensory properties of noodles made from low-protein wheat flour. Food Res. Int. 2013, 51, 879–885. [Google Scholar] [CrossRef]
- Sinclair-Waters, M.; Bradbury, I.R.; Morris, C.J.; Lien, S.; Kent, M.P.; Bentzen, P. Ancient chromosomal rearrangement associated with local adaptation of a postglacially colonized population of Atlantic Cod in the northwest Atlantic. Mol. Ecol. 2018, 27, 339–351. [Google Scholar] [CrossRef]
- Yu, S.; Wu, J.; Wang, M.; Shi, W.; Xia, G.; Jia, J.; Kang, Z.; Han, D. Haplotype variations in QTL for salt tolerance in Chinese wheat accessions identified by marker-based and pedigree-based kinship analyses. Crop J. 2020, 8, 1011–1024. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Appels, R.; Eversole, K.; Stein, N.; Feuillet, C.; Keller, B.; Rogers, J.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; Poland, J.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Hatta, A.; Periyannan, S.; Lagudah, E.; Wulff, B.B.H. Isolation of Wheat Genomic DNA for Gene Mapping and Cloning. In Wheat Rust Diseases. Methods in Molecular Biology; Periyannan, S., Ed.; Humana Press: New York, NY, USA, 2017; Volume 1659, pp. 207–213. [Google Scholar]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef] [Green Version]
- Law, C.N.; Wolfe, M.S. Location of genetic factors for mildew resistance and ear emergence time on chromosome 7B of wheat. Can. J. Genet. Cytol. 1966, 8, 462–470. [Google Scholar] [CrossRef]
- Li, S.; Wang, J.; Zhang, L. Inclusive Composite Interval Mapping of QTL by Environment Interactions in Biparental Populations. PLoS ONE 2015, 10, e0132414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosová, K.; Prášil, I.T.; Vítámvás, P. The relationship between vernalization-and photoperiodically-regulated genes and the development of frost tolerance in wheat and barley. Biol. Plant. 2008, 52, 601. [Google Scholar] [CrossRef]
- Yan, L.; Fu, D.; Li, C.; Blechl, A.; Tranquilli, G.; Bonafede, M.; Sanchez, A.; Valarik, M.; Yasuda, S.; Dubcovsky, J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. USA 2006, 103, 19581–19586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maccaferri, M.; Sanguineti, M.C.; Corneti, S.; Ortega, J.L.A.; Salem, M.B.; Bort, J.; DeAmbrogio, E.; del Moral, L.F.G.; Demontis, A.; El-Ahmed, A.; et al. Quantitative trait loci for grain yield and adaptation of durum wheat (Triticum durum Desf.) across a wide range of water availability. Genetics 2008, 178, 489–511. [Google Scholar] [CrossRef] [Green Version]
- Bennett, D.; Izanloo, A.; Edwards, J.; Kuchel, H.; Chalmers, K.; Tester, M.; Reynolds, M.; Schnurbusch, T.; Langridge, P. Identification of novel quantitative trait loci for days to ear emergence and flag leaf glaucousness in a bread wheat (Triticum aestivum L.) population adapted to southern Australian conditions. Theor. Appl. Genet. 2012, 124, 697–711. [Google Scholar] [CrossRef]
- Dubcovsky, J.; Lijavetzky, D.; Appendino, L.; Tranquilli, G.E. Comparative RFLP mapping of Triticum monococcum genes controlling vernalization requirement. Theor. Appl. Genet. 1998, 97, 968–975. [Google Scholar] [CrossRef] [Green Version]
- Chen, A.; Dubcovsky, J. Wheat tilling mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genet. 2012, 8, e1003134. [Google Scholar] [CrossRef] [Green Version]
- Yoo, S.Y.; Kim, Y.; Kim, S.Y.; Lee, J.S.; Ahn, J.H. Control of flowering time and cold response by a NAC-domain protein in Arabidopsis. PLoS ONE 2007, 2, e642. [Google Scholar] [CrossRef]
- Ando, E.; Ohnishi, M.; Wang, Y.; Matsushita, T.; Watanabe, A.; Hayashi, Y.; Fujii, M.; Ma, J.F.; Inoue, S.-I.; Kinoshita, T. TWIN SISTER OF FT, GIGANTEA, and CONSTANS Have a Positive but Indirect Effect on Blue Light-Induced Stomatal Opening in Arabidopsis. Plant Physiol. 2013, 162, 1529–1538. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Lu, S.; Wang, Y.; Zhang, X.; Lv, B.; Luo, L.; Xi, D.; Shen, J.; Ma, H.; Ming, F. OsNAC2 encoding a NAC transcription factor that affects plant height through mediating the gibberellic acid pathway in rice. Plant J. 2015, 82, 302–314. [Google Scholar] [CrossRef]
- Jang, K.; Gil Lee, H.; Jung, S.-J.; Paek, N.-C.; Seo, P.J. The E3 Ubiquitin Ligase COP1 Regulates Thermosensory Flowering by Triggering GI Degradation in Arabidopsis. Sci. Rep. 2015, 5, 12071. [Google Scholar] [CrossRef]
- Galvão, V.C.; Horrer, D.; Küttner, F.; Schmid, M. Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development 2012, 139, 4072–4082. [Google Scholar] [CrossRef] [Green Version]




| Development Stage | Long Day | Short Day | ||
|---|---|---|---|---|
| CASL7BS | CS | CASL7BS | CS | |
| Elongation stage | 8 | 12 | 13 | 16 |
| Single ridge stage | 10 | 12 | 15 | 18 |
| Two ridge stage | 18 | 24 | 12 | 16 |
| Glume protuberances stage | 3 | 7 | 3 | 5 |
| Floret differentiation stage | 3 | 3 | 2 | 7 |
| Pistil and stamen differentiation stage | 2 | 2 | 2 | 6 |
| Connective formation stage | 1 | 2 | 2 | 3 |
| Tetrad formation stage | 2 | 4 | 2 | 5 |
| Total | 47 | 66 | 51 | 76 |
| Year | Parents | F2 Population | |||||
|---|---|---|---|---|---|---|---|
| Mean ± SD (d) | Range (d) | CV (%) | Mean ± SD (d) | Range (d) | CV (%) | ||
| 2016–2017 (field) | CASL7BS | 142.5 ± 1.87 b | 140–145 | 1.31 | 146.5 ± 2.89 a | 141–154 | 2.0 |
| CS | 148.8 ± 2.48 a | 146–153 | 1.67 | ||||
| 2017 (growth room) | CASL7BS | 58.7 ± 2.16 f | 56–62 | 3.68 | 97.7 ± 9.87 c | 69–122 | 4.1 |
| CS | 80.2 ± 1.47 d | 78–82 | 1.83 | ||||
| Year | F2 Population | Interval | Distance (cM) | LOD | Additive | Phenotypic Contribution (%) |
|---|---|---|---|---|---|---|
| 2016–2017 (field) | P1 | C268–C390 | 8.94 | 7.68 | 1.09 | 9.14 |
| 2017 (growth room) | P2 | C272–C309 | 4.92 | 5.75 | 4.91 | 12.35 |
| F2:3 and Parents | F3 Plants | Range (d) | Mean ± SD (d) | CV% | Heading Date |
|---|---|---|---|---|---|
| C609 | 20 | 73–84 | 78.3 ± 3.3 | 4.22 | Segregation |
| C631 | 22 | 79–89 | 83.4 ± 3.0 | 3.59 | Late |
| C614 | 45 | 67–91 | 78.8 ± 6.4 | 8.07 | Segregation |
| C653 | 31 | 68–76 | 72.3 ± 2.4 | 3.33 | Early |
| C626 | 69 | 67–87 | 77.7 ± 5.9 | 7.57 | Segregation |
| C675 | 21 | 80–87 | 83.9 ± 3.2 | 3.82 | Late |
| C785 | 27 | 77–89 | 83.1 ± 3.5 | 4.15 | Late |
| 1278 | 34 | 68–89 | 78.2 ± 7.7 | 9.89 | Segregation |
| CASL7BS | 10 | 67–71 | 68.3 ± 1.1 | 1.54 | Early |
| CS | 10 | 95–100 | 97.3 ± 1.6 | 1.60 | Late |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, F.; Chen, M.; Zhao, Y.; Wu, S.; Yasir, M.; Zhang, H.; Hu, X.; Rong, J. Genetic Mapping and Candidate Gene Prediction of a QTL Related to Early Heading on Wild Emmer Chromosome 7BS in the Genetic Background of Common Wheat. Agronomy 2022, 12, 1089. https://doi.org/10.3390/agronomy12051089
Lu F, Chen M, Zhao Y, Wu S, Yasir M, Zhang H, Hu X, Rong J. Genetic Mapping and Candidate Gene Prediction of a QTL Related to Early Heading on Wild Emmer Chromosome 7BS in the Genetic Background of Common Wheat. Agronomy. 2022; 12(5):1089. https://doi.org/10.3390/agronomy12051089
Chicago/Turabian StyleLu, Fei, Manxia Chen, Yanhao Zhao, Shaoxin Wu, Muhammad Yasir, Hua Zhang, Xin Hu, and Junkang Rong. 2022. "Genetic Mapping and Candidate Gene Prediction of a QTL Related to Early Heading on Wild Emmer Chromosome 7BS in the Genetic Background of Common Wheat" Agronomy 12, no. 5: 1089. https://doi.org/10.3390/agronomy12051089
APA StyleLu, F., Chen, M., Zhao, Y., Wu, S., Yasir, M., Zhang, H., Hu, X., & Rong, J. (2022). Genetic Mapping and Candidate Gene Prediction of a QTL Related to Early Heading on Wild Emmer Chromosome 7BS in the Genetic Background of Common Wheat. Agronomy, 12(5), 1089. https://doi.org/10.3390/agronomy12051089






