Silage Grass Sward Nitrogen Concentration and Dry Matter Yield Estimation Using Deep Regression and RGB Images Captured by UAV
Abstract
:1. Introduction
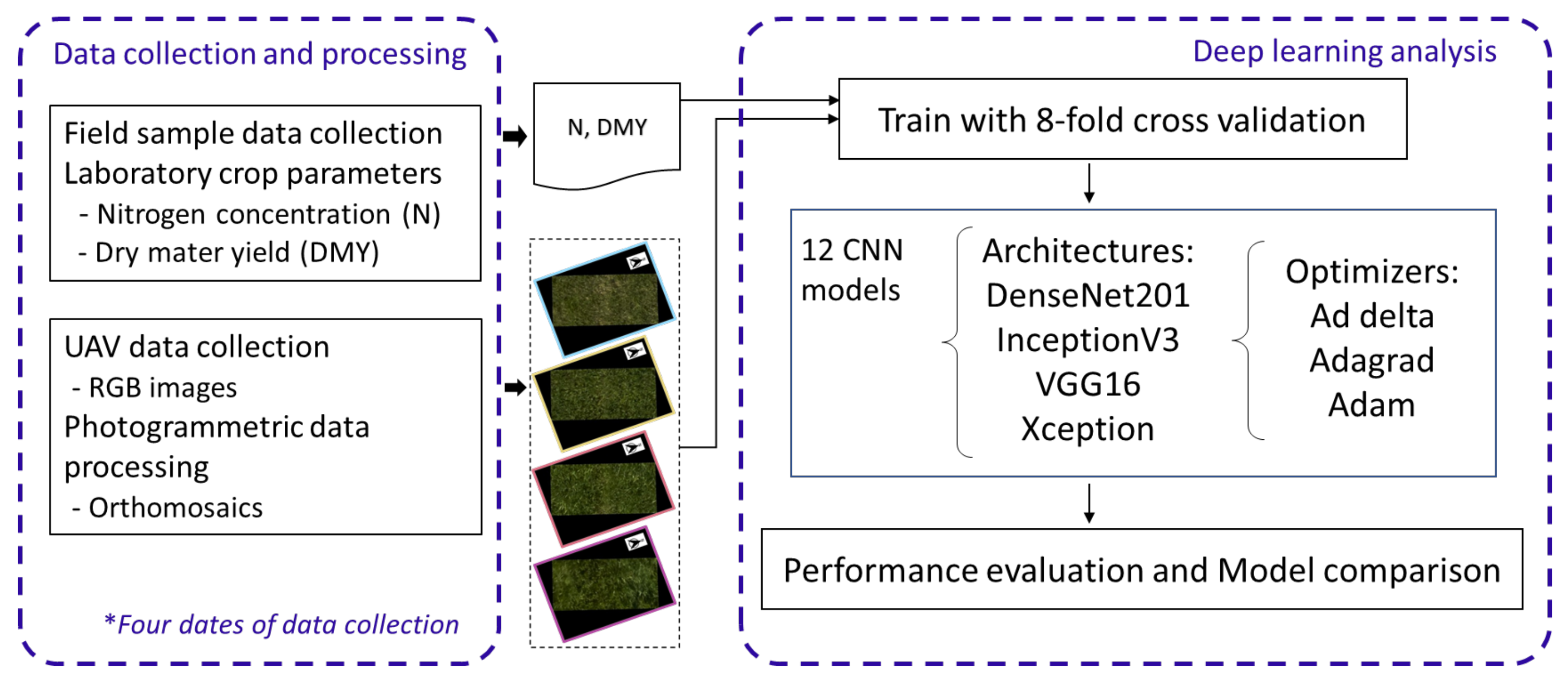
2. Materials and Methods
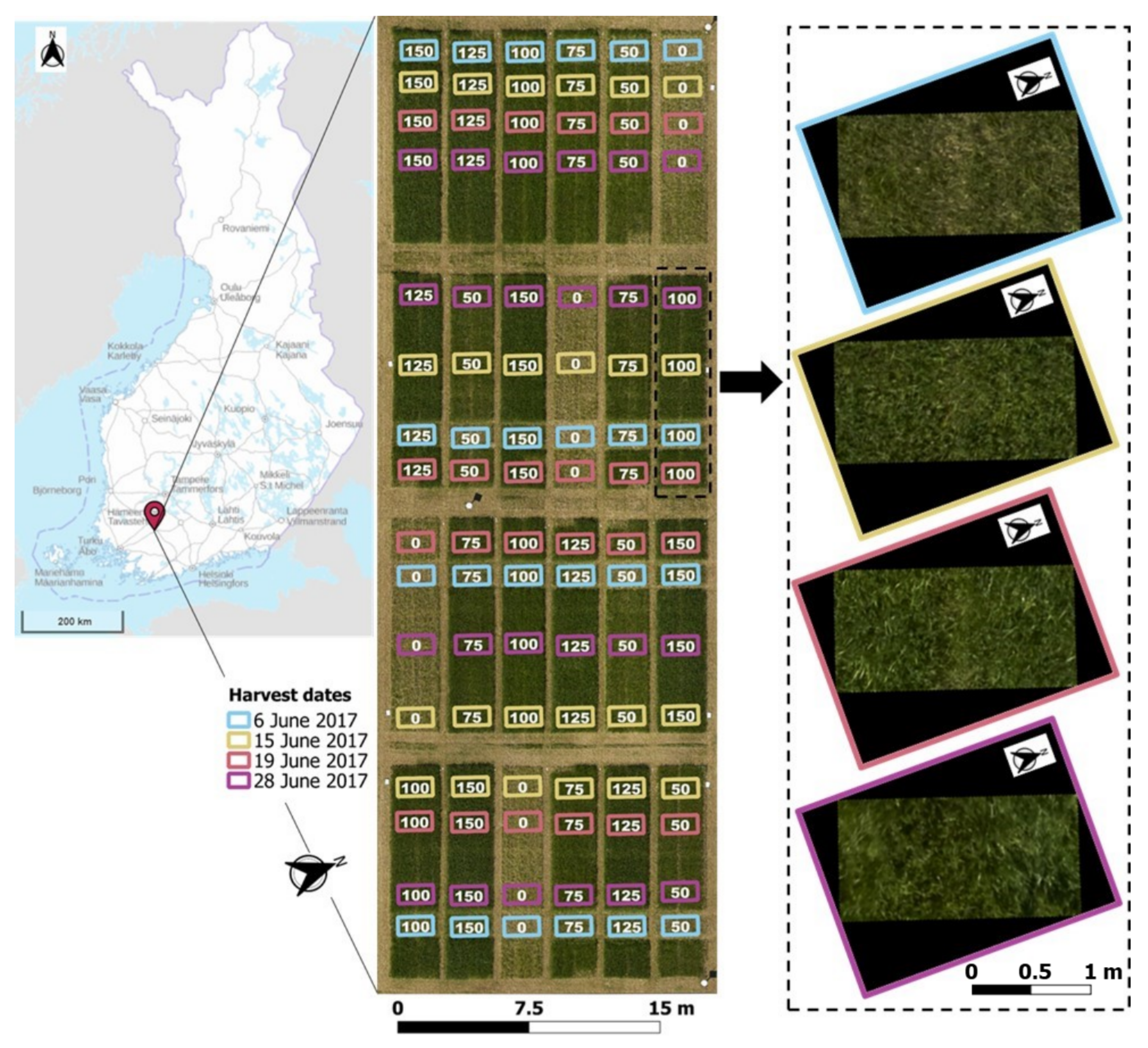
2.1. Study Area and Experimental Design
2.2. UAV Data Collection and Processing
2.3. Deep Regression Models and Optimizers
2.4. Experimental Setup
3. Results
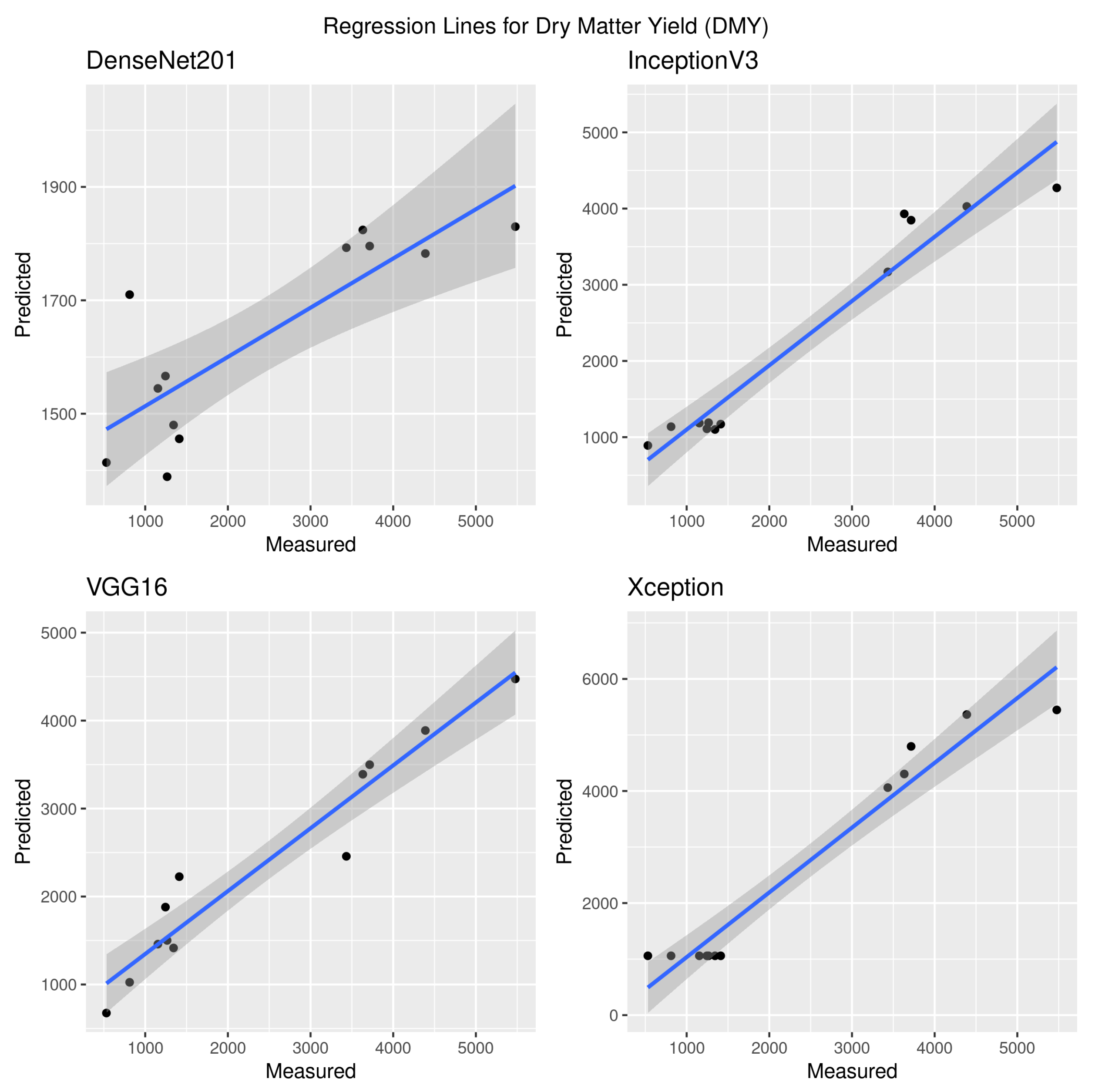
3.1. Dry Matter Yield
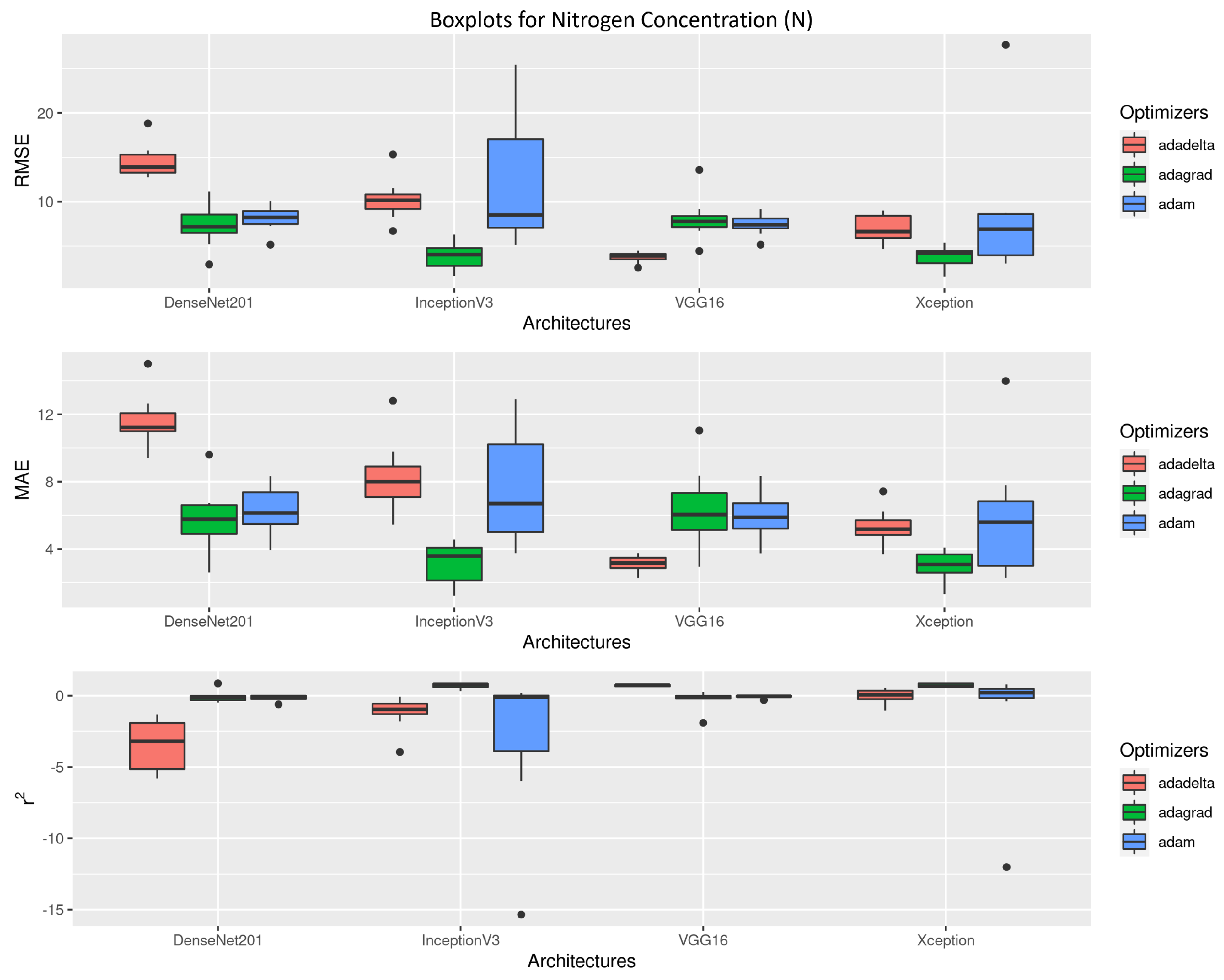
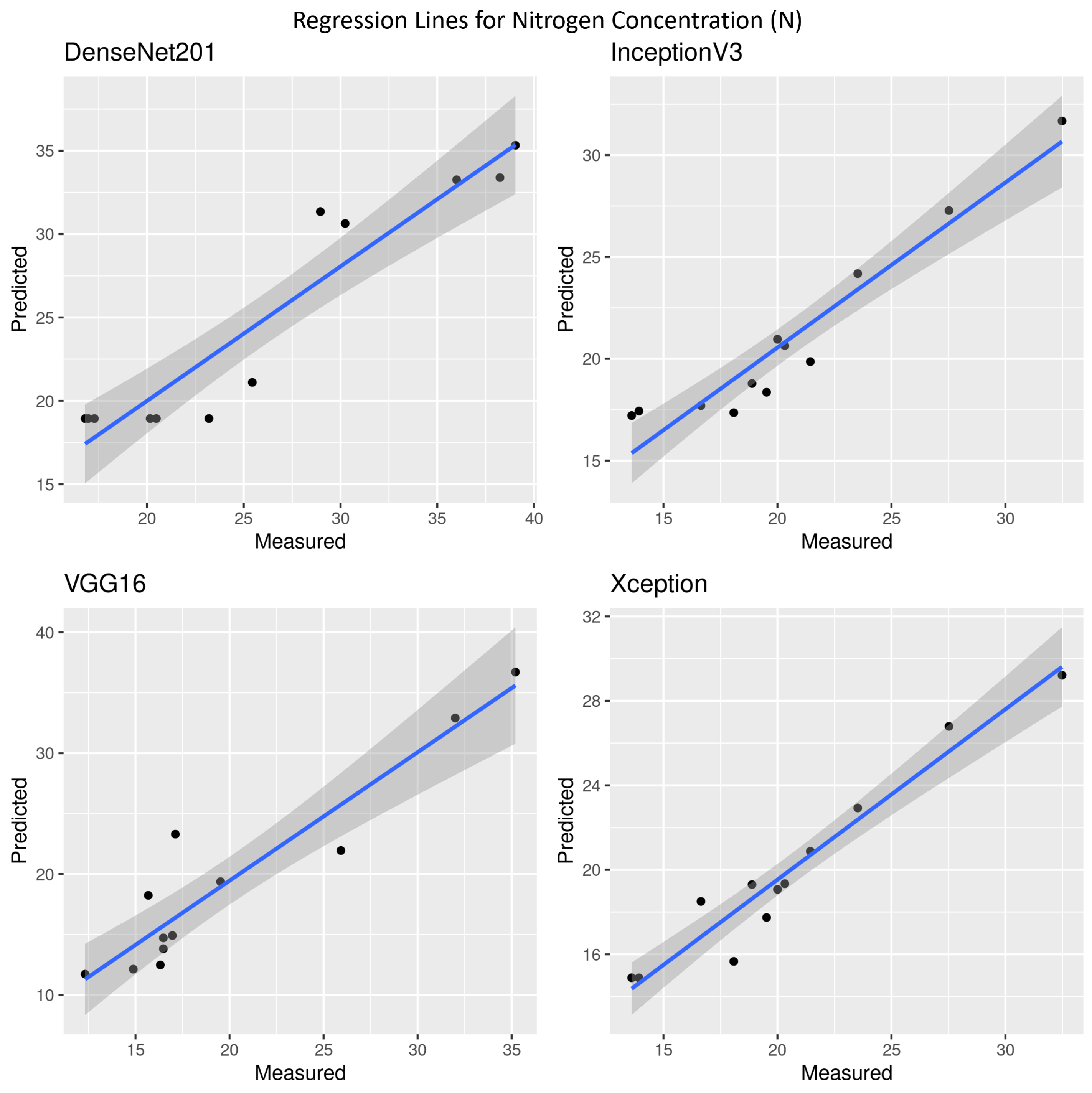
3.2. Nitrogen Concentration
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rockström, J.; Williams, J.; Daily, G.; Noble, A.; Matthews, N.; Gordon, L.; Wetterstrand, H.; DeClerck, F.; Shah, M.; Steduto, P.; et al. Sustainable intensification of agriculture for human prosperity and global sustainability. Ambio 2017, 46, 4–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Mara, F.P. The role of grasslands in food security and climate change. Ann. Bot. 2012, 110, 1263–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bengtsson, J.; Bullock, J.; Egoh, B.; Everson, C.; Everson, T.; O’Connor, T.; O’Farrell, P.; Smith, H.; Lindborg, R. Grasslands—More important for ecosystem services than you might think. Ecosphere 2019, 10, e02582. [Google Scholar] [CrossRef]
- Korhonen, P.; Palosuo, T.; Persson, T.; Höglind, M.; Jégo, G.; Van Oijen, M.; Gustavsson, A.M.; Bélanger, G.; Virkajärvi, P. Modelling grass yields in northern climates–a comparison of three growth models for timothy. Field Crops Res. 2018, 224, 37–47. [Google Scholar] [CrossRef]
- Reinermann, S.; Asam, S.; Kuenzer, C. Remote sensing of grassland production and management—A review. Remote Sens. 2020, 12, 1949. [Google Scholar] [CrossRef]
- Pajares, G. Overview and current status of remote sensing applications based on unmanned aerial vehicles (UAVs). Photogramm. Eng. Remote Sens. 2015, 81, 281–330. [Google Scholar] [CrossRef] [Green Version]
- Manfreda, S.; McCabe, M.F.; Miller, P.E.; Lucas, R.; Pajuelo Madrigal, V.; Mallinis, G.; Ben Dor, E.; Helman, D.; Estes, L.; Ciraolo, G.; et al. On the use of unmanned aerial systems for environmental monitoring. Remote Sens. 2018, 10, 641. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, R.A.; Näsi, R.; Niemeläinen, O.; Nyholm, L.; Alhonoja, K.; Kaivosoja, J.; Jauhiainen, L.; Viljanen, N.; Nezami, S.; Markelin, L.; et al. Machine learning estimators for the quantity and quality of grass swards used for silage production using drone-based imaging spectrometry and photogrammetry. Remote Sens. Environ. 2020, 246, 111830. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef] [Green Version]
- Osco, L.P.; Junior, J.M.; Ramos, A.P.M.; de Castro Jorge, L.A.; Fatholahi, S.N.; de Andrade Silva, J.; Matsubara, E.T.; Pistori, H.; Gonçalves, W.N.; Li, J. A review on deep learning in UAV remote sensing. Int. J. Appl. Earth Obs. Geoinf. 2021, 102, 102456. [Google Scholar] [CrossRef]
- Kattenborn, T.; Leitloff, J.; Schiefer, F.; Hinz, S. Review on Convolutional Neural Networks (CNN) in vegetation remote sensing. ISPRS J. Photogramm. Remote Sens. 2021, 173, 24–49. [Google Scholar] [CrossRef]
- Ma, J.; Li, Y.; Chen, Y.; Du, K.; Zheng, F.; Zhang, L.; Sun, Z. Estimating above ground biomass of winter wheat at early growth stages using digital images and deep convolutional neural network. Eur. J. Agron. 2019, 103, 117–129. [Google Scholar] [CrossRef]
- Nevavuori, P.; Narra, N.; Lipping, T. Crop yield prediction with deep convolutional neural networks. Comput. Electron. Agric. 2019, 163, 104859. [Google Scholar] [CrossRef]
- Yang, Q.; Shi, L.; Han, J.; Zha, Y.; Zhu, P. Deep convolutional neural networks for rice grain yield estimation at the ripening stage using UAV-based remotely sensed images. Field Crops Res. 2019, 235, 142–153. [Google Scholar] [CrossRef]
- Yang, W.; Nigon, T.; Hao, Z.; Paiao, G.D.; Fernández, F.G.; Mulla, D.; Yang, C. Estimation of corn yield based on hyperspectral imagery and convolutional neural network. Comput. Electron. Agric. 2021, 184, 106092. [Google Scholar] [CrossRef]
- Chen, Y.; Lee, W.S.; Gan, H.; Peres, N.; Fraisse, C.; Zhang, Y.; He, Y. Strawberry yield prediction based on a deep neural network using high-resolution aerial orthoimages. Remote Sens. 2019, 11, 1584. [Google Scholar] [CrossRef] [Green Version]
- Apolo-Apolo, O.; Martínez-Guanter, J.; Egea, G.; Raja, P.; Pérez-Ruiz, M. Deep learning techniques for estimation of the yield and size of citrus fruits using a UAV. Eur. J. Agron. 2020, 115, 126030. [Google Scholar] [CrossRef]
- Castro, W.; Marcato Junior, J.; Polidoro, C.; Osco, L.; Gonçalves, W.; Rodrigues, L.; Santos, M.; Jank, L.; Barrios, S.; Valle, C.; et al. Deep Learning Applied to Phenotyping of Biomass in Forages with UAV-Based RGB Imagery. Sensors 2020, 20, 4802. [Google Scholar] [CrossRef]
- de Oliveira, G.S.; Marcato Junior, J.; Polidoro, C.; Osco, L.P.; Siqueira, H.; Rodrigues, L.; Jank, L.; Barrios, S.; Valle, C.; Simeão, R.; et al. Convolutional Neural Networks to Estimate Dry Matter Yield in a Guineagrass Breeding Program Using UAV Remote Sensing. Sensors 2021, 21, 3971. [Google Scholar] [CrossRef]
- Berger, K.; Verrelst, J.; Feret, J.B.; Wang, Z.; Wocher, M.; Strathmann, M.; Danner, M.; Mauser, W.; Hank, T. Crop nitrogen monitoring: Recent progress and principal developments in the context of imaging spectroscopy missions. Remote Sens. Environ. 2020, 242, 111758. [Google Scholar] [CrossRef]
- Geipel, J.; Link, J.; Wirwahn, J.A.; Claupein, W. A programmable aerial multispectral camera system for in-season crop biomass and nitrogen content estimation. Agriculture 2016, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Cheng, T.; Zhu, Y.; Tian, Y.; Cao, W.; Yao, X.; Wang, N. Comparative analysis of vegetation indices, non-parametric and physical retrieval methods for monitoring nitrogen in wheat using UAV-based multispectral imagery. In Proceedings of the 2016 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Beijing, China, 10–15 July 2016; pp. 7362–7365. [Google Scholar]
- Näsi, R.; Honkavaara, E.; Blomqvist, M.; Lyytikäinen-Saarenmaa, P.; Hakala, T.; Viljanen, N.; Kantola, T.; Holopainen, M. Remote sensing of bark beetle damage in urban forests at individual tree level using a novel hyperspectral camera from UAV and aircraft. Urban For. Urban Green. 2018, 30, 72–83. [Google Scholar] [CrossRef]
- Capolupo, A.; Kooistra, L.; Berendonk, C.; Boccia, L.; Suomalainen, J. Estimating plant traits of grasslands from UAV-acquired hyperspectral images: A comparison of statistical approaches. ISPRS Int. J. Geo-Inf. 2015, 4, 2792–2820. [Google Scholar] [CrossRef]
- Askari, M.S.; McCarthy, T.; Magee, A.; Murphy, D.J. Evaluation of grass quality under different soil management scenarios using remote sensing techniques. Remote Sens. 2019, 11, 1835. [Google Scholar] [CrossRef] [Green Version]
- Michez, A.; Philippe, L.; David, K.; Sébastien, C.; Christian, D.; Bindelle, J. Can low-cost unmanned aerial systems describe the forage quality heterogeneity? Insight from a Timothy Pasture case study in Southern Belgium. Remote Sens. 2020, 12, 1650. [Google Scholar] [CrossRef]
- Barnetson, J.; Phinn, S.; Scarth, P. Estimating plant pasture biomass and quality from UAV imaging across Queensland’s Rangelands. AgriEngineering 2020, 2, 523–543. [Google Scholar] [CrossRef]
- Peel, M.C.; Finlayson, B.L.; McMahon, T.A. Updated world map of the Köppen-Geiger climate classification. Hydrol. Earth Syst. Sci. 2007, 11, 1633–1644. [Google Scholar] [CrossRef] [Green Version]
- Viljanen, N.; Honkavaara, E.; Näsi, R.; Hakala, T.; Niemeläinen, O.; Kaivosoja, J. A novel machine learning method for estimating biomass of grass swards using a photogrammetric canopy height model, images and vegetation indices captured by a drone. Agriculture 2018, 8, 70. [Google Scholar] [CrossRef] [Green Version]
- Huang, G.; Liu, Z.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. arXiv 2016, arXiv:1610.02357. [Google Scholar]
- Zeiler, M.D. ADADELTA: An Adaptive Learning Rate Method. arXiv 2012, arXiv:1212.5701. [Google Scholar]
- Duchi, J.; Hazan, E.; Singer, Y. Adaptive subgradient methods for online learning and stochastic optimization. J. Mach. Learn. Res. 2011, 12, 2121–2159. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization, 2014. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Togeiro de Alckmin, G.; Lucieer, A.; Roerink, G.; Rawnsley, R.; Hoving, I.; Kooistra, L. Retrieval of crude protein in perennial ryegrass using spectral data at the Canopy level. Remote Sens. 2020, 12, 2958. [Google Scholar] [CrossRef]
- Pullanagari, R.; Dehghan-Shoar, M.; Yule, I.J.; Bhatia, N. Field spectroscopy of canopy nitrogen concentration in temperate grasslands using a convolutional neural network. Remote Sens. Environ. 2021, 257, 112353. [Google Scholar] [CrossRef]
- Berger, K.; Verrelst, J.; Féret, J.B.; Hank, T.; Wocher, M.; Mauser, W.; Camps-Valls, G. Retrieval of aboveground crop nitrogen content with a hybrid machine learning method. Int. J. Appl. Earth Obs. Geoinf. 2020, 92, 102174. [Google Scholar] [CrossRef]

| Architecture | Trainable Parameters | Reference | |
|---|---|---|---|
| #1 | DenseNet201 | 66,525,569 | Huang et al. [30] |
| #2 | InceptionV3 | 48,246,433 | Szegedy et al. [31] |
| #3 | VGG16 | 27,823,425 | Simonyan et al. [32] |
| #4 | Xception | 72,450,857 | Chollet [33] |
| Hyperparameter | Value | |
|---|---|---|
| #1 | Training Epochs | 200 |
| #2 | Early Stop Patience | 40 |
| #3 | Early Stop Monitor | Loss |
| #4 | Loss Function | MAE |
| #5 | Checkpoint Saving | True |
| #6 | Initial Learning Rate | 0.01 |
| #7 | Validation Split | 20% |
| #8 | Neurons Fully Connected (FC) | 512 |
| #9 | Dropout FC Layer | 50% |
| #10 | Data Augmentation | None |
| #11 | Test Set Sampling | 8-Fold |
| #12 | Transfer Learning | ImageNet |
| #13 | Fine Tuning | True |
| Optimizer | Reference | |
|---|---|---|
| #1 | Adadelta | Zeiler [34] |
| #2 | Adagrad | Duchi et al. [35] |
| #3 | Adam | Kingma and Ba [36] |
| Architecture | Optimizer | RMSE (kg DM/ha) | MAE (kg DM/ha) | nMAE (%) | |
|---|---|---|---|---|---|
| DenseNet201 | Adadelta | 2278.95 (608.69) | 1858.41 (442.83) | 70.06 | −1.49 (1.32) |
| DenseNet201 | Adagrad | 1494.85 (414.68) | 1141.18 (334.93) | 43.02 | −0.06 (0.48) |
| DenseNet201 | Adam | 1984.89 (508.92) | 1634.87 (443.55) | 61.63 | −0.89 (1.04) |
| InceptionV3 | Adadelta | 1739.52 (242.13) | 1413.80 (172.38) | 53.30 | −0.43 (0.57) |
| InceptionV3 | Adagrad | 801.72 (303.11) | 606.96 (212.71) | 22.88 | 0.66 (0.29) |
| InceptionV3 | Adam | 8830.16 (10,405.33) | 3615.15 (3486.56) | 136.29 | −72.43 (115.08) |
| VGG16 | Adadelta | 668.79 (144.07) | 538.07 (111.12) | 20.28 | 0.79 (0.10) |
| VGG16 | Adagrad | 1759.99 (368.30) | 1293.27 (348.31) | 48.75 | −0.46 (0.71) |
| VGG16 | Adam | 2064.42 (420.08) | 1610.65 (460.89) | 60.72 | −1.03 (1.03) |
| Xception | Adadelta | 1066.17 (264.53) | 819.55 (213.82) | 30.90 | 0.46 (0.26) |
| Xception | Adagrad | 915.36 (366.36) | 667.72 (265.45) | 25.17 | 0.59 (0.29) |
| Xception | Adam | 997.50 (411.66) | 724.23 (307.57) | 27.30 | 0.45 (0.47) |
| Architecture | Optimizer | RMSE (g N/kg DM) | MAE (g N/kg DM) | nMAE (%) | |
|---|---|---|---|---|---|
| DenseNet201 | Adadelta | 14.66 (2.11) | 11.68 (1.75) | 52.94 | −3.49 (1.87) |
| DenseNet201 | Adagrad | 7.23 (2.45) | 5.76 (2.10) | 26.11 | −0.07 (0.43) |
| DenseNet201 | Adam | 8.02 (1.70) | 6.27 (1.59) | 28.42 | −0.17 (0.24) |
| InceptionV3 | Adadelta | 10.28 (2.53) | 8.32 (2.25) | 37.71 | −1.21 (1.24) |
| InceptionV3 | Adagrad | 3.87 (1.59) | 3.11 (1.31) | 14.10 | 0.70 (0.20) |
| InceptionV3 | Adam | 12.23 (7.59) | 7.74 (3.57) | 35.08 | −3.06 (5.43) |
| VGG16 | Adadelta | 3.72 (0.66) | 3.11 (0.53) | 14.10 | 0.73 (0.07) |
| VGG16 | Adagrad | 8.11 (2.60) | 6.45 (2.44) | 29.24 | −0.29 (0.67) |
| VGG16 | Adam | 7.45 (1.32) | 5.99 (1.41) | 27.15 | −0.07 (0.10) |
| Xception | Adadelta | 6.91 (1.58) | 5.30 (1.17) | 24.02 | −0.01 (0.53) |
| Xception | Adagrad | 3.80 (1.30) | 2.96 (0.94) | 13.42 | 0.71 (0.14) |
| Xception | Adam | 8.65 (8.01) | 5.94 (3.80) | 26.92 | −1.27 (4.36) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alves Oliveira, R.; Marcato Junior, J.; Soares Costa, C.; Näsi, R.; Koivumäki, N.; Niemeläinen, O.; Kaivosoja, J.; Nyholm, L.; Pistori, H.; Honkavaara, E. Silage Grass Sward Nitrogen Concentration and Dry Matter Yield Estimation Using Deep Regression and RGB Images Captured by UAV. Agronomy 2022, 12, 1352. https://doi.org/10.3390/agronomy12061352
Alves Oliveira R, Marcato Junior J, Soares Costa C, Näsi R, Koivumäki N, Niemeläinen O, Kaivosoja J, Nyholm L, Pistori H, Honkavaara E. Silage Grass Sward Nitrogen Concentration and Dry Matter Yield Estimation Using Deep Regression and RGB Images Captured by UAV. Agronomy. 2022; 12(6):1352. https://doi.org/10.3390/agronomy12061352
Chicago/Turabian StyleAlves Oliveira, Raquel, José Marcato Junior, Celso Soares Costa, Roope Näsi, Niko Koivumäki, Oiva Niemeläinen, Jere Kaivosoja, Laura Nyholm, Hemerson Pistori, and Eija Honkavaara. 2022. "Silage Grass Sward Nitrogen Concentration and Dry Matter Yield Estimation Using Deep Regression and RGB Images Captured by UAV" Agronomy 12, no. 6: 1352. https://doi.org/10.3390/agronomy12061352
APA StyleAlves Oliveira, R., Marcato Junior, J., Soares Costa, C., Näsi, R., Koivumäki, N., Niemeläinen, O., Kaivosoja, J., Nyholm, L., Pistori, H., & Honkavaara, E. (2022). Silage Grass Sward Nitrogen Concentration and Dry Matter Yield Estimation Using Deep Regression and RGB Images Captured by UAV. Agronomy, 12(6), 1352. https://doi.org/10.3390/agronomy12061352







