Impact of Azospirillum sp. B510 on the Rhizosphere Microbiome of Rice under Field Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Field Experiments
2.2. Evaluation of Plant Growth and Yield
2.3. Physicochemical Properties of Soil
2.4. DNA Extraction and Amplicon Analysis
2.5. Statistical Analysis
3. Results
3.1. The Effect of Azospirillum sp. B510 Inoculation on the Growth and Yield of Rice
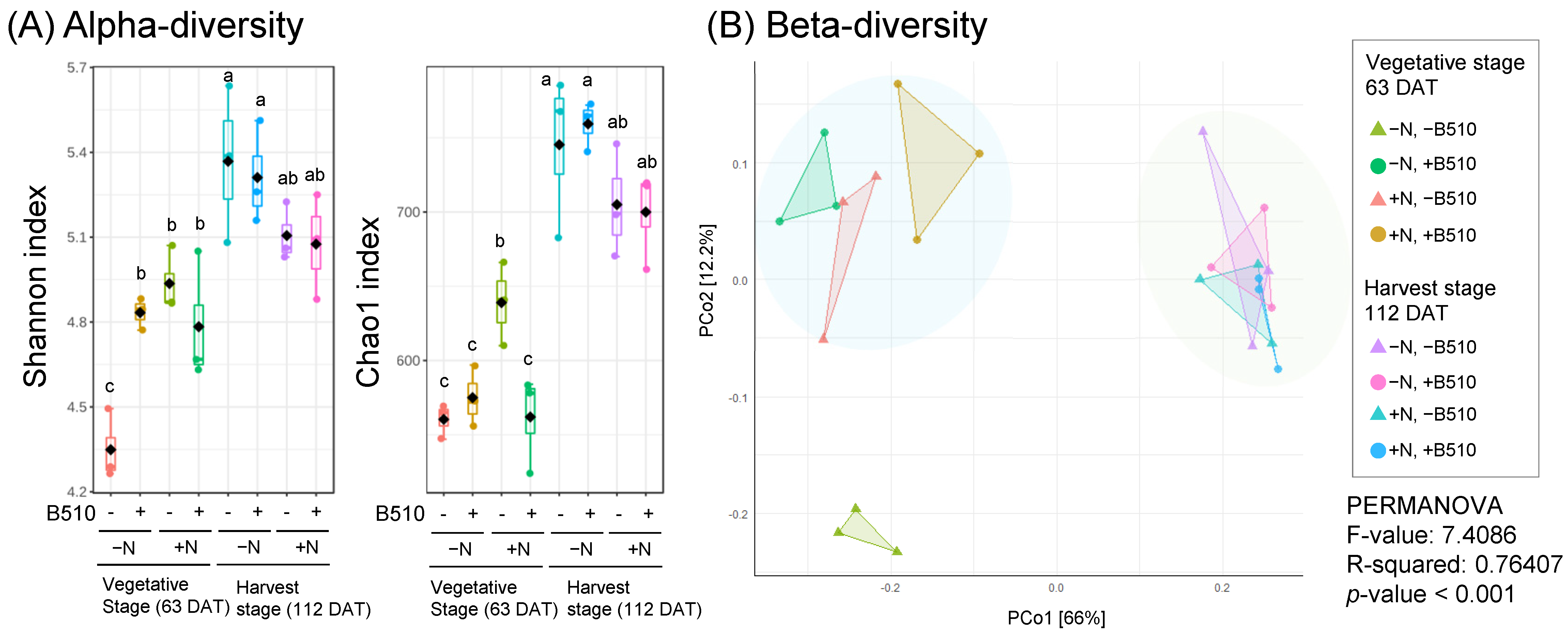
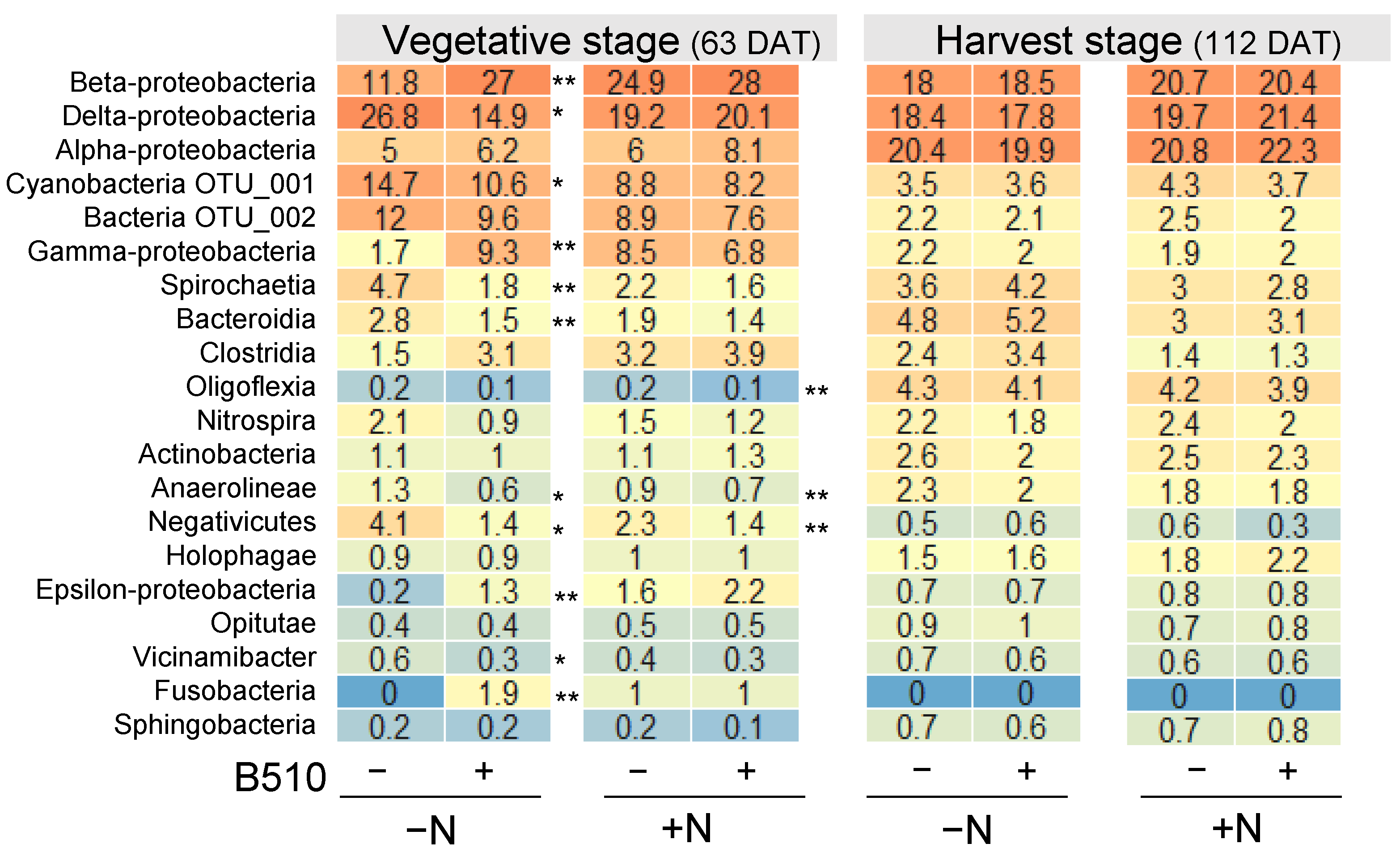
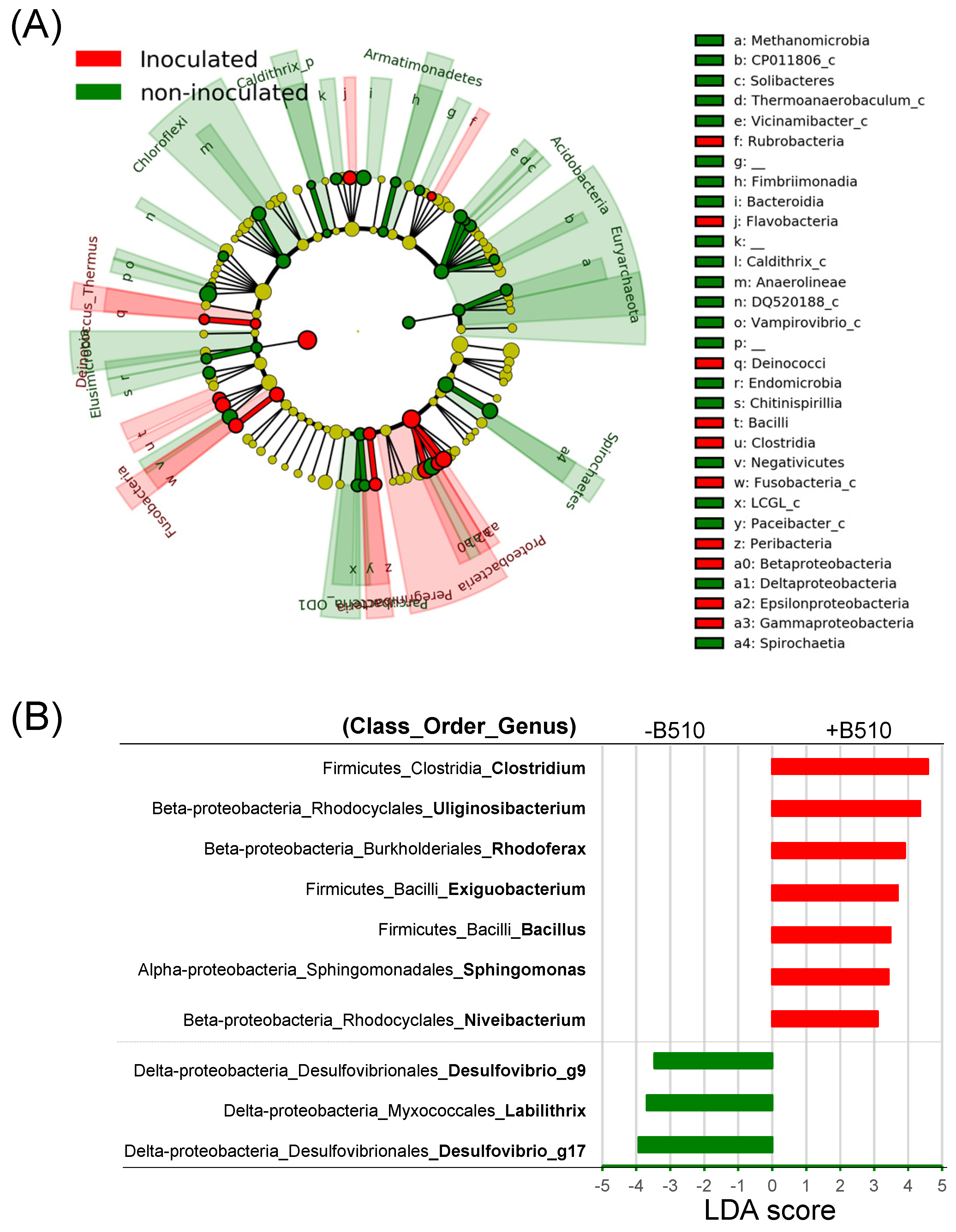
3.2. The Effect of B510 Inoculation on Bacterial Community Structure
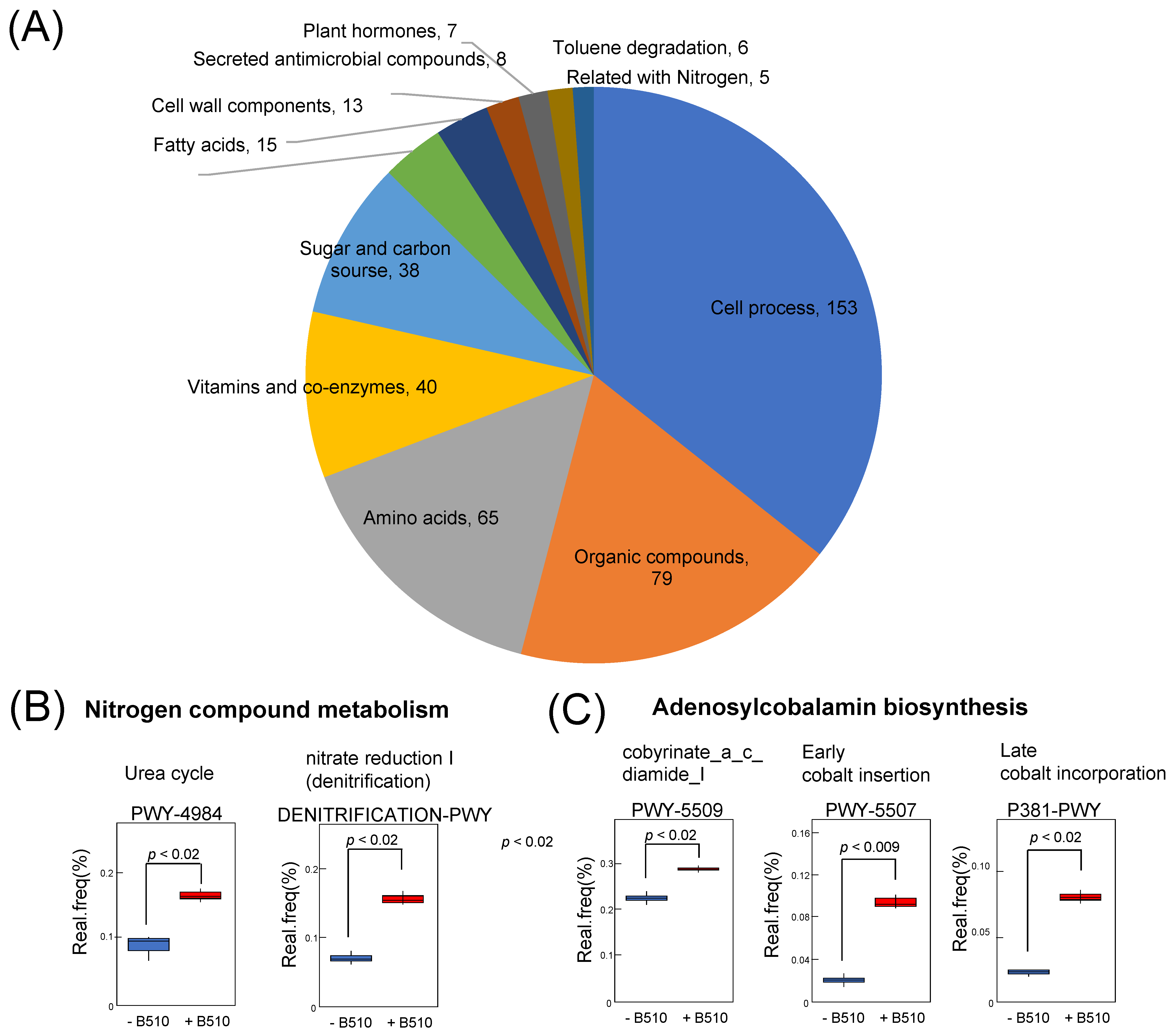
3.3. The Predicted Function of Bacteria Influenced by B510 Inoculation
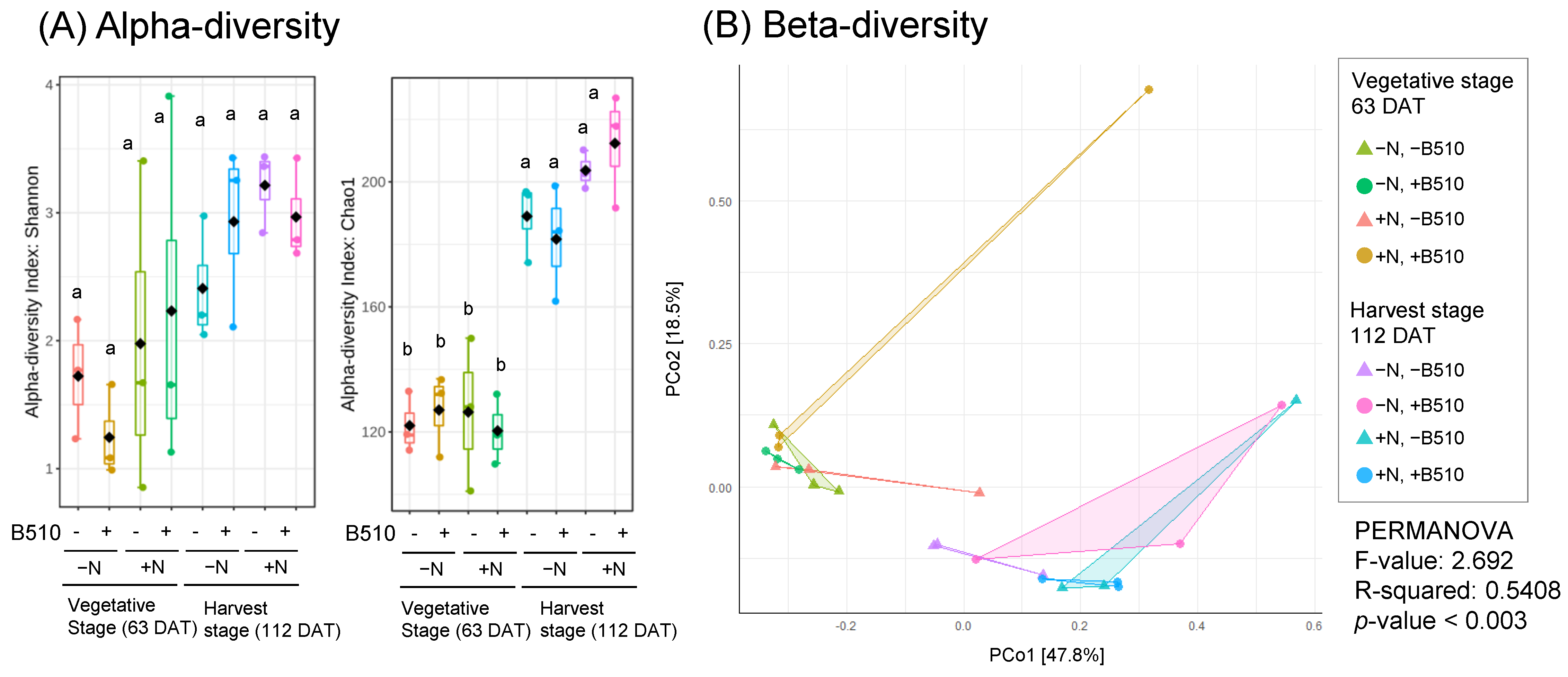
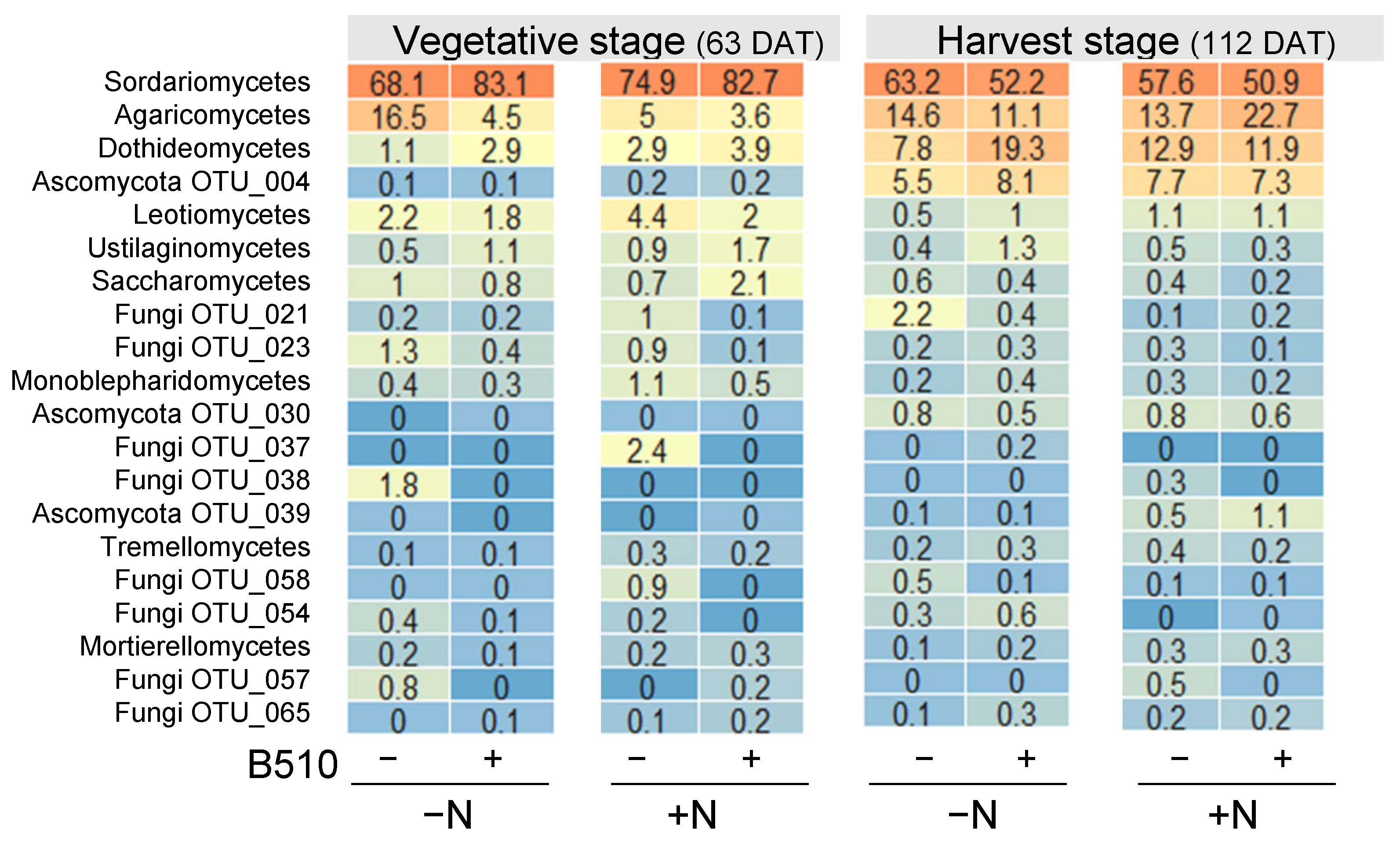
3.4. The Effect of B510 Inoculation on Fungal Community Structure
4. Discussion
4.1. Impact of Azospirillum Inoculation on Fields
4.2. Inoculation with Azospirillum sp. B510 Increased Nitrogen-Fixing Bacteria
4.3. The Function of Cobalamin in the Rhizosphere Community
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maheshwari, D.K. (Ed.) Bacteria in Agrobiology: Crop Ecosystems; Springer Berlin/Heidelberg: Berlin/Heidelberg, Germany, 2011; ISBN 978-3-642-18356-0. [Google Scholar]
- Cassán, F.; Coniglio, A.; López, G.; Molina, R.; Nievas, S.; de Carlan, C.L.N.; Donadio, F.; Torres, D.; Rosas, S.; Pedrosa, F.O.; et al. Everything You Must Know about Azospirillum and Its Impact on Agriculture and Beyond. Biol. Fertil. Soils 2020, 56, 461–479. [Google Scholar] [CrossRef]
- Bao, Z.; Sasaki, K.; Okubo, T.; Ikeda, S.; Anda, M.; Hanzawa, E.; Kakizaki, K.; Sato, T.; Mitsui, H.; Minamisawa, K. Impact of Azospirillum sp. B510 Inoculation on Rice-Associated Bacterial Communities in a Paddy Field. Microb. Environ. 2013, 28, 487–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horn, S.; Hempel, S.; Verbruggen, E.; Rillig, M.C.; Caruso, T. Linking the Community Structure of Arbuscular Mycorrhizal Fungi and Plants: A Story of Interdependence? ISME J. 2017, 11, 1400–1411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Win, K.T.; Okazaki, K.; Ohkama-Ohtsu, N.; Yokoyama, T.; Ohwaki, Y. Short-Term Effects of Biochar and Bacillus Pumilus TUAT-1 on the Growth of Forage Rice and Its Associated Soil Microbial Community and Soil Properties. Biol. Fertil. Soils 2020, 56, 481–497. [Google Scholar] [CrossRef]
- Liu, Q.; Pang, Z.; Yang, Z.; Nyumah, F.; Hu, C.; Lin, W.; Yuan, Z. Bio-Fertilizer Affects Structural Dynamics, Function, and Network Patterns of the Sugarcane Rhizospheric Microbiota. Microb. Ecol. 2021. [Google Scholar] [CrossRef]
- Han, Q.; Ma, Q.; Chen, Y.; Tian, B.; Xu, L.; Bai, Y.; Chen, W.; Li, X. Variation in Rhizosphere Microbial Communities and Its Association with the Symbiotic Efficiency of Rhizobia in Soybean. ISME J. 2020, 14, 1915–1928. [Google Scholar] [CrossRef] [Green Version]
- Reynders, L.; Vlassak, K. Use OfAzospirillum Brasilense as Biofertilizer in Intensive Wheat Cropping. Plant Soil 1982, 66, 217–223. [Google Scholar] [CrossRef]
- Fallik, E.; Okon, Y. The Response of Maize (Zea Mays) to Azospirillum Inoculation in Various Types of Soils in the Field. World J. Microbiol. Biotechnol. 1996, 12, 511–515. [Google Scholar] [CrossRef]
- Heulin, T.; GUCKERT, A.; Balandreau, J. Stimulation of Root Exudation of Rice Seedlings by Azospirillum Strains: Carbon Budget under Gnotobiotic Conditions. Biol. Fertil. Soils 1987, 4, 9–14. [Google Scholar] [CrossRef]
- Bashan, Y.; de-Bashan, L.E. How the Plant Growth-Promoting Bacterium Azospirillum Promotes Plant Growth—A Critical Assessment. In Advances in Agronomy; Elsevier: Amsterdam, The Netherlands, 2010; Volume 108, pp. 77–136. ISBN 978-0-12-381031-1. [Google Scholar]
- Janzen, R.A.; Rood, S.B.; Dormaar, J.F.; McGill, W.B. Azospirillum Brasilense Produces Gibberellin in Pure Culture on Chemically-Defined Medium and in Co-Culture on Straw. Soil Biol. Biochem. 1992, 24, 1061–1064. [Google Scholar] [CrossRef]
- Lin, W.; Okon, Y.; Hardy, R.W.F. Enhanced Mineral Uptake by Zea Mays and Sorghum Bicolor Roots Inoculated with Azospirillum Brasilense. Appl. Environ. Microbiol. 1983, 45, 1775–1779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murty, M.; Ladha, J. Influence of Azospirillum Inoculation on the Mineral Uptake and Growth of Rice under Hydroponic Conditions. Plant Soil 1988, 108, 281–285. [Google Scholar] [CrossRef]
- Elbeltagy, A.; Nishioka, K.; Sato, T.; Suzuki, H.; Ye, B.; Hamada, T.; Isawa, T.; Mitsui, H.; Minamisawa, K. Endophytic Colonization and In Planta Nitrogen Fixation by a Herbaspirillum sp. Isolated from Wild Rice Species. Appl. Environ. Microbiol. 2001, 67, 5285–5293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chamam, A.; Sanguin, H.; Bellvert, F.; Meiffren, G.; Comte, G.; Wisniewski-Dyé, F.; Bertrand, C.; Prigent-Combaret, C. Plant Secondary Metabolite Profiling Evidences Strain-Dependent Effect in the Azospirillum–Oryza Sativa Association. Phytochemistry 2013, 87, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Isawa, T.; Yasuda, M.; Awazaki, H.; Minamisawa, K.; Shinozaki, S.; Nakashita, H. Azospirillum sp. Strain B510 Enhances Rice Growth and Yield. Microb. Environ. 2010, 25, 58–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasaki, K.; Ikeda, S.; Eda, S.; Mitsui, H.; Hanzawa, E.; Kisara, C.; Kazama, Y.; Kushida, A.; Shinano, T.; Minamisawa, K.; et al. Impact of Plant Genotype and Nitrogen Level on Rice Growth Response to Inoculation with Azospirillum sp. Strain B510 under Paddy Field Conditions. Soil Sci. Plant Nutr. 2010, 56, 636–644. [Google Scholar] [CrossRef] [Green Version]
- Asiloglu, R.; Shiroishi, K.; Suzuki, K.; Turgay, O.C.; Murase, J.; Harada, N. Protist-Enhanced Survival of a Plant Growth Promoting Rhizobacteria, Azospirillum sp. B510, and the Growth of Rice (Oryza sativa L.) Plants. Appl. Soil Ecol. 2020, 154, 103599. [Google Scholar] [CrossRef]
- Kaneko, T.; Minamisawa, K.; Isawa, T.; Nakatsukasa, H.; Mitsui, H.; Kawaharada, Y.; Nakamura, Y.; Watanabe, A.; Kawashima, K.; Ono, A.; et al. Complete Genomic Structure of the Cultivated Rice Endophyte Azospirillum sp. B510. DNA Res. 2010, 17, 37–50. [Google Scholar] [CrossRef] [Green Version]
- Drogue, B.; Sanguin, H.; Chamam, A.; Mozar, M.; Llauro, C.; Panaud, O.; Prigent-Combaret, C.; Picault, N.; Wisniewski-Dyé, F. Plant Root Transcriptome Profiling Reveals a Strain-Dependent Response during Azospirillum-Rice Cooperation. Front. Plant Sci. 2014, 5, 607. [Google Scholar] [CrossRef] [Green Version]
- Yasuda, M.; Isawa, T.; Shinozaki, S.; Minamisawa, K.; Nakashita, H. Effects of Colonization of a Bacterial Endophyte, Azospirillum sp. B510, on Disease Resistance in Rice. Biosci. Biotechnol. Biochem. 2009, 73, 2595–2599. [Google Scholar] [CrossRef] [Green Version]
- Kusajima, M.; Shima, S.; Fujita, M.; Minamisawa, K.; Che, F.-S.; Yamakawa, H.; Nakashita, H. Involvement of Ethylene Signaling in Azospirillum sp. B510-Induced Disease Resistance in Rice. Biosci. Biotechnol. Biochem. 2018, 82, 1522–1526. [Google Scholar] [CrossRef] [PubMed]
- Sugihara, S.; Funakawa, S.; Kilasara, M.; Kosaki, T. Effect of Land Management and Soil Texture on Seasonal Variations in Soil Microbial Biomass in Dry Tropical Agroecosystems in Tanzania. Appl. Soil Ecol. 2010, 44, 80–88. [Google Scholar] [CrossRef]
- Ikeda, S.; Watanabe, K.N.; Minamisawa, K.; Ytow, N. Evaluation of Soil DNA from Arable Land in Japan Using a Modified Direct-Extraction Method. Microbes Environ. 2004, 19, 301–309. [Google Scholar] [CrossRef] [Green Version]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A Web-Based Tool for Comprehensive Statistical, Visual and Meta-Analysis of Microbiome Data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douglas, G.M.; Mafei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for Prediction of Metagenome Functions. Nat. Biotechnol. 2020, 38, 669–688. [Google Scholar] [CrossRef] [PubMed]
- Naher, K.; Miwa, H.; Okazaki, S.; Yasuda, M. Effects of Different Sources of Nitrogen on Endophytic Colonization of Rice Plants by Azospirillum sp. B510. Microbes Environ. 2018, 33, 301–308. [Google Scholar] [CrossRef] [Green Version]
- Herschkovitz, Y.; Lerner, A.; Davidov, Y.; Okon, Y.; Jurkevitch, E. Azospirillum Brasilense Does Not Affect Population Structure of Specific Rhizobacterial Communities of Inoculated Maize (Zea mays). Environ. Microbiol. 2005, 7, 1847–1852. [Google Scholar] [CrossRef]
- Renoud, S.; Abrouk, D.; Prigent-Combaret, C.; Wisniewski-Dyé, F.; Legendre, L.; Moënne-Loccoz, Y.; Muller, D. Effect of Inoculation Level on the Impact of the PGPR Azospirillum Lipoferum CRT1 on Selected Microbial Functional Groups in the Rhizosphere of Field Maize. Microorganisms 2022, 10, 325. [Google Scholar] [CrossRef]
- Zeiller, M.; Rothballer, M.; Iwobi, A.N.; Böhnel, H.; Gessler, F.; Hartmann, A.; Schmid, M. Systemic Colonization of Clover (Trifolium repens) by Clostridium Botulinum Strain 2301. Front. Microbiol. 2015, 6, 1207. [Google Scholar] [CrossRef]
- Polyanskaya, L.M.; Vedina, O.T.; Lysak, L.V.; Zvyagintsev, D.G. The Growth-Promoting Effect of Beijerinckia Mobilis and Clostridium sp. Cultures on Some Agricultural Crops. Microbiology 2002, 71, 109–115. [Google Scholar] [CrossRef]
- Lu, Y.; Dirk, R.; Werner, L.; Ralf, C. Structure and Activity of Bacterial Community Inhabiting Rice Roots and the Rhizosphere. Environ. Microbiol. 2006, 8, 1351–1360. [Google Scholar] [CrossRef] [PubMed]
- Addison, S.L.; McDonald, I.R.; Lloyd-Jones, G. Identifying Diazotrophs by Incorporation of Nitrogen from 15N2 into RNA. Appl. Microbiol. Biotechnol. 2010, 87, 2313–2322. [Google Scholar] [CrossRef] [PubMed]
- Videira, S.S.; de Araujo, J.L.S.; da Silva Rodrigues, L.; Baldani, V.L.D.; Baldani, J.I. Occurrence and Diversity of Nitrogen-Fixing Sphingomonas Bacteria Associated with Rice Plants Grown in Brazil. FEMS Microbiol. Lett. 2009, 293, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, C.-H.; Yokota, A. Sphingomonas azotifigens sp. nov., a Nitrogen-Fixing Bacterium Isolated from the Roots of Oryza Sativa. Int. J. Syst. Evol. Microbiol. 2006, 56, 889–893. [Google Scholar] [CrossRef] [PubMed]
- Weon, H.-Y.; Kim, B.-Y.; Yoo, S.-H.; Kwon, S.-W.; Go, S.-J.; Stackebrandt, E. Uliginosibacterium Gangwonense Gen. Nov., Sp. Nov., Isolated from a Wetland, Yongneup, in Korea. Int. J. Syst. Evol. Microbiol. 2008, 58, 131–135. [Google Scholar] [CrossRef] [Green Version]
- Okamoto, T.; Shinjo, R.; Nishihara, A.; Uesaka, K.; Tanaka, A.; Sugiura, D.; Kondo, M. Genotypic Variation of Endophytic Nitrogen-Fixing Activity and Bacterial Flora in Rice Stem Based on Sugar Content. Front. Plant Sci. 2021, 12, 719259. [Google Scholar] [CrossRef]
- Ilangumaran, G.; Schwinghamer, T.D.; Smith, D.L. Rhizobacteria From Root Nodules of an Indigenous Legume Enhance Salinity Stress Tolerance in Soybean. Front. Sustain. Food Syst. 2021, 4, 617978. [Google Scholar] [CrossRef]
- Bharti, N.; Yadav, D.; Barnawal, D.; Maji, D.; Kalra, A. Exiguobacterium Oxidotolerans, a Halotolerant Plant Growth Promoting Rhizobacteria, Improves Yield and Content of Secondary Metabolites in Bacopa monnieri (L.) Pennell under Primary and Secondary Salt Stress. World J. Microbiol. Biotechnol. 2013, 29, 379–387. [Google Scholar] [CrossRef]
- Correa-Galeote, D.; Bedmar, E.J.; Arone, G.J. Maize Endophytic Bacterial Diversity as Affected by Soil Cultivation History. Front. Microbiol. 2018, 9, 484. [Google Scholar] [CrossRef]
- Kimbrel, J.A.; Givan, S.A.; Halgren, A.B.; Creason, A.L.; Mills, D.I.; Banowetz, G.M.; Armstrong, D.J.; Chang, J.H. An Improved, High-Quality Draft Genome Sequence of the Germination-Arrest Factor-Producing Pseudomonas Fluorescens WH6. BMC Genom. 2010, 11, 522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramaprasad, E.V.V.; Sasikala, C.; Ramana, C.V. Roseomonas oryzae sp. Nov., Isolated from Paddy Rhizosphere Soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 3535–3540. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.L.; Bishop-Lilly, K.A.; Ladner, J.T.; Daligault, H.E.; Davenport, K.W.; Jaissle, J.; Frey, K.G.; Koroleva, G.I.; Bruce, D.C.; Coyne, S.R.; et al. Complete Genome Sequences for 59 Burkholderia Isolates, Both Pathogenic and Near Neighbor. Genome Announc. 2015, 3, e00159-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iguchi, H.; Yurimoto, H.; Sakai, Y. Stimulation of Methanotrophic Growth in Cocultures by Cobalamin Excreted by Rhizobia. Appl. Env. Microbiol. 2011, 77, 8509–8515. [Google Scholar] [CrossRef] [Green Version]
- Taga, M.E.; Walker, G.C. Sinorhizobium meliloti Requires a Cobalamin-Dependent Ribonucleotide Reductase for Symbiosis With Its Plant Host. Mol. Plant-Microbe Interact. 2010, 23, 1643–1654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawrence, A.D.; Nemoto-Smith, E.; Deery, E.; Baker, J.A.; Schroeder, S.; Brown, D.G.; Tullet, J.M.A.; Howard, M.J.; Brown, I.R.; Smith, A.G.; et al. Construction of Fluorescent Analogs to Follow the Uptake and Distribution of Cobalamin (Vitamin B12) in Bacteria, Worms, and Plants. Cell Chem. Biol. 2018, 25, 941–951.e6. [Google Scholar] [CrossRef]
- Rakoff-Nahoum, S.; Foster, K.R.; Comstock, L.E. The Evolution of Cooperation within the Gut Microbiota. Nature 2016, 533, 255–259. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Xu, Z.; Xie, J.; Hesselberg-Thomsen, V.; Tan, T.; Zheng, D.; Strube, M.L.; Dragoš, A.; Shen, Q.; Zhang, R.; et al. Bacillus Velezensis Stimulates Resident Rhizosphere Pseudomonas Stutzeri for Plant Health through Metabolic Interactions. ISME J. 2022, 16, 774–787. [Google Scholar] [CrossRef]
- Rudrappa, T.; Czymmek, K.J.; Paré, P.W.; Bais, H.P. Root-Secreted Malic Acid Recruits Beneficial Soil Bacteria. Plant Physiol. 2008, 148, 1547–1556. [Google Scholar] [CrossRef] [Green Version]
| Vegetative Stage (63 DAT) | Non-Applied Nitrogen Fertilizer (−N) | Applied Nitogen Fertilizer (+N) | ||
| Non-Inoculated | Inoculated with B510 | Non-Inoculated | Inoculated with B510 | |
| Shoot length (cm/plant) | 86 ± 5.1 ab | 83 ± 3.8 b | 91 ± 3.5 a | 87 ± 3.3 a |
| Shoot weight (g/plant) | 319 ± 47 abd | 297 ± 113 b | 368 ± 50 ab | 387 ± 65 a |
| Root length (g/plant) | 25.3 ± 4.1 a | 22.6 ± 4.1 a | 24.7 ± 2.4 a | 23.2 ± 2.89 a |
| Root weight (g/plant) | 123 ± 29 a | 113 ± 48 a | 121 ± 22 a | 140 ± 20 a |
| Harvest Stage (112 DAT) | Non-Applied Nitrogen Fertilizer (−N) | Applied Nitogen Fertilizer (+N) | ||
| Non-Inoculated | Inoculated with B510 | Non-Inoculated | Inoculated with B510 | |
| Shoot length (cm/plant) | 103 ± 5.5 a | 105 ± 4.4 a | 108 ± 4.4 a | 105 ± 4.5 a |
| Shoot weight (g/plant) | 257 ± 57.4 b | 307 ± 33.2 ab | 348 ± 64.8 ab | 370 ± 63.4 a |
| Root weight (g/plant) | 84 ± 25.5 a | 96 ± 12.7 a | 91 ± 15.6 a | 102 ± 31.6 a |
| Tiller number (number/plant) | 20.7± 2.5 b | 25.7 ± 2.8 a | 27.8 ± 4.4 a | 30.7 ± 7.05 a |
| Panicle number (number/plant) | 19.9 ± 4.5 c | 24.7 ± 3.3 b | 25.9 ± 3.8 ab | 29.9 ± 6.5 a |
| Grain weight (g/plant) | 53.6 ± 14.4 b | 69.3 ± 10.0 ab | 69.2 ± 16.4 a | 73.5 ± 14.2 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yasuda, M.; Dastogeer, K.M.G.; Sarkodee-Addo, E.; Tokiwa, C.; Isawa, T.; Shinozaki, S.; Okazaki, S. Impact of Azospirillum sp. B510 on the Rhizosphere Microbiome of Rice under Field Conditions. Agronomy 2022, 12, 1367. https://doi.org/10.3390/agronomy12061367
Yasuda M, Dastogeer KMG, Sarkodee-Addo E, Tokiwa C, Isawa T, Shinozaki S, Okazaki S. Impact of Azospirillum sp. B510 on the Rhizosphere Microbiome of Rice under Field Conditions. Agronomy. 2022; 12(6):1367. https://doi.org/10.3390/agronomy12061367
Chicago/Turabian StyleYasuda, Michiko, Khondoker M. G. Dastogeer, Elsie Sarkodee-Addo, Chihiro Tokiwa, Tsuyoshi Isawa, Satoshi Shinozaki, and Shin Okazaki. 2022. "Impact of Azospirillum sp. B510 on the Rhizosphere Microbiome of Rice under Field Conditions" Agronomy 12, no. 6: 1367. https://doi.org/10.3390/agronomy12061367
APA StyleYasuda, M., Dastogeer, K. M. G., Sarkodee-Addo, E., Tokiwa, C., Isawa, T., Shinozaki, S., & Okazaki, S. (2022). Impact of Azospirillum sp. B510 on the Rhizosphere Microbiome of Rice under Field Conditions. Agronomy, 12(6), 1367. https://doi.org/10.3390/agronomy12061367






