Profiling the Diversity of Sweet Pepper ‘Peperone Cornetto di Pontecorvo’ PDO (Capsicum annuum) through Multi-Phenomic Approaches and Sequencing-Based Genotyping
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Field Trial
2.2. Morpho-Agronomic Characterization
2.3. Chemical and Fruit Colour Traits
2.4. Digital Imaging for Fruit Morphological Features
2.5. DNA Isolation
2.6. Double-Digest Restriction-Site-Associated (ddRad) DNA Sequencing
2.7. Alignment, Variant Calling, and SNP Filtering
2.8. Genomic Diversity Analysis
2.9. Data Analysis
3. Results
3.1. Phenotypic Variability
3.2. Agronomic, Morphological and Qualitative Performances
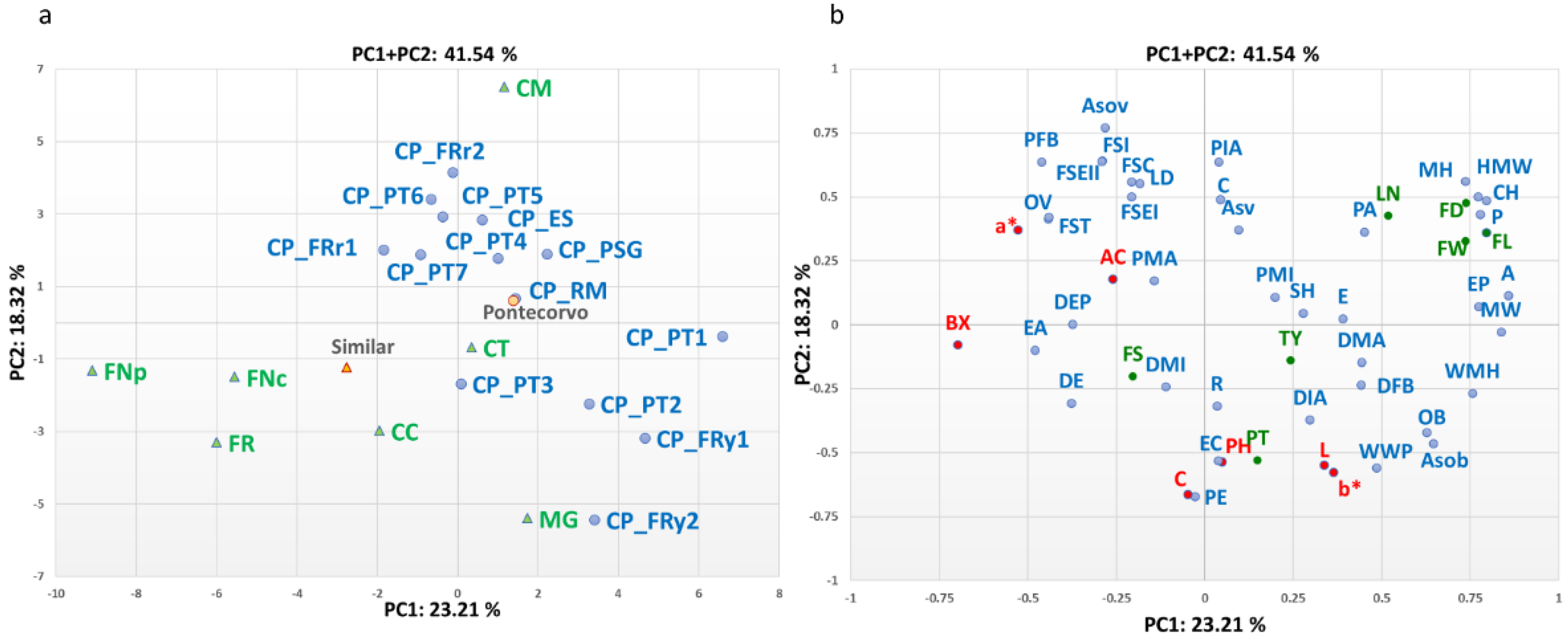
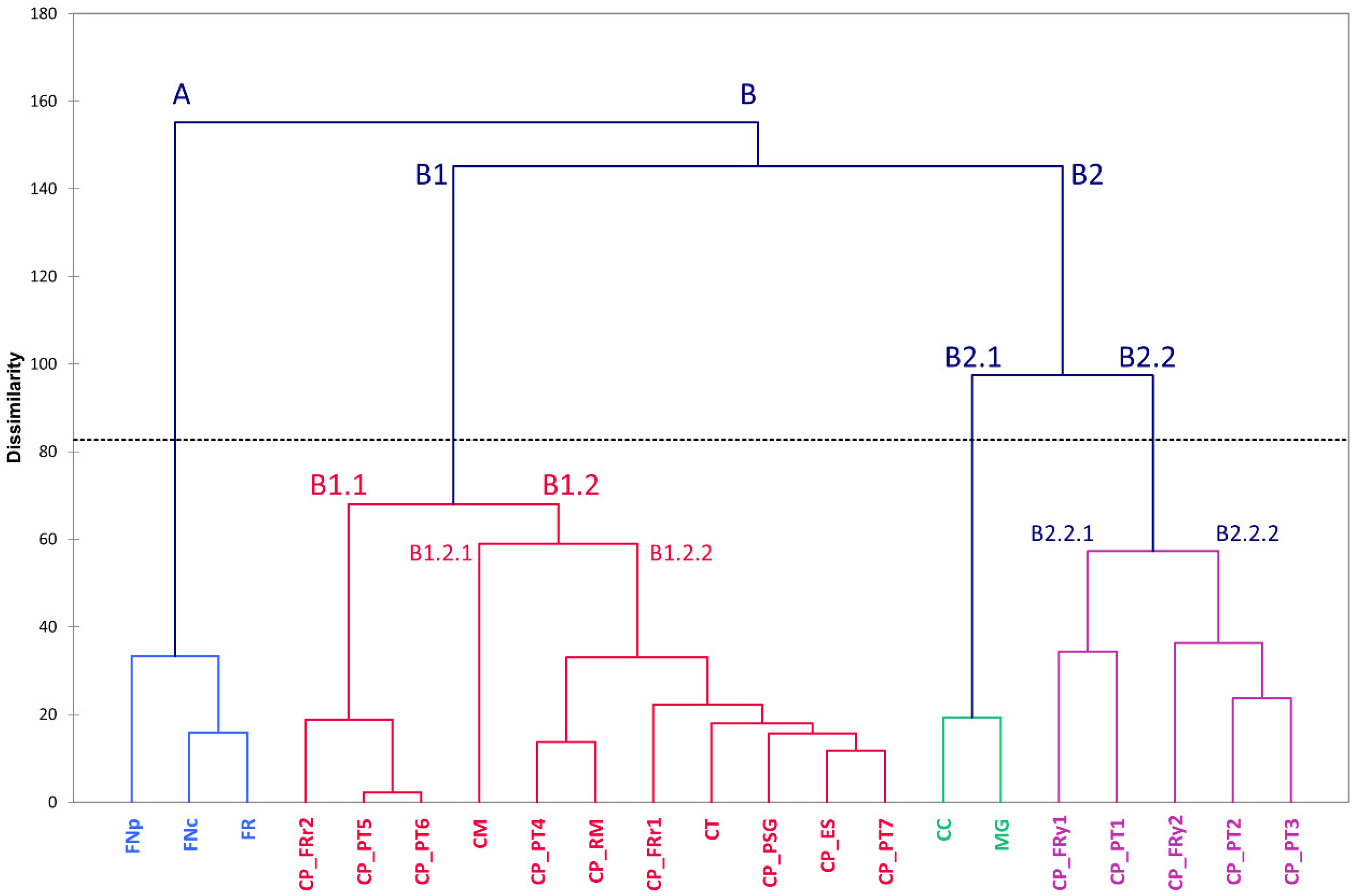
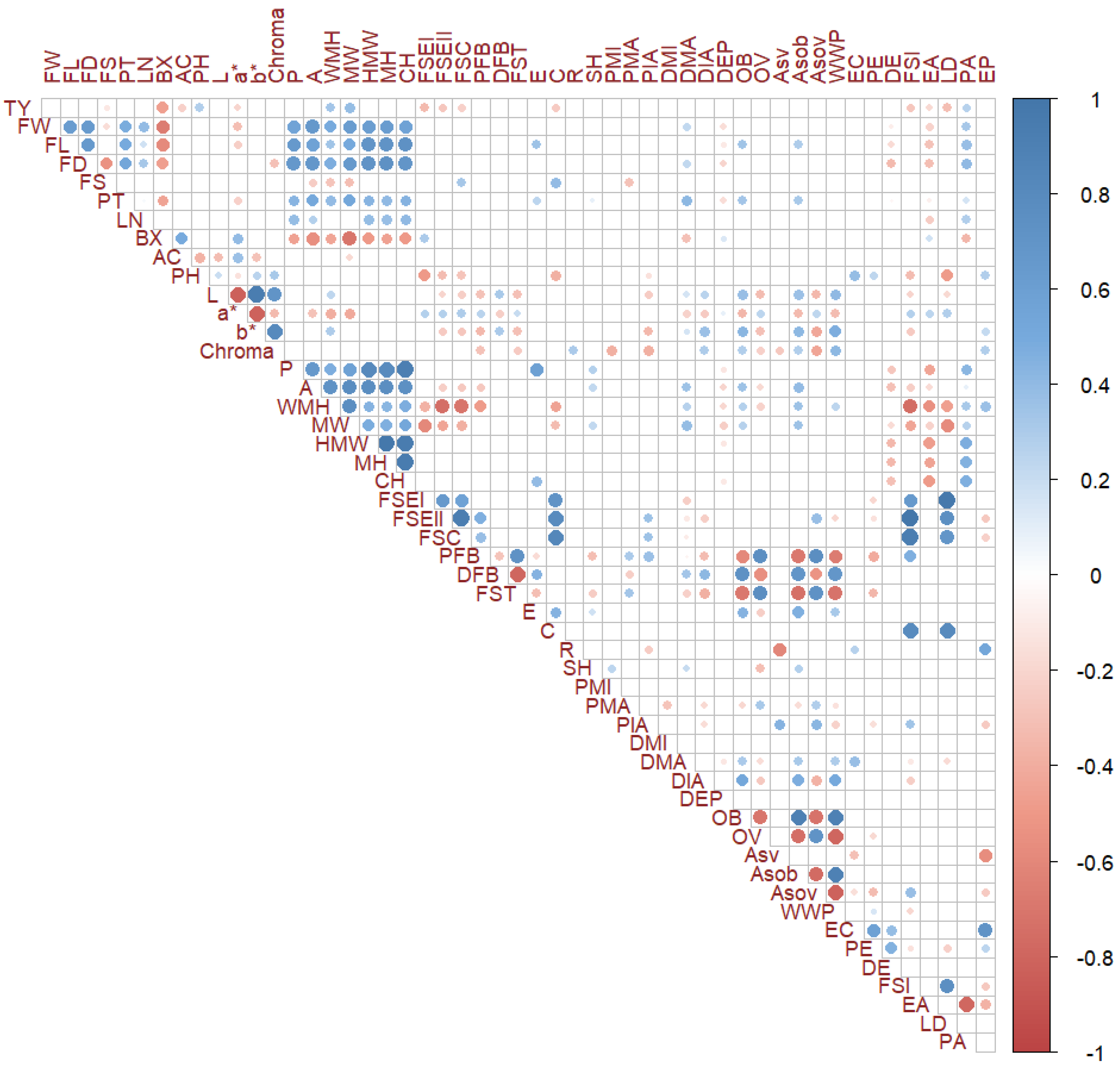
3.3. Multivariate Analysis and Correlations between Traits
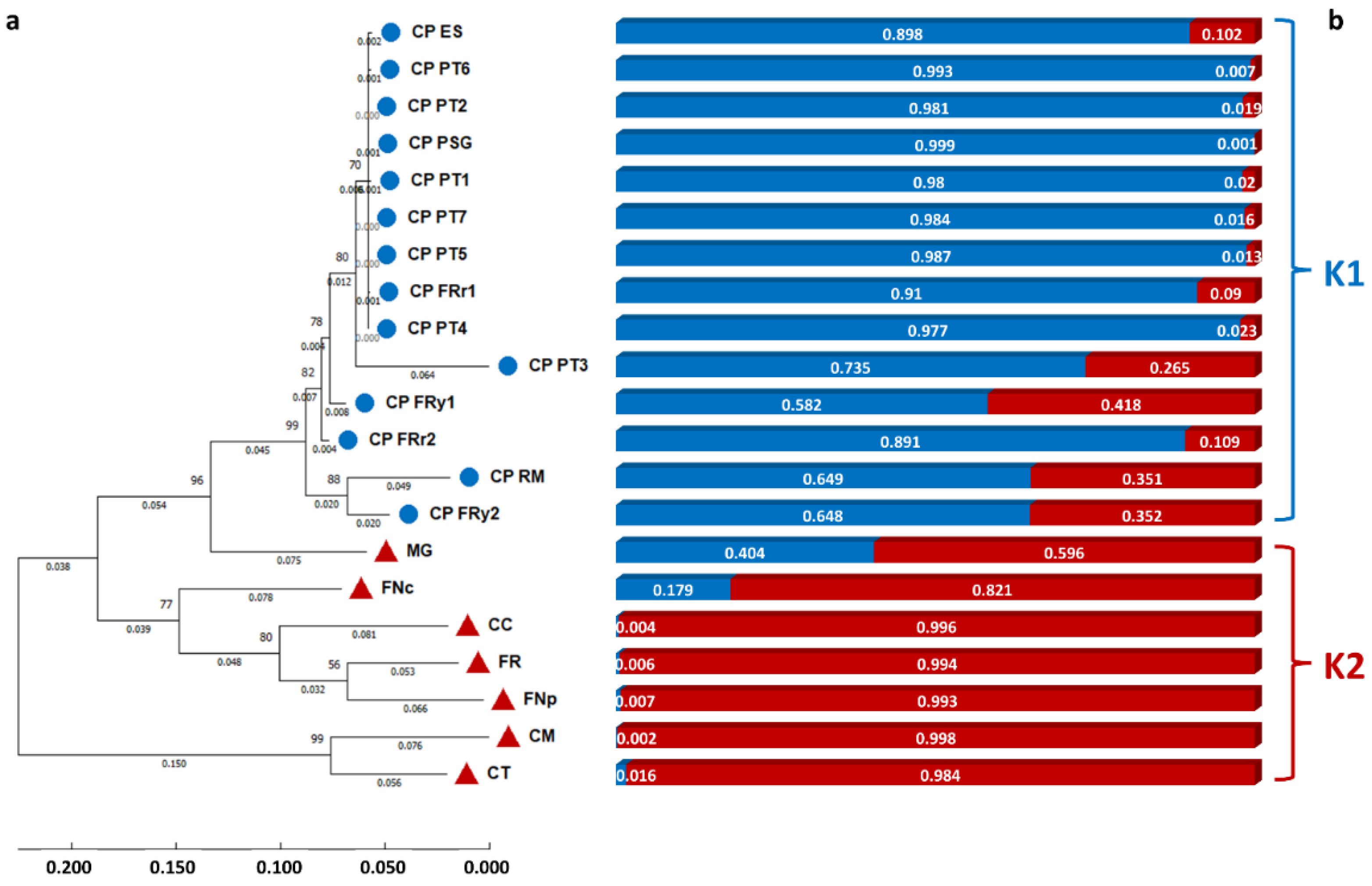
3.4. Genomic Diversity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jarret, R.L.; Barboza, G.E.; Costa Batista, F.; Berke, T.; Chou, Y.; Hulse-Kemp, A.; Ochoa-Alejo, N.; Tripodi, P.; Veres, A.; Garcia, C.C.; et al. Capsicum—An abbreviated compendium. J. Am. Soc. Hortic. Sci. 2019, 144, 3–22. [Google Scholar] [CrossRef] [Green Version]
- FAOSTAT. 2021. Available online: http://www.fao.org/faostat/en/#home (accessed on 26 May 2022).
- Tripodi, P.; Francese, G.; Sanajà, V.O.; di Cesare, C.; Festa, G.; D’Alessandro, A.; Mennella, G. A multi-methodological approach to study genomic footprints and environmental influence on agronomic and metabolic profiles in a panel of Italian traditional sweet pepper varieties. J. Food Compos. Anal. 2021, 103, 104116. [Google Scholar] [CrossRef]
- Sobolev, A.P.; Mannina, L.; Capitani, D.; Sanzò, G.; Ingallina, C.; Botta, B.; Fornarini, S.; Crestoni, M.E.; Chiavarino, B.; Carradori, S.; et al. A multi-methodological approach in the study of Italian PDO “Cornetto di Pontecorvo” red sweet pepper. Food Chem. 2018, 255, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Di Sotto, A.; Vecchiato, M.; Abete, L.; Toniolo, C.; Giusti, A.M.; Mannina, L.; Locatelli, M.; Nicoletti, M.; Di Giacomo, S. Capsicum annuum L. var. Cornetto di Pontecorvo PDO: Polyphenolic profile and in vitro biological activities. J. Funct. Foods 2018, 40, 679–691. [Google Scholar] [CrossRef]
- Mennella, G.; D’Alessandro, A.; Francese, G.; Fontanella, D.; Parisi, M.; Tripodi, P. Occurrence of variable levels of health-promoting fruit compounds in horn-shaped Italian sweet pepper varieties assessed by a comprehensive approach. J. Sci. Food Agric. 2018, 98, 3280–3289. [Google Scholar] [CrossRef]
- Registro Volontario Regionale. Available online: https://www.arsial.it/biodiversita/registro-volontario-regionale/ (accessed on 26 May 2022).
- Costanza, M.T.; Miracolo, I.B.; Taviani, P.; Paoletti, S.; Rea, R.; Lelli, L.; Garzia, J.H.; Porfiri, O.; Nardi, P.; Tanca, M. On Farm Conservation of Plant Genetic Resources in Lazio Region, Italy. Implementation of the Regional Act 1st March 2000 N°15. In Agrobiodiversity Conservation: Securing the Diversity of Crop Wild Relatives and Landraces; Maxted, N., Dulloo, E.M., Ford-Lloyd, B.V., Frese, L., Iriondo, J., de Carvalho, M.A.A.P., Eds.; CABI: Wallingford, UK, 2012; Volume 365, pp. 161–172. ISBN 9781845938512. [Google Scholar]
- Union. The Council of the European Union. Council Regulation (EC) No 510/2006 of 20 March 2006 on the Protection of Geographical Indications and Designations of Origin for Agricultural Products and Foodstuffs. 2006. Available online: https://eur-lex.europa.eu/eli/reg/2006/510/oj (accessed on 26 May 2022).
- Dias, C.; Mendes, L. Protected Designation of Origin (PDO), Protected Geographical Indication (PGI) and Traditional Speciality Guaranteed (TSG): A bibiliometric analysis. Food Res. Int. 2018, 103, 492–508. [Google Scholar] [CrossRef]
- Arfini, F.; Bellassen, V. (Eds.) Sustainability of European Food Quality Schemes: Multi-Performance, Structure, and Governance of PDO, PGI, and Organic Agri-Food Systems; Springer Nature: Basel, Switzerland, 2019. [Google Scholar]
- Scheben, A.; Batley, J.; Edwards, D. Revolution in Genotyping Platforms for Crop Improvement. In Advances in Biochemical Engineering Biotechnology; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- D’Agostino, N.; Tripodi, P. NGS-based genotyping, high-throughput phenotyping and genome-wide association studies laid the foundations for next-generation breeding in horticultural crops. Diversity 2017, 9, 38. [Google Scholar] [CrossRef] [Green Version]
- Basak, M.; Uzun, B.; Yol, E. Genetic diversity and population structure of the Mediterranean sesame core collection with use of genome-wide SNPs developed by double digest RAD-Seq. PLoS ONE 2019, 14, e0223757. [Google Scholar] [CrossRef]
- De Woody, J.; Nason, J.D.; Hipkins, V.D. Mitigating scoring errors in microsatellite data from wild populations. Mol. Ecol. Notes 2006, 6, 951–957. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.Q.; Chen, Y.M.; Wang, J.P.; Guo, C.; Zhao, L.; Wang, X.Y.; Guo, Y.; Li, L.; Li, D.Z.; Guo, Z.H. Development of a universal and simplified ddRAD library preparation approach for SNP discovery and genotyping in angiosperm plants. Plant Methods 2016, 12, 39. [Google Scholar] [CrossRef] [Green Version]
- Tripodi, P.; Greco, B. Large scale phenotyping provides insight into the diversity of vegetative and reproductive organs in a wide collection of wild and domesticated peppers (Capsicum spp.). Plants 2018, 7, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Tarasov, A.; Vilella, A.J.; Cuppen, E.; Nijman, I.J.; Prins, P. Sambamba: Fast processing of NGS alignment formats. Bioinformatics 2015, 31, 2032–2034. [Google Scholar] [CrossRef]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Structure Harverster. Available online: http://taylor0.biology.ucla.edu/structureHarvester/ (accessed on 26 May 2022).
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- R Core Team. The R Project for Statistical Computing. 2020. Available online: https://www.R-project.org/ (accessed on 26 May 2022).
- Wei, T.; Simko, V. R package “corrplot”: Visualization of a Correlation Matrix. Available online: https://github.com/taiyun/corrplot (accessed on 26 May 2022).
- Mantel, N.A. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
- XLSTAT. Data Analysis and Statistical Solutions for Microsoft Excel; Addinsoft: Paris, France, 2012. [Google Scholar]
- Tripodi, P.; Ficcadenti, N.; Rotino, G.L.; Festa, G.; Bertone, A.; Pepe, A.; Caramanico, R.; Migliori, C.A.; Spadafora, D.; Schiavi, M.; et al. Genotypic and environmental effects on the agronomic, health-related compounds and antioxidant properties of chilli peppers for diverse market destinations. J. Sci. Food Agric. 2019, 99, 4550–4560. [Google Scholar] [CrossRef]
- UPOV. General Introduction to the Examination of Distinctness, Uniformity and Stability and the Development of Harmonized Descriptions of New Varieties of Plants, TG/1/3. Available online: https://www.upov.int/en/publications/tg-rom/tg001/tg_1_3.pdf(2002) (accessed on 13 June 2022).
- Ji, M.; Deng, J.; Yao, B.; Chen, R.; Fan, Z.; Guan, J.; Li, X.; Wu, F.; Niklas, K.J. Ecogeographical variation of 12 morphological traits within Pinus tabulaeformis: The effects of environmental factors and demographic histories. J. Plant Ecol. 2017, 10, 386–396. [Google Scholar]
- Amiteye, S. Basic concepts and methodologies of DNA marker systems in plant molecular breeding. Heliyon 2021, 7, e080932. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Falcón, J.E.; Vilanova, S.; Plazas, M.; Prohens, J. Diversity, relationships, and genetic fingerprinting of the Listada de Gandía eggplant landrace using genomic SSRS and EST-SSRs. Sci. Hortic. 2011, 129, 238–246. [Google Scholar] [CrossRef] [Green Version]
- Caramante, M.; Rao, R.; Monti, L.M.; Corrado, G. Discrimination of San Marzano accessions: A comparison of minisatellite, CAPS and SSR markers in relation to morphological traits. Sci. Hortic. 2009, 120, 560–564. [Google Scholar] [CrossRef]
- Tripodi, P.; Pepe, R.; Francese, G.; Rosaria, M.; Onofaro Sanajà, V.; Di Cesare, C.; Festa, G.; D’Alessandro, A.; Mennella, G. Biochemical Characterisation and Genetic Structure Provide Insight into the Diversity of the Mediterranean Tomato Ancient Varieties ‘San Marzano’ and ‘Re Fiascone’: New Resources for Breeding. Agronomy 2022, 12, 18. [Google Scholar] [CrossRef]
- Ganopoulos, I.; Argiriou, A.; Tsaftaris, A. Microsatellite high resolution melting (SSR-HRM) analysis for authenticity testing of protected designation of origin (PDO) sweet cherry products. Food Control 2011, 22, 532–541. [Google Scholar] [CrossRef]
- Pusadee, T.; Jamjod, S.; Chiang, Y.C.; Rerkasem, B.; Schaal, B.A. Genetic structure and isolation by distance in a landrace of Thai rice. Proc. Natl. Acad. Sci. USA 2009, 106, 13880–13885. [Google Scholar] [CrossRef] [Green Version]


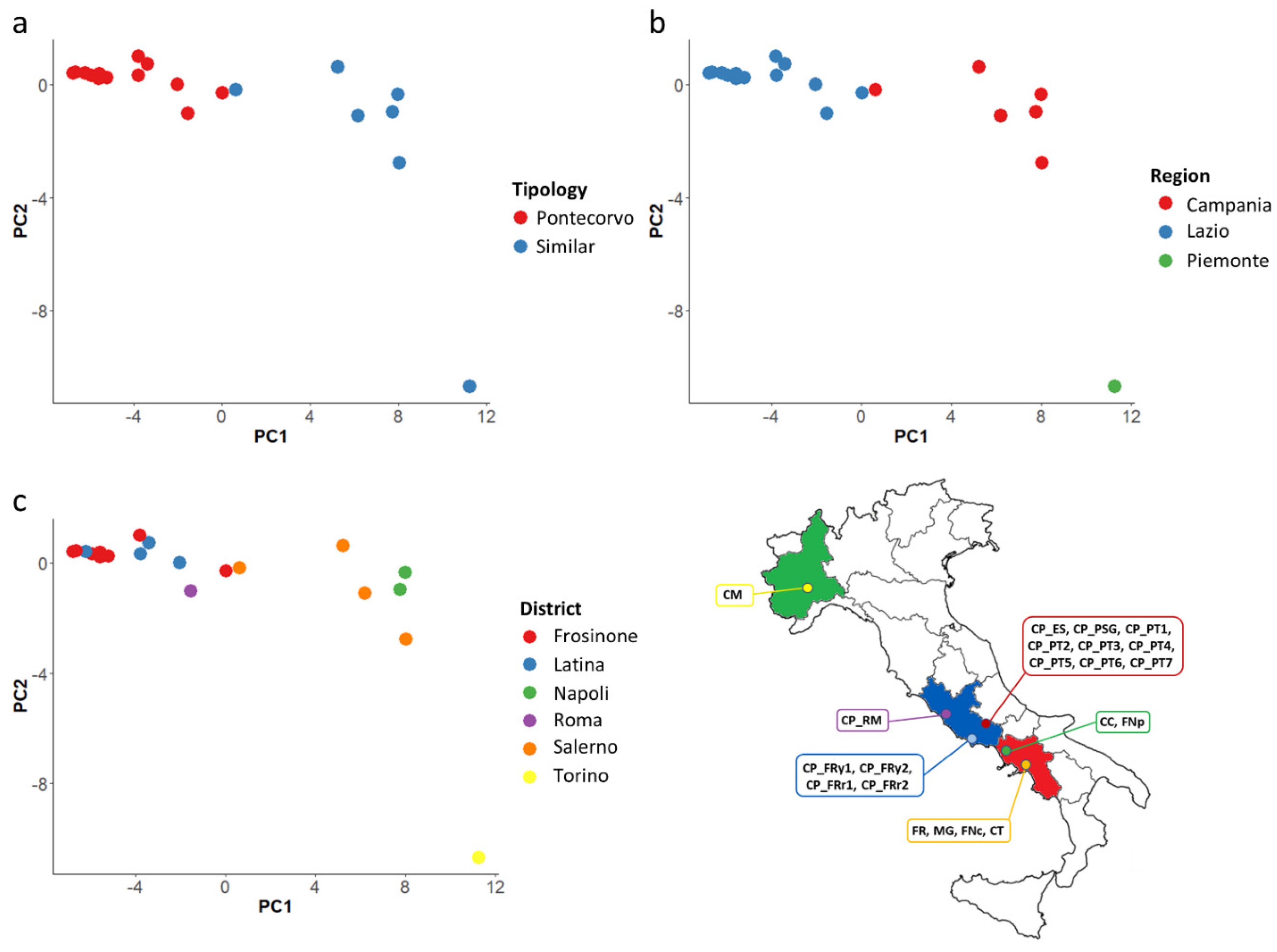
| Code | Name | Cultivation Area | District | Region | Coordinate | M.a.s.l. |
|---|---|---|---|---|---|---|
| CP_ES | Cornetto di Pontecorvo | Esperia | Frosinone | Latium | 41°23′ N; 13°41′ E | 370 |
| CP_FRy1 | Cornetto di Pontecorvo ‘giallo’ | Formia | Latina | Latium | 42°15′ N; 13°37′ E | 19 |
| CP_FRy2 | Cornetto di Pontecorvo ‘giallo’ | Formia | Latina | Latium | 42°15′ N; 13°37′ E | 19 |
| CP_FRr1 | Cornetto di Pontecorvo Rosso | Formia | Latina | Latium | 42°15′ N; 13°37′ E | 19 |
| CP_FRr2 | Cornetto di Pontecorvo ‘linea 49 rosso’ | Formia | Latina | Latium | 42°15′ N; 13°37′ E | 19 |
| CP_PSG | Cornetto di Pontecorvo | Piedimonte San Germano | Frosinone | Latium | 41°29′ N; 13°45′ E | 115 |
| CP_PT1 | Cornetto di Pontecorvo | Pontecorvo | Frosinone | Latium | 42°27′ N; 13°40′ E | 97 |
| CP_PT2 | Cornetto di Pontecorvo | Pontecorvo | Frosinone | Latium | 42°27′ N; 13°40′ E | 97 |
| CP_PT3 | Cornetto di Pontecorvo | Pontecorvo | Frosinone | Latium | 42°27′ N; 13°40′ E | 97 |
| CP_PT4 | Cornetto di Pontecorvo | Pontecorvo | Frosinone | Latium | 42°27′ N; 13°40′ E | 97 |
| CP_PT5 | Cornetto di Pontecorvo | Pontecorvo | Frosinone | Latium | 42°27′ N; 13°40′ E | 97 |
| CP_PT6 | Cornetto di Pontecorvo | Pontecorvo | Frosinone | Latium | 42°27′ N; 13°40′ E | 97 |
| CP_PT7 | Corno Pontecorvo | Pontecorvo | Frosinone | Latium | 42°27′ N; 13°40′ E | 97 |
| CP_RM | Cornetto di Pontecorvo | Roma | Roma | Latium | 41°54′ N; 12°28′ E | 21 |
| CM | Corno tipo Carmagnola | Carmagnola | Torino | Piedmont | 44°50′ N; 07°43′ E | 240 |
| FR | Frigitello | Sarno | Salerno | Campania | 40°49′ N; 14°37′ E | 30 |
| CC | Corno di Capra | Acerra | Napoli | Campania | 40°57′ N; 14°22′ E | 26 |
| MG | Marconi Giallo | Sarno | Salerno | Campania | 40°49′ N; 14°37′ E | 30 |
| FNp | Friariello Napoli | Torre Del Greco | Napoli | Campania | 40°47′ N; 14°23′ E | 40 |
| FNc | Friariello Nocerese | Nocera | Salerno | Campania | 40°45′ N; 14°38′ E | 43 |
| CT | Corno di Toro Giallo | Piana del Sele | Salerno | Campania | 41°37′ N; 14°59′ E | 72 |
| Multivariate Test (Factor ‘Cultivar Groups’). | Multivariate Test (Factor ‘Genotype’) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect | Test Statistic | Value | F | df Hypothesis | df Error | Sig. (p) | Value | F | df Hypothesis | df Error | Sig. (p) |
| Intercept | Pillai’s trace | 1.00 | 3113.43 | 52 | 10 | <0.001 | 1.00 | 114,074.88 | 42 | 1 | <0.01 |
| Wilks’s lambda | 0.00 | 3113.43 | 52 | 10 | <0.001 | 0.00 | 114,074.88 | 42 | 1 | <0.01 | |
| Category | Pillai’s trace | 0.98 | 9.05 | 52 | 10 | <0.001 | 17.61 | 3.50 | 840 | 400 | <0.001 |
| Wilks’s lambda | 0.02 | 9.05 | 52 | 10 | <0.001 | 0.00 | 18.94 | 840 | 132 | <0.001 | |
| Trait | Acronyms | Rsquare | F Ratio | Prob > F | Pontecorvo | Similar | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Range | Mean | CV | Range | Mean | CV | |||||
| Agronomic Traits | ||||||||||
| Total Yield | TY | 0.00 | 0.16 | ns | 2.24–0.87 | 1.42 | 21.83 | 2.21–0.93 | 1.38 | 26.09 |
| Fruit Weight | FW | 0.55 | 73.86 | *** | 186.18–91.71 | 136.37 | 15.93 | 132.62–23.11 | 71.45 | 53.63 |
| Fruit Length | FL | 0.34 | 31.53 | *** | 19.50–9.00 | 15.08 | 14.32 | 17.00–6.10 | 11.41 | 25.77 |
| Fruit Width | FD | 0.16 | 11.32 | ** | 5.50–3.40 | 4.70 | 10.43 | 6.30–1.60 | 3.92 | 34.18 |
| Fruit Shape | FS | 0.01 | 0.52 | ns | 4.44–2.18 | 3.23 | 15.79 | 4.89–2.08 | 3.11 | 26.37 |
| Pericarp Thickness | PT | 0.14 | 10.17 | ** | 6.41–2.50 | 4.59 | 20.26 | 7.06–2.29 | 3.71 | 33.42 |
| Locules Number | LN | 0.07 | 4.94 | * | 4.00–3.00 | 3.50 | 12.29 | 4.00–3.00 | 3.26 | 10.43 |
| Quality Traits | ||||||||||
| Brix Degree | BX | 0.38 | 38.12 | *** | 6.90–4.60 | 5.94 | 10.10 | 9.30–4.90 | 7.56 | 19.58 |
| Total Acidity | AC | 0.00 | 0.16 | ns | 0.21–0.10 | 0.17 | 17.65 | 0.25–0.10 | 0.18 | 22.22 |
| pH | PH | 0.01 | 0.31 | ns | 5.90–5.09 | 5.31 | 2.82 | 6.36–4.76 | 5.35 | 6.92 |
| L | L | 0.02 | 1.25 | ns | 56.68–26.85 | 38.14 | 19.06 | 57.73–32.27 | 40.47 | 21.70 |
| a | a* | 0.00 | 0.25 | ns | 38.82–3.34 | 29.42 | 27.97 | 58.60–3.11 | 27.93 | 56.14 |
| b | b* | 0.02 | 1.23 | ns | 55.81–13.39 | 23.88 | 49.37 | 58.90–14.44 | 27.70 | 53.43 |
| Chroma | C | 0.07 | 4.34 | * | 56.50–32.56 | 39.96 | 16.14 | 62.28–27.05 | 43.89 | 18.60 |
| Category | Trait | Acronyms | R-Squared | F Ratio | Prob > F | Pontecorvo | Similar | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Range | Mean | CV | Range | Mean | CV | ||||||
| Fruit size | Perimeter | P | 0.289 | 24.853 | *** | 109.42–50.85 | 63.33 | 19.04 | 79.69–28.64 | 46.71 | 28.45 |
| Area | A | 0.286 | 24.424 | *** | 96.54–58.28 | 80.19 | 12.89 | 89.35–23.70 | 59.6 | 38.34 | |
| Width Mid-height | WMH | 0.049 | 3.137 | ns | 7.95–3.73 | 5.29 | 17.01 | 7.24–2.29 | 4.76 | 30.67 | |
| Maximum Width | MW | 0.197 | 14.953 | *** | 8.70–5.88 | 7.12 | 8.43 | 8.33–2.92 | 6.04 | 26.66 | |
| Height Mid-width | HMW | 0.246 | 19.876 | *** | 23.02–16.40 | 18.7 | 9.20 | 25.61–10.61 | 15.43 | 26.64 | |
| Maximum Height | MH | 0.233 | 18.492 | *** | 29.04–16.72 | 19.78 | 7.84 | 28.42–11.57 | 16.38 | 28.51 | |
| Curved Height | CH | 0.248 | 20.108 | *** | 29.78–17.29 | 20.86 | 11.36 | 27.97–12.11 | 17.09 | 25.34 | |
| Fruit shape index | Fruit Shape Index External I | FSEI | 0.000 | 0.000 | ns | 3.45–2.20 | 2.8 | 11.43 | 4.09–1.61 | 2.8 | 26.07 |
| Fruit Shape Index External II | FSEII | 0.023 | 1.414 | ns | 4.52–2.61 | 3.62 | 16.02 | 4.83–1.96 | 3.39 | 27.14 | |
| Curved Fruit Shape Index | FSC | 0.067 | 4.351 | * | 5.04–2.98 | 4.02 | 15.42 | 5.35–2.11 | 3.6 | 26.39 | |
| Blockiness | Proximal Fruit Blockiness | PFB | 0.000 | 0.004 | ns | 1.45–0.57 | 1.07 | 26.17 | 1.49–0.14 | 1.08 | 23.15 |
| Distal Fruit Blockiness | DFB | 0.257 | 21.073 | *** | 1.39–0.52 | 0.89 | 24.72 | 0.85–0.42 | 0.65 | 20.00 | |
| Fruit Shape Triangle | FST | 0.127 | 8.896 | ** | 2.42–0.46 | 1.31 | 38.17 | 2.81–0.19 | 1.72 | 32.56 | |
| Homogeneity | Ellipsoid | E | 0.059 | 3.832 | ns | 0.24–0.08 | 0.11 | 27.27 | 0.14–0.08 | 0.1 | 20.00 |
| Circular | C | 0.122 | 8.450 | ** | 0.43–0.31 | 0.35 | 8.57 | 0.40–0.18 | 0.32 | 18.75 | |
| Rectangular | R | 0.000 | 0.001 | ns | 0.61–0.30 | 0.48 | 16.67 | 0.60–0.34 | 0.48 | 14.58 | |
| Proximal fruit end-shape | Shoulder Height | SH | 0.004 | 0.239 | ns | 0.11–0.00 | 0.03 | 66.67 | 0.09–0.00 | 0.02 | 100.00 |
| Proximal Angle Micro | PMI | 0.170 | 12.537 | *** | 359.50–9.30 | 206.6 | 47.69 | 296.40–0.40 | 116.9 | 74.10 | |
| Proximal Angle Macro | PMA | 0.021 | 1.325 | ns | 219.60–4.09 | 82.56 | 70.54 | 209.70–1.50 | 100.29 | 56.13 | |
| Proximal Indentation Area | PIA | 0.107 | 7.317 | ** | 0.29–0.00 | 0.08 | 100.00 | 0.07–0.00 | 0.03 | 66.67 | |
| Distal fruit end-shape | Distal Angle Micro | DMI | 0.033 | 2.113 | ns | 348.10–1.80 | 117.14 | 62.38 | 342.70–0.00 | 88.81 | 81.78 |
| Distal Angle Macro | DMA | 0.056 | 3.650 | ns | 152.00–8.00 | 71.05 | 39.52 | 112.00–10.00 | 57.32 | 42.24 | |
| Distal Indentation Area | DIA | 0.089 | 5.954 | * | 0.18–0.00 | 0.03 | 166.67 | 0.04–0.00 | 0.01 | 100.00 | |
| Distal End Protrusion | DEP | 0.002 | 0.139 | ns | 0.99–0.00 | 0.08 | 250.00 | 0.81–0.00 | 0.1 | 220.00 | |
| Asymmetry | Obovoid | OB | 0.107 | 7.337 | ** | 0.64–0.00 | 0.13 | 169.23 | 0.00–-0.01 | 0 | 0.00 |
| Ovoid | OV | 0.018 | 1.097 | ns | 0.68–0.00 | 0.31 | 80.65 | 0.63–0.00 | 0.38 | 36.84 | |
| V. Asymmetry | Asv | 0.125 | 8.720 | ** | 1.21–0.19 | 0.52 | 48.08 | 0.61–0.14 | 0.35 | 31.43 | |
| H. Asymmetry.ob | Asob | 0.086 | 5.737 | * | 2.81–0.00 | 0.59 | 166.10 | 1.32–0.00 | 0.06 | 483.33 | |
| H. Asymmetry.ov | Asov | 0.001 | 0.046 | ns | 2.92–0.00 | 1.39 | 71.22 | 2.48–0.00 | 1.33 | 48.12 | |
| Width Widest Pos | WWP | 0.027 | 1.670 | ns | 0.89–0.06 | 0.34 | 94.12 | 0.50–0.08 | 0.25 | 40.00 | |
| Internal eccentricity | Eccentricity | EC | 0.012 | 0.730 | ns | 0.80–0.69 | 0.76 | 3.95 | 0.80–0.61 | 0.75 | 6.67 |
| Proximal Eccentricity | PE | 0.001 | 0.090 | ns | 1.00–0.84 | 0.89 | 3.37 | 1.04–0.77 | 0.89 | 5.62 | |
| Distal Eccentricity | DE | 0.005 | 0.293 | ns | 1.00–0.83 | 0.89 | 3.37 | 0.96–0.74 | 0.89 | 5.62 | |
| Fruit Shape Index Internal | FSI | 0.022 | 1.365 | ns | 4.52–2.62 | 3.62 | 16.02 | 4.84–1.96 | 3.4 | 27.06 | |
| Eccentricity Area Index | EA | 0.005 | 0.323 | ns | 0.55–0.13 | 0.38 | 26.32 | 0.50–-0.02 | 0.36 | 33.33 | |
| Latitudinal section | Lobedness Degree | LD | 0.011 | 0.686 | ns | 45.73–26.40 | 35.55 | 14.37 | 53.51–13.62 | 33.79 | 34.89 |
| Pericarp Area | PA | 0.005 | 0.282 | ns | 0.80–0.57 | 0.67 | 10.45 | 1.10–0.56 | 0.66 | 19.70 | |
| Epicarp Thickness | EP | 0.001 | 0.043 | ns | 0.25–0.21 | 0.23 | 4.35 | 0.24–0.19 | 0.23 | 4.35 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tripodi, P.; D’Alessandro, R.; Festa, G.; Taviani, P.; Rea, R. Profiling the Diversity of Sweet Pepper ‘Peperone Cornetto di Pontecorvo’ PDO (Capsicum annuum) through Multi-Phenomic Approaches and Sequencing-Based Genotyping. Agronomy 2022, 12, 1433. https://doi.org/10.3390/agronomy12061433
Tripodi P, D’Alessandro R, Festa G, Taviani P, Rea R. Profiling the Diversity of Sweet Pepper ‘Peperone Cornetto di Pontecorvo’ PDO (Capsicum annuum) through Multi-Phenomic Approaches and Sequencing-Based Genotyping. Agronomy. 2022; 12(6):1433. https://doi.org/10.3390/agronomy12061433
Chicago/Turabian StyleTripodi, Pasquale, Rosa D’Alessandro, Giovanna Festa, Paola Taviani, and Roberto Rea. 2022. "Profiling the Diversity of Sweet Pepper ‘Peperone Cornetto di Pontecorvo’ PDO (Capsicum annuum) through Multi-Phenomic Approaches and Sequencing-Based Genotyping" Agronomy 12, no. 6: 1433. https://doi.org/10.3390/agronomy12061433
APA StyleTripodi, P., D’Alessandro, R., Festa, G., Taviani, P., & Rea, R. (2022). Profiling the Diversity of Sweet Pepper ‘Peperone Cornetto di Pontecorvo’ PDO (Capsicum annuum) through Multi-Phenomic Approaches and Sequencing-Based Genotyping. Agronomy, 12(6), 1433. https://doi.org/10.3390/agronomy12061433







