GWAS and Identification of Candidate Genes Associated with Seed Soluble Sugar Content in Vegetable Soybean
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Experimental Conditions
2.2. The Total of Soluble Sugar Measurement in Vegetable Soybean Fresh Seeds
2.3. Genome-Wide Association Study (GWAS) Analysis
2.4. Survey of Candidate Genes
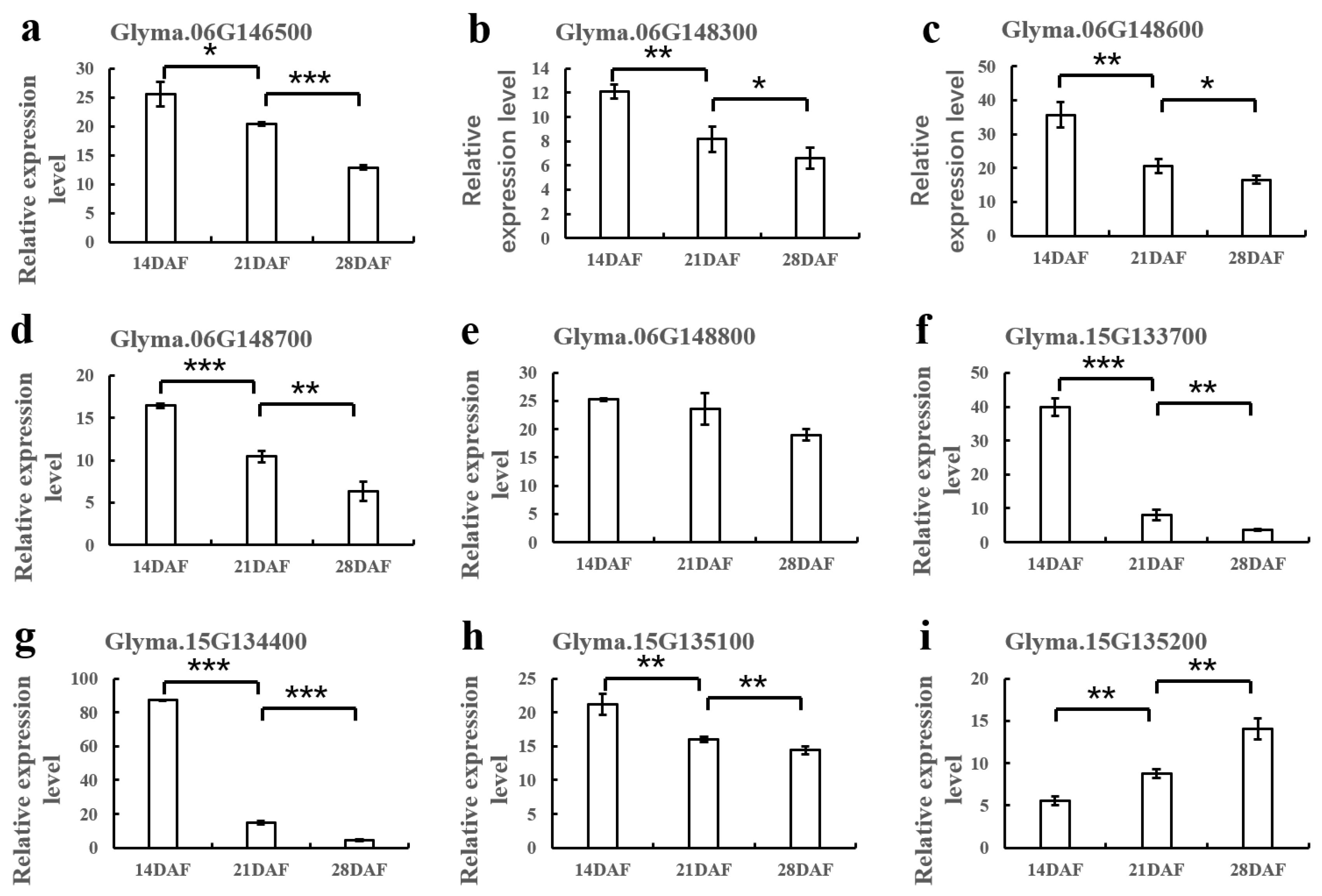
2.5. RT-PCR for Candidate Genes
3. Results
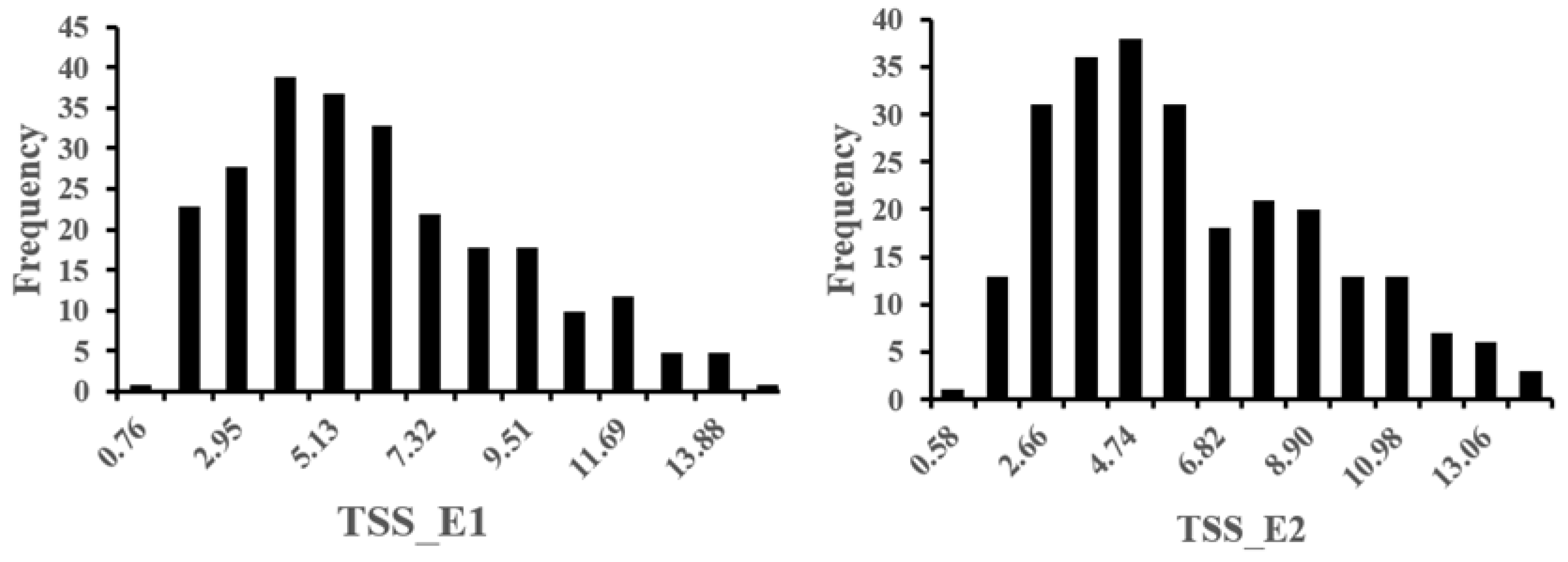
3.1. Phenotypic Variation
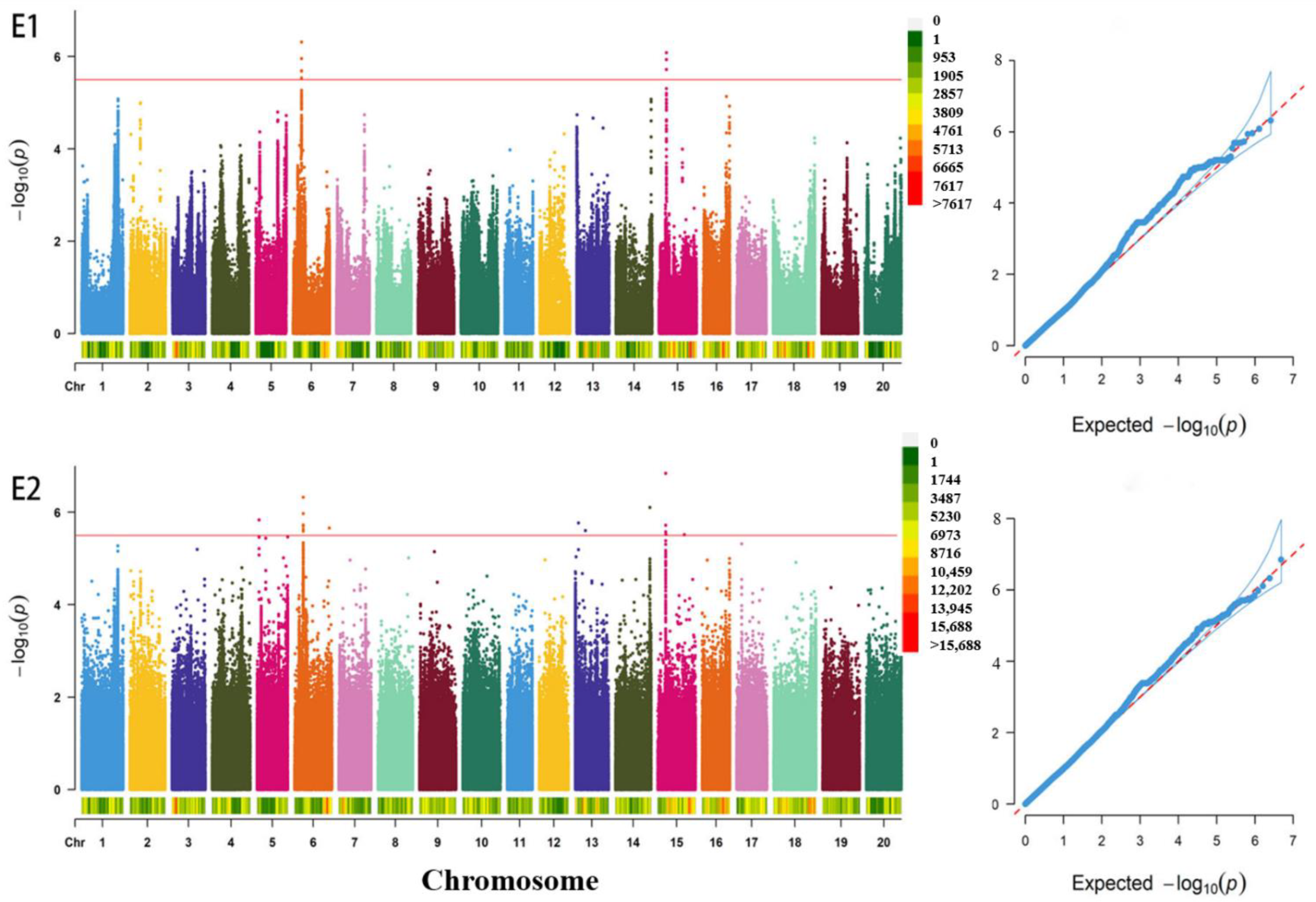
3.2. GWAS for the Total Soluble Sugar
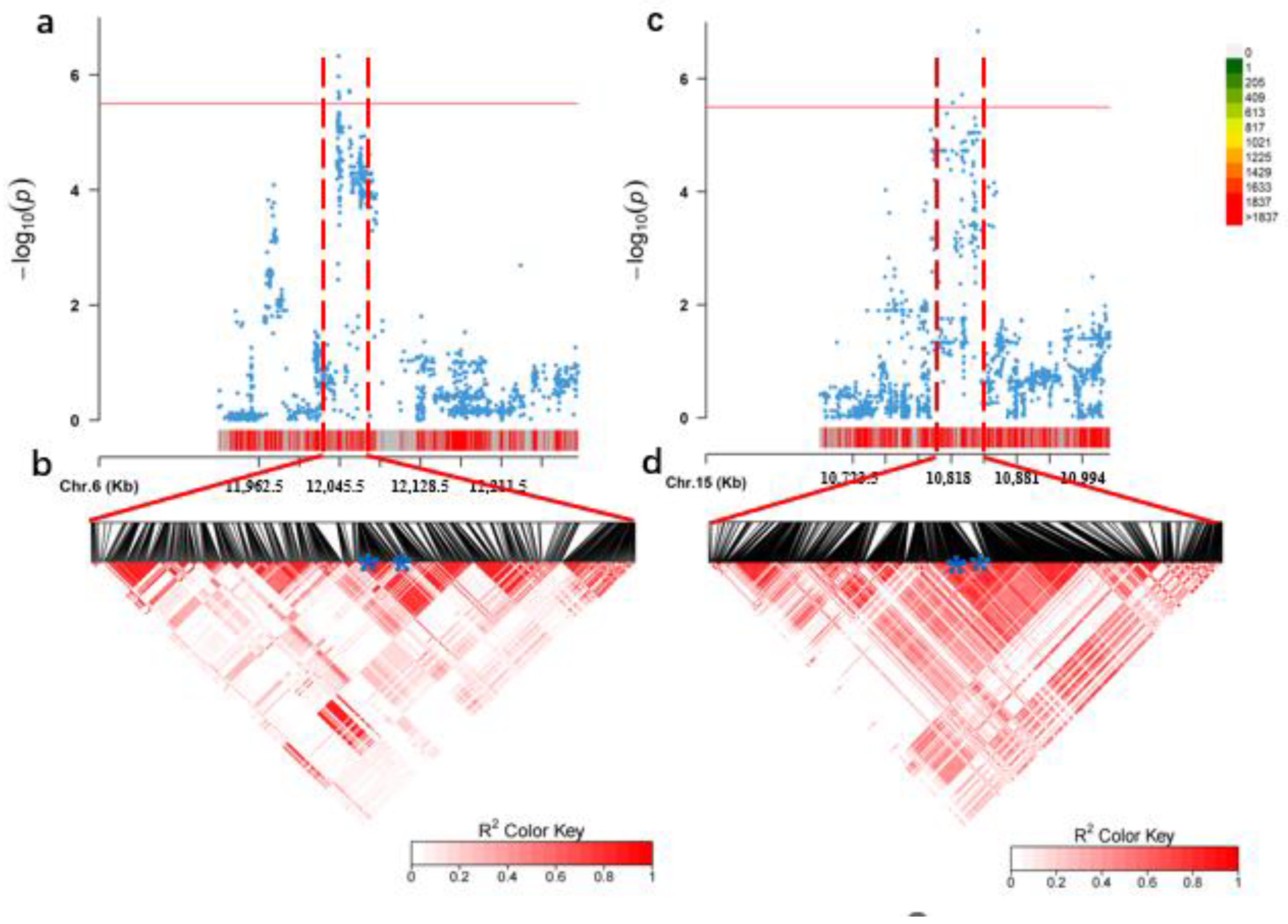
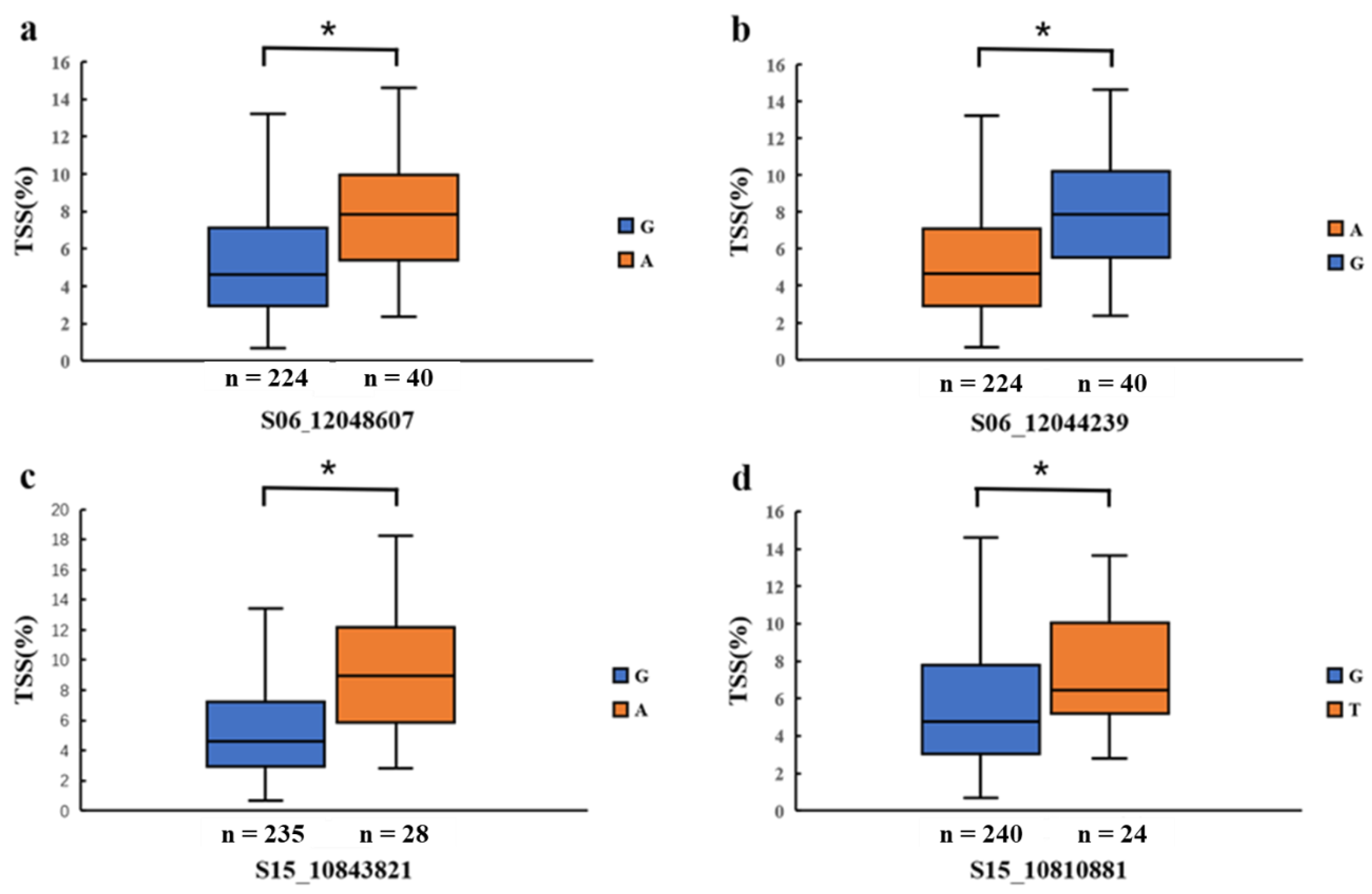
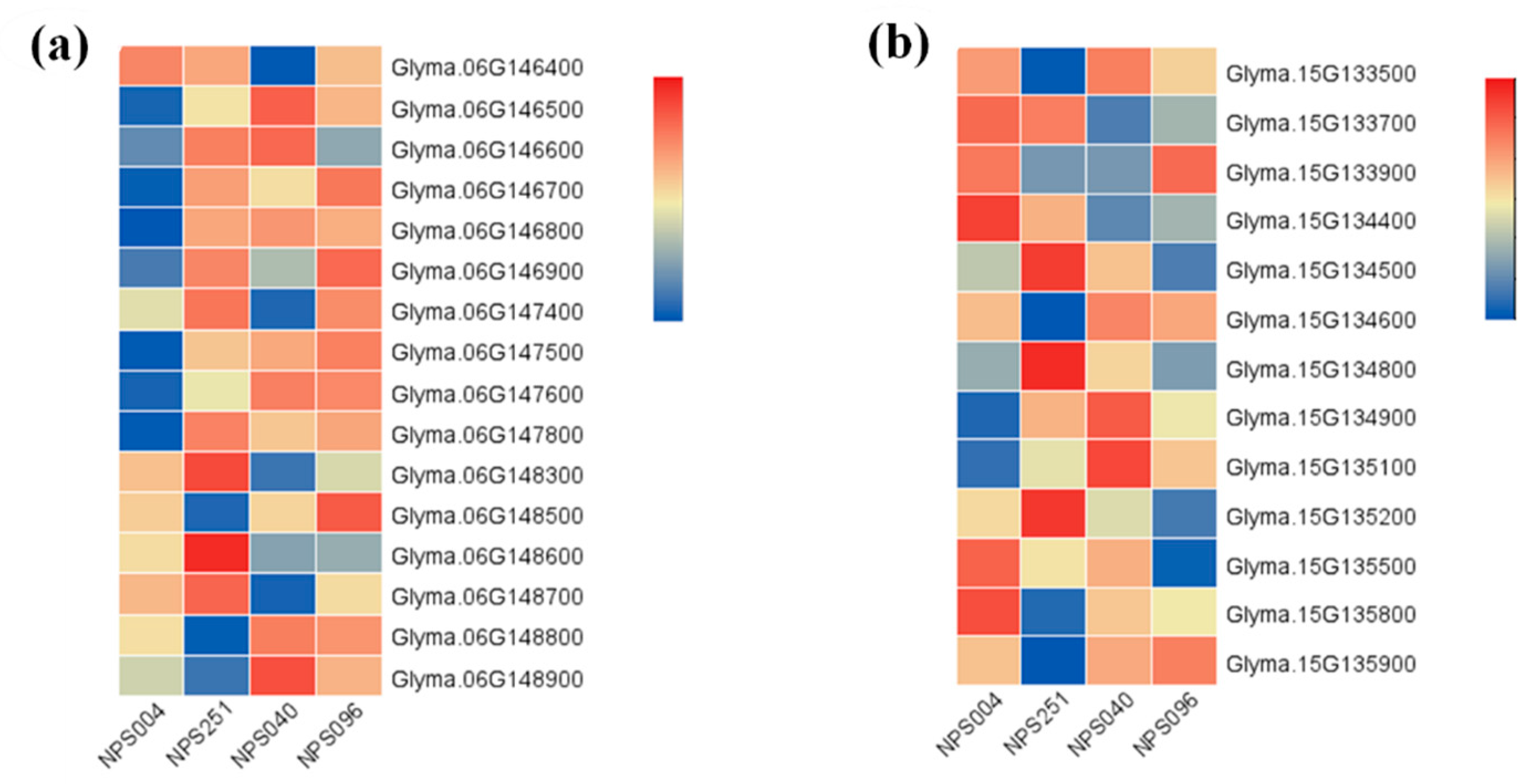
3.3. Identification of Candidate Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Young, G.; Mebrahtu, T.; Johnson, J. Acceptability of green soybeans as a vegetable entity. Mater. Veg. 2000, 55, 323–333. [Google Scholar] [CrossRef]
- Hou, A.; Chen, P.; Shi, A.; Alloatti, J.; Zhang, B.; Wang, Y.-J. Sugar Variation in Soybean Seed Assessed with a Rapid Extraction and Quantification Method. Int. J. Agron. 2008, 2009, 484571. [Google Scholar] [CrossRef] [Green Version]
- Umer, M.J.; Bin Safdar, L.; Gebremeskel, H.; Zhao, S.; Yuan, P.; Zhu, H.; Kaseb, M.O.; Anees, M.; Lu, X.; He, N.; et al. Identification of key gene networks controlling organic acid and sugar metabolism during watermelon fruit development by integrating metabolic phenotypes and gene expression profiles. Hortic. Res. 2020, 7, 193–206. [Google Scholar] [CrossRef]
- Moretti, A.; Arias, C.L.; Mozzoni, L.A.; Chen, P.; McNeece, B.T.; Mian, M.A.R.; McHale, L.K.; Alonso, A.P. Workflow for the Quantification of Soluble and Insoluble Carbohydrates in Soybean Seed. Molecules 2020, 25, 3806. [Google Scholar] [CrossRef]
- Shangguan, L.; Song, C.; Leng, X.; Kayesh, E.; Sun, X.; Fang, J. Mining and comparison of the genes encoding the key enzymes involved in sugar biosynthesis in apple, grape, and sweet orange. Sci. Hortic. 2014, 165, 311–318. [Google Scholar] [CrossRef]
- Zhang, S.; Du, H.; Ma, Y.; Li, H.; Kan, G.; Yu, D. Linkage and association study discovered loci and candidate genes for glycinin and β-conglycinin in soybean (Glycine max L. Merr.). Theor. Appl. Genet. 2021, 134, 1201–1215. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Ma, Y.; Wu, S.; Liu, Z.; Wang, Z.; Yang, R.; Hu, G.; Zhou, Z.; Yu, H.; Zhang, M.; et al. Genome-wide association studies dissect the genetic networks underlying agronomical traits in soybean. Genome Biol. 2017, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Kumawat, G.; Xu, D.H. A major and stable quantitative trait locus qSS2 for seed size and shape traits in a soybean RIL popu-lation. Front. Genet. 2021, 12, 646102. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Wani, S.H.; Collins, P.J.; Wen, Z.; Li, W.; Zhang, N.; McCoy, A.G.; Bi, Y.; Tan, R.; Zhang, S.; et al. QTL mapping and GWAS for identification of loci conferring partial resistance to Pythium sylvaticum in soybean (Glycine max (L.) Merr.). Mol. Breed. 2020, 40, 54. [Google Scholar] [CrossRef]
- Wu, D.P.; Zhan, Y.H.; Sun, Q.X.; Xu, L.X.; Lian, M.; Zhao, X.; Han, Y.P.; Li, W.B. Identification of quantitative trait loci underlying soybean (Glycine max [L.] Merr.) seed weight including main, epistatic and QTL × environment effects in different regions of Northeast China. Plant Breed. 2018, 137, 194–202. [Google Scholar] [CrossRef]
- Zhang, W.; Liao, X.; Cui, Y.; Ma, W.; Zhang, X.; Du, H.; Ma, Y.; Ning, L.; Wang, H.; Huang, F.; et al. A cation diffusion facilitator, GmCDF1, negatively regulates salt tolerance in soybean. PLoS Genet. 2019, 15, e1007798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Zhou, Y.; Bu, Y.; Wang, X.; Zhang, Y.; Guo, N.; Zhao, J.; Xing, H. Genome-wide association analysis for yield-related traits at the R6 stage in a Chinese soybean mini core collection. Genes Genom. 2021, 43, 897–912. [Google Scholar] [CrossRef] [PubMed]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slatkin, M. Linkage disequilibrium—Understanding the evolutionary past and mapping the medical future. Nat. Rev. Genet. 2008, 9, 477–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, T.; Kim, K.; Kim, J.-M.; Shin, I.; Heo, J.; Jung, J.; Lee, J.; Moon, J.-K.; Kang, S. Genome-Wide Association Study for Ultraviolet-B Resistance in Soybean (Glycine max L.). Plants 2021, 10, 1335. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, L.; Zhang, D.; Cheng, H.; Wang, Q.; Yang, H.; Yu, D. GWAS identifies two novel loci for photosynthetic traits related to phosphorus efficiency in soybean. Mol. Breed. 2020, 40, 29. [Google Scholar] [CrossRef]
- Zhou, Z.; Jiang, Y.; Wang, Z.; Gou, Z.; Lyu, J.; Li, W.; Yu, Y.; Shu, L.; Zhao, Y.; Ma, Y.; et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 2015, 33, 408–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ficht, A.; Bruce, R.; Torkamaneh, D.; Grainger, C.M.; Eskandari, M.; Rajcan, I. Genetic analysis of sucrose concentration in soybean seeds using a historical soybean genomic panel. Theor. Appl. Genet. 2022, 135, 1375–1383. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, W.; Zhang, H.; Liu, X.; Cui, X.; Li, S.; Song, L.; Zhu, Y.; Chen, X.; Chen, H. Comparative selective signature analysis and high-resolution GWAS reveal a new candidate gene controlling seed weight in soybean. Theor. Appl. Genet. 2021, 134, 1329–1341. [Google Scholar] [CrossRef]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F.G. A Colorimetric Method for the Determination of Sugars. Nature 1951, 168, 167. [Google Scholar] [CrossRef]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Ersoz, E.; Lai, C.-Q.; Todhunter, R.J.; Tiwari, H.K.; Gore, M.A.; Bradbury, P.J.; Yu, J.; Arnett, D.K.; Ordovas, J.M.; et al. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010, 42, 355–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X.; et al. rMVP: A Memory-efficient, Visualization-enhanced, and Parallel-accelerated Tool for Genome-wide Association Study. Genom. Proteom. Bioinform. 2021, 19, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cao, K.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Zhao, P.; Guo, J.; Ding, T.; Guan, L.; et al. Genomic analyses of an extensive collection of wild and cultivated accessions provide new insights into peach breeding history. Genome Biol. 2019, 20, 36. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Wu, T.; Wang, L.; Jiang, B.; Zhen, C.; Yuan, S.; Hou, W.; Wu, C.; Han, T.; Sun, S. A Combined Linkage and GWAS Analysis Identifies QTLs Linked to Soybean Seed Protein and Oil Content. Int. J. Mol. Sci. 2019, 20, 5915. [Google Scholar] [CrossRef] [Green Version]
- Shin, J.; Blay, S.; McNeney, B.; Graham, J. LDheatmap: An R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J. Stat. Softw. 2006, 16, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Saldivar, X.; Wang, Y.-J.; Chen, P.; Mauromoustakos, A. Effects of blanching and storage conditions on soluble sugar contents in vegetable soybean. LWT 2010, 43, 1368–1372. [Google Scholar] [CrossRef]
- Hou, A.; Chen, P.; Alloatti, J.; Li, D.; Mozzoni, L.; Zhang, B.; Shi, A. Genetic Variability of Seed Sugar Content in Worldwide Soybean Germplasm Collections. Crop Sci. 2009, 49, 903–912. [Google Scholar] [CrossRef]
- Li, Y.-S.; Du, M.; Zhang, Q.-Y.; Wang, G.-H.; Hashemi, M.; Liu, X.-B. Greater differences exist in seed protein, oil, total soluble sugar and sucrose content of vegetable soybean genotypes [Glycine max (L.) Merrill] in Northeast China. Aust. J. Crop Sci. 2012, 6, 1681–1686. [Google Scholar] [CrossRef]
- Yu, X.; Yuan, F.; Fu, X.; Zhu, D. Profiling and relationship of water-soluble sugar and protein compositions in soybean seeds. Food Chem. 2016, 196, 776–782. [Google Scholar] [CrossRef] [PubMed]
- Ju, S.L.; Kim, S.M.; Kang, S. Fine mapping of quantitative trait loci for sucrose and oligosaccharide contents in soybean [Glycine max (L.) Merr.] using 180K Axiom® SoyaSNP genotyping platform. Euphytica 2016, 208, 195–203. [Google Scholar]
- Cregan, P.; Jarvik, T.; Bush, A.L.; Shoemaker, R.C.; Lark, K.G.; Kahler, A.L.; Kaya, N.; VanToai, T.T.; Lohnes, D.G.; Chung, J.; et al. An Integrated Genetic Linkage Map of the Soybean Genome. Crop Sci. 1999, 39, 1464–1490. [Google Scholar] [CrossRef] [Green Version]
- Hyten, D.L.; Cannon, S.B.; Song, Q.; Weeks, N.; Fickus, E.W.; Shoemaker, R.C.; Specht, J.E.; Farmer, A.D.; May, G.D.; Cregan, P.B. High-throughput SNP discovery through deep resequencing of a reduced representation library to anchor and orient scaffolds in the soybean whole genome sequence. BMC Genom. 2010, 11, 38. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ju, H.-W.; Huang, P.; Chung, J.-S.; Kim, C.S. Functional identification of AtSKIP as a regulator of the cell cycle signaling pathway in Arabidopsis thaliana. J. Plant Biol. 2012, 55, 481–488. [Google Scholar] [CrossRef]
- Zhang, X.; Min, J.-H.; Huang, P.; Chung, J.-S.; Lee, K.-H.; Kim, C.S. AtSKIP functions as a mediator between cytokinin and light signaling pathway in Arabidopsis thaliana. Plant Cell Rep. 2014, 33, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Book, A.J.; Smalle, J.; Lee, K.-H.; Yang, P.; Walker, J.M.; Casper, S.; Holmes, J.H.; Russo, L.A.; Buzzinotti, Z.W.; Jenik, P.D.; et al. The RPN5 Subunit of the 26s Proteasome Is Essential for Gametogenesis, Sporophyte Development, and Complex Assembly in Arabidopsis. Plant Cell 2009, 21, 460–478. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Dubey, R.C.; Maheshwari, D.K.; Bajpa, V.K. ACC deaminase producing Rhizobium leguminosarum RPN5 isolated from root nodules of Phaseolus vulgaris L. Bangladesh J. Bot. 2016, 45, 477–484. [Google Scholar]
- Parsons, H.T.; Christiansen, K.; Knierim, B.; Carroll, A.; Ito, J.; Batth, T.S.; Smith-Moritz, A.M.; Morrison, S.; McInerney, P.; Hadi, M.Z.; et al. Isolation and Proteomic Characterization of the Arabidopsis Golgi Defines Functional and Novel Components Involved in Plant Cell Wall Biosynthesis. Plant Physiol. 2012, 159, 12–26. [Google Scholar] [CrossRef] [Green Version]
- Da Costa, K.S.; Galúcio, J.M.P.; Leonardo, E.S.; Cardoso, G.; Leal, É.; Conde, G.; Lameira, J. Structural and evolutionary analysis of Leishmania Alba proteins. Mol. Biochem. Parasitol. 2017, 217, 23–31. [Google Scholar] [CrossRef]
- Mao, B.; Liu, X.; Hu, D.; Li, D. Co-expression of RCH10 and AGLU1 confers rice resistance to fungal sheath blight Rhizoctonia solani and blast Magnorpathe oryzae and reveals impact on seed germination. World J. Microbiol. Biotechnol. 2014, 30, 1229–1238. [Google Scholar] [CrossRef] [PubMed]
- Ashlin, T.G.; Blunsom, N.J.; Cockcroft, S. Courier service for phosphatidylinositol: PITPs deliver on demand. Biochim. Et Bio-Phys. Acta (BBA) Mol. Cell Biol. Lipids 2021, 1866, 158985. [Google Scholar] [CrossRef] [PubMed]
- Schrumpfová, P.P.; Vychodilová, I.; Hapala, J.; Schořová, Š.; Dvořáček, V.; Fajkus, J. Telomere binding protein TRB1 is associated with promoters of translation machinery genes in vivo. Plant Mol. Biol. 2016, 90, 189–206. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Xu, R.R.; Liu, X.; Zhang, J.C.; Wang, X.F.; You, C.X.; Hao, Y.J. Jasmonate induces biosynthesis of anthocyanin and proanthocy-anidin in apple by mediating the JAZ1–TRB1–MYB9 complex. Plant J. 2021, 106, 1414–1430. [Google Scholar] [CrossRef] [PubMed]
- Dunkley, T.P.J.; Hester, S.; Shadforth, I.P.; Runions, J.; Weimar, T.; Hanton, S.L.; Griffin, J.L.; Bessant, C.; Brandizzi, F.; Hawes, C.; et al. Mapping the Arabidopsis organelle proteome. Proc. Natl. Acad. Sci. USA 2006, 103, 6518–6523. [Google Scholar] [CrossRef] [Green Version]
- Regnault, T.; Davière, J.M.; Heintz, D.; Lange, T.; Achard, P. The gibberellin biosynthetic genes AtKAO1 and AtKAO2 have overlapping roles throughout Arabidopsis development. Plant J. 2014, 80, 462–474. [Google Scholar] [CrossRef]
| Year | Chr. | SNP ID | Position (bp) | p-Value | R2 |
|---|---|---|---|---|---|
| 2019 | 6 | S06_12048607 | 12048607 | 4.86 × 10−7 | 0.156 |
| 6 | S06_12049391 | 12049391 | 2.06 × 10−6 | 0.156 | |
| 6 | S06_12049420 | 12049420 | 1.10 × 10−6 | 0.154 | |
| 6 | S06_12060255 | 12060255 | 2.06 × 10−6 | 0.156 | |
| 6 | S06_12060477 | 12060477 | 2.02 × 10−6 | 0.158 | |
| 6 | S06_12060545 | 12060545 | 2.06 × 10−6 | 0.156 | |
| 15 | S15_10796464 | 10796464 | 1.90 × 10−6 | 0.112 | |
| 15 | S15_10801915 | 10801915 | 1.17 × 10−6 | 0.110 | |
| 15 | S15_10810881 | 10810881 | 8.21 × 10−7 | 0.112 | |
| 2020 | 5 | S05_2722069 | 2722069 | 1.46 × 10−6 | 0.168 |
| 6 | S06_12044204 | 12044204 | 1.97 × 10−6 | 0.158 | |
| 6 | S06_12044233 | 12044233 | 1.06 × 10−6 | 0.156 | |
| 6 | S06_12044239 | 12044239 | 4.71 × 10−7 | 0.158 | |
| 6 | S06_12044278 | 12044278 | 2.56 × 10−6 | 0.156 | |
| 6 | S06_12045115 | 12045115 | 2.34 × 10−6 | 0.121 | |
| 6 | S06_12045118 | 12045118 | 2.34 × 10−6 | 0.121 | |
| 6 | S06_12055116 | 12055116 | 1.97 × 10−6 | 0.158 | |
| 6 | S06_12055338 | 12055338 | 1.88 × 10−6 | 0.160 | |
| 6 | S06_12055406 | 12055406 | 1.97 × 10−6 | 0.158 | |
| 6 | S06_47846602 | 47846602 | 2.19 × 10−6 | 0.133 | |
| 13 | S13_4260712 | 4260712 | 1.69 × 10−6 | 0.136 | |
| 13 | S13_13999262 | 13999262 | 2.48 × 10−6 | 0.188 | |
| 14 | S14_47665362 | 47665362 | 7.82 × 10−7 | 0.087 | |
| 15 | S15_10819276 | 10819276 | 2.64 × 10−6 | 0.109 | |
| 15 | S15_10828362 | 10828362 | 1.90 × 10−6 | 0.111 | |
| 15 | S15_10843821 | 10843821 | 1.43 × 10−7 | 0.111 | |
| 15 | S15_11110657 | 11110657 | 3.01 × 10−6 | 0.162 |
| Chr. | Start (bp) | End (bp) | Candidate Gene ID | Homologous Gene ID | Function Annotation |
|---|---|---|---|---|---|
| 6 | 12044239 | 12048607 | Glyma.06g146500 | AT2G21950 | SKP1 interacting partner 6 (SKIP6) |
| Glyma.06g148300 | AT5G09900 | 26S proteasome regulatory subunit, putative(RPN5) (EMB2107,MSA,RPN5A) | |||
| Glyma.06g148600 | AT1G43130 | Like COV 2 | |||
| Glyma.06g148700 | AT5G09880 | Splicing factor, CC1-like | |||
| Glyma.06g148800 | AT1G76010 | Alba DNA/RNA-binding protein | |||
| 15 | 10810881 | 10843821 | Glyma.15g133700 | AT5G11720 | Glycosyl hydrolases family 31 protein |
| Glyma.15g134400 | AT1G30690 | Sec14p-like phosphatidylinositol transfer family protein | |||
| Glyma.15g135100 | AT1G49950 | Telomere repeat binding factor 1 (ATTRB1,TRB1) | |||
| Glyma.15g135200 | AT5G11720 | Glycosyl hydrolases family 31 protein |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, W.; Liu, H.; Li, S.; Zhang, W.; Wang, Q.; Zhang, H.; Liu, X.; Cui, X.; Chen, X.; Tang, W.; et al. GWAS and Identification of Candidate Genes Associated with Seed Soluble Sugar Content in Vegetable Soybean. Agronomy 2022, 12, 1470. https://doi.org/10.3390/agronomy12061470
Xu W, Liu H, Li S, Zhang W, Wang Q, Zhang H, Liu X, Cui X, Chen X, Tang W, et al. GWAS and Identification of Candidate Genes Associated with Seed Soluble Sugar Content in Vegetable Soybean. Agronomy. 2022; 12(6):1470. https://doi.org/10.3390/agronomy12061470
Chicago/Turabian StyleXu, Wenjing, Hui Liu, Songsong Li, Wei Zhang, Qiong Wang, Hongmei Zhang, Xiaoqing Liu, Xiaoyan Cui, Xin Chen, Wei Tang, and et al. 2022. "GWAS and Identification of Candidate Genes Associated with Seed Soluble Sugar Content in Vegetable Soybean" Agronomy 12, no. 6: 1470. https://doi.org/10.3390/agronomy12061470
APA StyleXu, W., Liu, H., Li, S., Zhang, W., Wang, Q., Zhang, H., Liu, X., Cui, X., Chen, X., Tang, W., Li, Y., Zhu, Y., & Chen, H. (2022). GWAS and Identification of Candidate Genes Associated with Seed Soluble Sugar Content in Vegetable Soybean. Agronomy, 12(6), 1470. https://doi.org/10.3390/agronomy12061470







