Understanding R Gene Evolution in Brassica
Abstract
:1. Introduction
2. Evolutionary Origin of R Genes
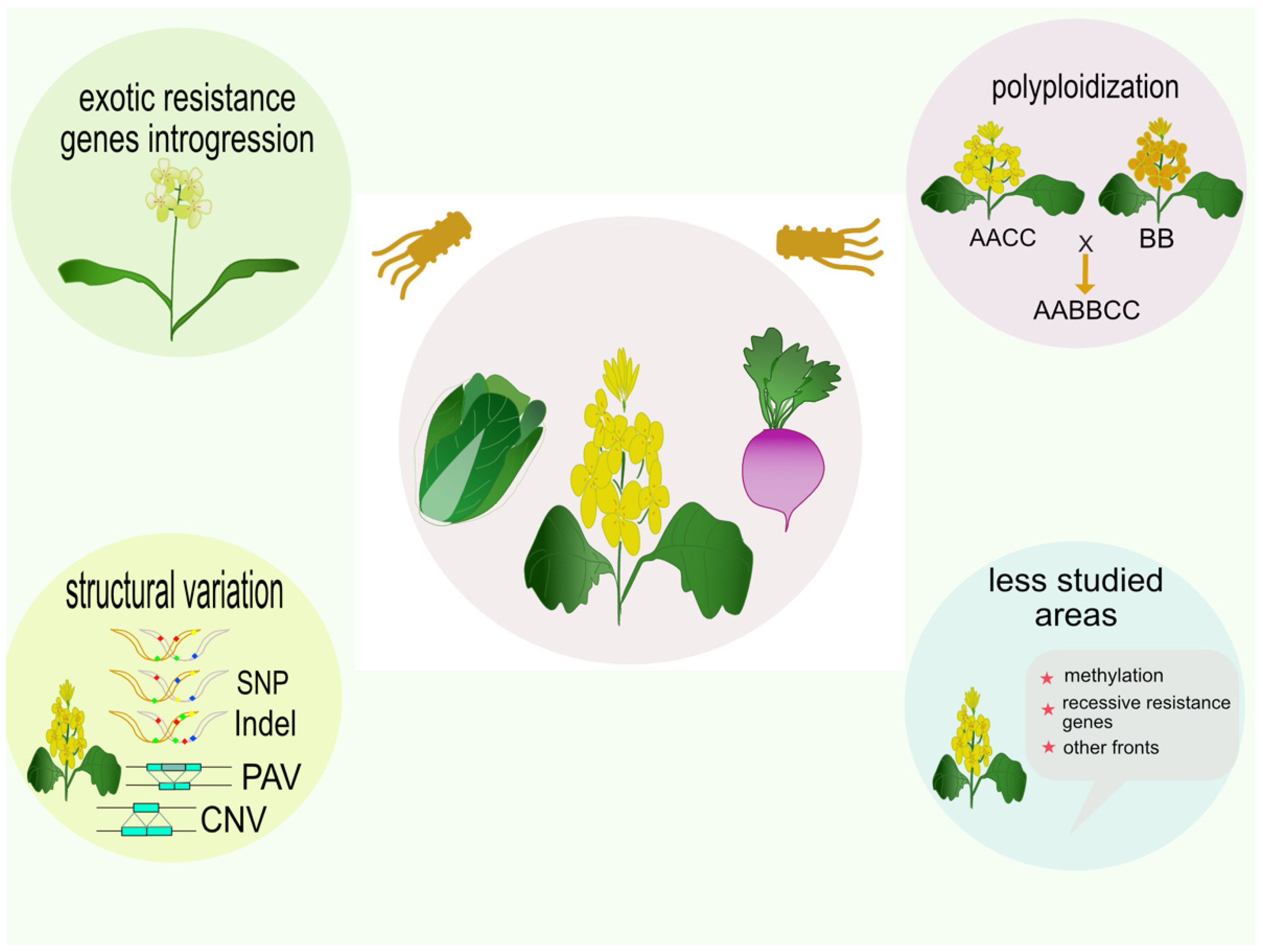
2.1. Polyploid Ancestry
2.2. Disease Resistance Genes from Introgression Lines
2.3. Studying Disease Resistance Genes from Close Relatives of Brassica
2.4. Structural Variation of Brassica Resistance Genes
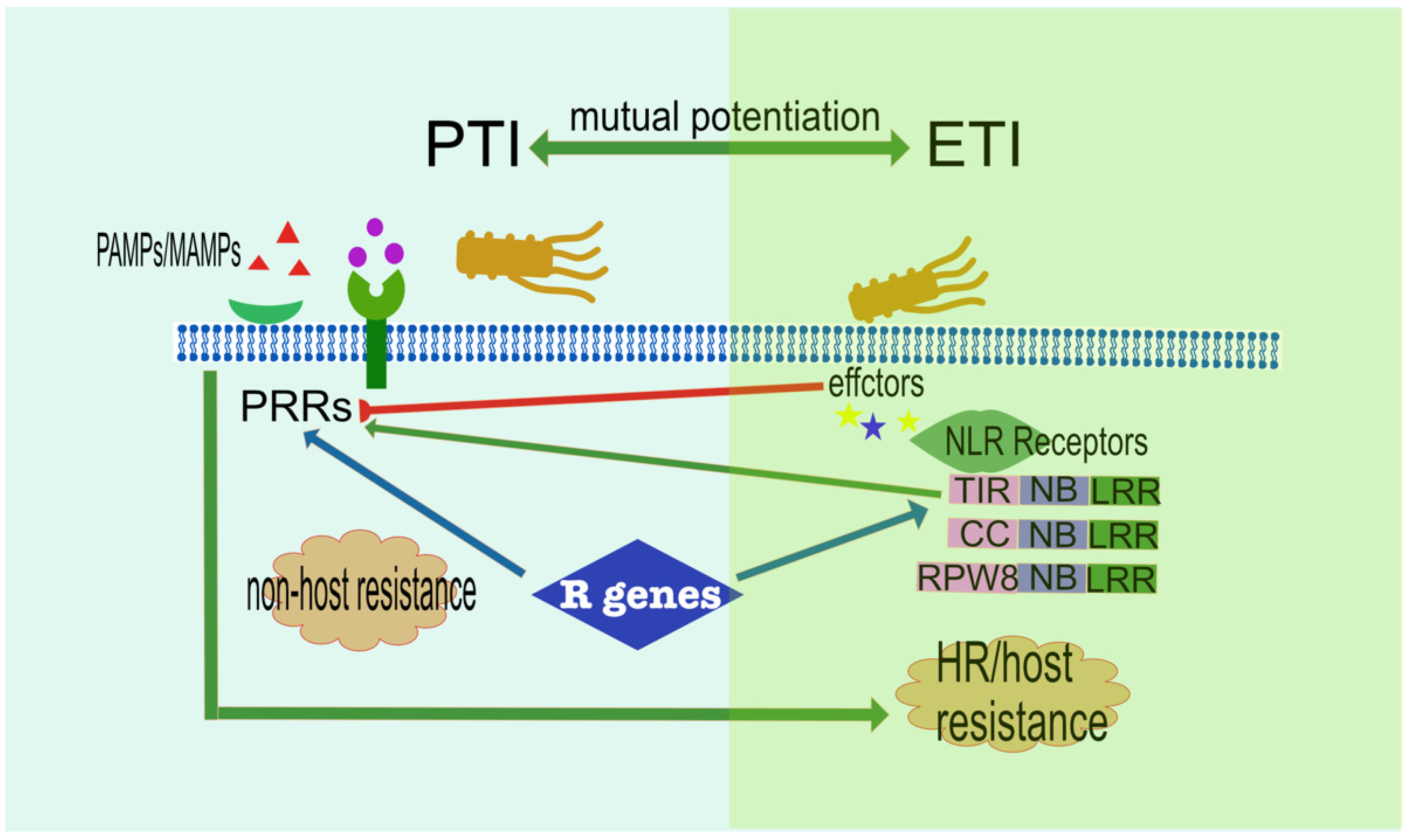
2.5. Complex Host-Pathogen Interaction
2.6. Complex Signalling Network Influencing Plant Immunity
2.7. Epigenetics and R Gene Evolution
2.8. Recessive Resistance Genes
3. R Gene Evolution in the Blackleg B. napus–L. maculans Pathosystem and the Impact on Disease Management
4. Technologies to Study Evolutionary Origins of Brassica
4.1. Genome Level
4.2. Pangenome Level
4.3. Epigenetic Level
4.4. High-Throughput Phenotyping
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Żyła, N.; Fidler, J.; Babula-Skowrońska, D. Economic and academic importance of Brassica oleracea. In The Brassica oleracea Genome; Liu, S., Snowdon, R., Kole, C., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 1–6. [Google Scholar]
- Nagaharu, U. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 1935, 7, 389–452. [Google Scholar]
- Hohmann, N.; Wolf, E.M.; Lysak, M.A.; Koch, M.A. A time-calibrated road map of Brassicaceae species radiation and evolutionary history. Plant Cell 2015, 27, 2770–2784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, Y.; Mabry, M.E.; Edger, P.P.; Freeling, M.; Zheng, C.; Jin, L.; VanBuren, R.; Colle, M.; An, H.; Abrahams, R.S.; et al. The contributions from the progenitor genomes of the mesopolyploid Brassiceae are evolutionarily distinct but functionally compatible. Genome Res. 2021, 31, 799–810. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef] [Green Version]
- Cheng, F.; Wu, J.; Wang, X. Genome triplication drove the diversification of Brassica plants. Hortic. Res. 2014, 1, 14024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, X.; An, H.; Hall, T.E.; Di, C.; Blischak, P.D.; McKibben, M.T.W.; Hao, Y.; Conant, G.C.; Pires, J.C.; Barker, M.S. Genes derived from ancient polyploidy have higher genetic diversity and are associated with domestication in Brassica rapa. New Phytol. 2021, 230, 372–386. [Google Scholar] [CrossRef]
- Araújo, A.C.d.; Fonseca, F.C.D.A.; Cotta, M.G.; Alves, G.S.C.; Miller, R.N.G. Plant NLR receptor proteins and their potential in the development of durable genetic resistance to biotic stresses. Biotechnol. Res. Innov. 2019, 3, 80–94. [Google Scholar] [CrossRef]
- Yu, X.; Feng, B.M.; He, P.; Shan, L.B. From chaos to harmony: Responses and signaling upon microbial pattern recognition. In Annual Review of Phytopathology; Leach, J.E., Lindow, S.E., Eds.; Annual Reviews Inc.: Palo Alto, CA, USA, 2017; Volume 55, pp. 109–137. [Google Scholar]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef]
- Cui, H.T.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. In Annual Review of Plant Biology; Merchant, S.S., Ed.; Annual Reviews Inc.: Palo Alto, CA, USA, 2015; Volume 66, pp. 487–511. [Google Scholar]
- Zhou, J.M.; Zhang, Y.L. Plant immunity: Danger perception and signaling. Cell 2020, 181, 978–989. [Google Scholar] [CrossRef]
- Monaghan, J.; Zipfel, C. Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 2012, 15, 349–357. [Google Scholar] [CrossRef]
- Shao, Z.Q.; Xue, J.Y.; Wang, Q.; Wang, B.; Chen, J.Q. Revisiting the origin of plant NBS-LRR genes. Trends Plant Sci. 2019, 24, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Sekhwal, M.K.; Li, P.C.; Lam, I.; Wang, X.E.; Cloutier, S.; You, F.M. Disease resistance gene analogs (RGAs) in plants. Int. J. Mol. Sci. 2015, 16, 19248–19290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, J.D.G.; Vance, R.E.; Dangl, J.L. Intracellular innate immune surveillance devices in plants and animals. Science 2016, 354, aaf6395. [Google Scholar] [CrossRef] [Green Version]
- Tirnaz, S.; Bayer, P.; Inturrisi, F.; Zhang, F.; Yang, H.; Dolatabadian, A.; Neik, T.X.; Severn-Ellis, A.; Patel, D.; Ibrahim, M.I.; et al. Resistance gene analogs in the Brassicaceae: Identification, characterization, distribution, and evolution. Plant Physiol. 2020, 184, 909–922. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.N.; Wu, Z.S.; Chen, S.Y.; Ao, K.V.; Huang, W.J.; Yaghmaiean, H.; Sun, T.J.; Xu, F.; Zhang, Y.N.; Wang, S.C.; et al. Activation of TIR signalling boosts pattern-triggered immunity. Nature 2021, 598, 500–503. [Google Scholar] [CrossRef]
- Ngou, B.P.M.; Ahn, H.K.; Ding, P.T.; Jones, J.D.G. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 2021, 592, 110–115. [Google Scholar] [CrossRef]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.-M.; He, S.Y.; Xin, X.-F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, T.; Li, X.; Duan, M.; Wang, J.; Qiu, Y.; Wang, H.; Song, J.; Shen, D. Interspecific hybridization, polyploidization, and backcross of Brassica oleracea var. alboglabra with B. rapa var. purpurea morphologically recapitulate the evolution of Brassica vegetables. Sci. Rep. 2016, 6, 18618. [Google Scholar] [CrossRef]
- McAlvay, A.C.; Ragsdale, A.P.; Mabry, M.E.; Qi, X.; Bird, K.A.; Velasco, P.; An, H.; Pires, J.C.; Emshwiller, E. Brassica rapa domestication: Untangling wild and feral forms and convergence of crop morphotypes. Mol. Biol. Evol. 2021, 38, 3358–3372. [Google Scholar] [CrossRef]
- Mabry, M.E.; Turner-Hissong, S.D.; Gallagher, E.Y.; McAlvay, A.C.; An, H.; Edger, P.P.; Moore, J.D.; Pink, D.A.C.; Teakle, G.R.; Stevens, C.J.; et al. The evolutionary history of wild, domesticated, and feral Brassica oleracea (Brassicaceae). Mol. Biol. Evol. 2021, 38, 4419–4434. [Google Scholar] [CrossRef]
- Kang, L.; Qian, L.; Zheng, M.; Chen, L.; Chen, H.; Yang, L.; You, L.; Yang, B.; Yan, M.; Gu, Y.; et al. Genomic insights into the origin, domestication and diversification of Brassica juncea. Nat. Genet. 2021, 53, 1392–1402. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.-Y.; Wang, Y.; Chen, M.; Dong, S.; Shao, Z.-Q.; Liu, Y. Maternal inheritance of U’s triangle and evolutionary process of brassica mitochondrial genomes. Front. Plant Sci. 2020, 11, 805. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Wei, L.; Li, X.; Wang, Y.; Wu, J.; Liu, M.; Zhang, C.; Chen, Z.; Xiao, Z.; Jian, H.; et al. Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat. Commun. 2019, 10, 1154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rashid, M.H.; Hausner, G.; Fernando, W.G.D. Molecular and phenotypic identification of B-genome introgression linked to Leptosphaeria maculans resistant gene Rlm6 in Brassica napus × B. juncea interspecific hybrids. Euphytica 2018, 214, 205. [Google Scholar] [CrossRef]
- Fredua-Agyeman, R.; Coriton, O.; Huteau, V.; Parkin, I.A.P.; Chèvre, A.-M.; Rahman, H. Molecular cytogenetic identification of B genome chromosomes linked to blackleg disease resistance in Brassica napus × B. carinata interspecific hybrids. Theor. Appl. Genet. 2014, 127, 1305–1318. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Lydiate, D.J.; Gugel, R.K.; Sharpe, A.G.; Rimmer, S.R. Introgression of Brassica rapa subsp. sylvestris blackleg resistance into B. napus. Mol. Breed. 2012, 30, 1495–1506. [Google Scholar] [CrossRef]
- Hasan, M.J.; Shaikh, R.; Basu, U.; Rahman, H. Mapping clubroot resistance of Brassica rapa introgressed into Brassica napus and development of molecular markers for the resistance. Crop Sci. 2021, 61, 4112–4127. [Google Scholar] [CrossRef]
- Yu, F.; Zhang, Y.; Wang, J.; Chen, Q.; Karim, M.M.; Gossen, B.D.; Peng, G. Identification of two major QTLs in Brassica napus lines with introgressed clubroot resistance from turnip cultivar ECD01. Front. Plant Sci. 2022, 12, 785989. [Google Scholar]
- Rana, K.; Atri, C.; Akhatar, J.; Kaur, R.; Goyal, A.; Singh Mohini, P.; Kumar, N.; Sharma, A.; Sandhu Prabhjodh, S.; Kaur, G.; et al. Detection of first marker trait associations for resistance against Sclerotinia sclerotiorum in Brassica juncea–Erucastrum cardaminoides introgression lines. Front. Plant Sci. 2019, 10, 1015. [Google Scholar] [CrossRef] [Green Version]
- Mei, J.; Shao, C.; Yang, R.; Feng, Y.; Gao, Y.; Ding, Y.; Li, J.; Qian, W. Introgression and pyramiding of genetic loci from wild Brassica oleracea into B. napus for improving Sclerotinia resistance of rapeseed. Theor. Appl. Genet. 2020, 133, 1313–1319. [Google Scholar] [CrossRef]
- Katche, E.; Quezada-Martinez, D.; Katche, E.I.; Vasquez-Teuber, P.; Mason, A.S. Interspecific hybridization for Brassica crop improvement. Crop Breed. Genet. Genom. 2019, 1, e190007. [Google Scholar] [CrossRef] [Green Version]
- Gaebelein, R.; Mason, A.S. Allohexaploids in the Genus Brassica. Crit. Rev. Plant Sci. 2018, 37, 422–437. [Google Scholar] [CrossRef]
- Gaebelein, R.; Alnajar, D.; Koopmann, B.; Mason, A.S. Hybrids between Brassica napus and B. nigra show frequent pairing between the B and A/C genomes and resistance to blackleg. Chromosome Res. 2019, 27, 221–236. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Ji, R.; Havlickova, L.; Wang, L.; Li, Y.; Lee, H.T.; Song, J.; Koh, C.; Yang, J.; Zhang, M.; et al. Genome structural evolution in Brassica crops. Nat. Plants 2021, 7, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Pathak, R.K.; Baunthiyal, M.; Pandey, D.; Kumar, A. Computational analysis of microarray data of Arabidopsis thaliana challenged with Alternaria brassicicola for identification of key genes in Brassica. J. Genet. Eng. Biotechnol. 2020, 18, 17. [Google Scholar] [CrossRef] [PubMed]
- Cevik, V.; Boutrot, F.; Apel, W.; Robert-Seilaniantz, A.; Furzer, O.J.; Redkar, A.; Castel, B.; Kover, P.X.; Prince, D.C.; Holub, E.B.; et al. Transgressive segregation reveals mechanisms of Arabidopsis; immunity to Brassica-infecting races of white rust (Albugo candida). Proc. Natl. Acad. Sci. USA 2019, 116, 2767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Chhapekar, S.S.; Lu, L.; Oh, S.; Singh, S.; Kim, C.S.; Kim, S.; Choi, G.J.; Lim, Y.P.; Choi, S.R. Genome-wide identification and characterization of NBS-encoding genes in Raphanus sativus L. and their roles related to Fusarium oxysporum resistance. BMC Plant Biol. 2021, 21, 47. [Google Scholar] [CrossRef]
- Chen, H.; Wang, T.; He, X.; Cai, X.; Lin, R.; Liang, J.; Wu, J.; King, G.; Wang, X. BRAD V3.0: An upgraded Brassicaceae database. Nucleic Acids Res. 2022, 50, D1432–D1441. [Google Scholar] [CrossRef]
- Zhang, Y.; Thomas, W.; Bayer, P.E.; Edwards, D.; Batley, J. Frontiers in dissecting and managing Brassica diseases: From reference-based RGA candidate identification to building Pan-RGAomes. Int. J. Mol. Sci. 2020, 21, 8964. [Google Scholar] [CrossRef]
- Van de Weyer, A.-L.; Monteiro, F.; Furzer, O.J.; Nishimura, M.T.; Cevik, V.; Witek, K.; Jones, J.D.G.; Dangl, J.L.; Weigel, D.; Bemm, F. A species-wide inventory of NLR genes and alleles in Arabidopsis thaliana. Cell 2019, 178, 1260–1272.e1214. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Tong, C.; Zhang, X.; Song, A.; Hu, M.; Dong, W.; Chen, F.; Wang, Y.; Tu, J.; Liu, S.; et al. A high-quality Brassica napus genome reveals expansion of transposable elements, subgenome evolution and disease resistance. Plant Biotechnol. J. 2021, 19, 615–630. [Google Scholar] [CrossRef]
- Dolatabadian, A.; Bayer, P.E.; Tirnaz, S.; Hurgobin, B.; Edwards, D.; Batley, J. Characterization of disease resistance genes in the Brassica napus pangenome reveals significant structural variation. Plant Biotechnol. J. 2019, 18, 969–982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurgobin, B.; Golicz, A.A.; Bayer, P.E.; Chan, C.-K.K.; Tirnaz, S.; Dolatabadian, A.; Schiessl, S.V.; Samans, B.; Montenegro, J.D.; Parkin, I.A.P.; et al. Homoeologous exchange is a major cause of gene presence/absence variation in the amphidiploid Brassica napus. Plant Biotechnol. J. 2018, 16, 1265–1274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayer, P.E.; Golicz, A.A.; Tirnaz, S.; Chan, C.-K.K.; Edwards, D.; Batley, J. Variation in abundance of predicted resistance genes in the Brassica oleracea pangenome. Plant Biotechnol. J. 2019, 17, 789–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayer, P.E.; Scheben, A.; Golicz, A.A.; Yuan, Y.; Faure, S.; Lee, H.; Chawla, H.S.; Anderson, R.; Bancroft, I.; Raman, H.; et al. Modelling of gene loss propensity in the pangenomes of three Brassica species suggests different mechanisms between polyploids and diploids. Plant Biotechnol. J. 2021, 19, 2488–2500. [Google Scholar] [CrossRef]
- Fu, Y.; Zhang, Y.; Mason, A.S.; Lin, B.; Zhang, D.; Yu, H.; Fu, D. NBS-encoding genes in Brassica napus evolved rapidly after allopolyploidization and Co-localize with known disease resistance loci. Front. Plant Sci. 2019, 10, 26. [Google Scholar] [CrossRef]
- Kopec, P.M.; Mikolajczyk, K.; Jajor, E.; Perek, A.; Nowakowska, J.; Obermeier, C.; Chawla, H.S.; Korbas, M.; Bartkowiak-Broda, I.; Karlowski, W.M. Local duplication of TIR-NBS-LRR gene marks clubroot resistance in Brassica napus cv. Tosca. Front. Plant Sci. 2021, 12, 528. [Google Scholar] [CrossRef]
- Hatakeyama, K.; Niwa, T.; Kato, T.; Ohara, T.; Kakizaki, T.; Matsumoto, S. The tandem repeated organization of NB-LRR genes in the clubroot-resistant CRb locus in Brassica rapa L. Mol. Genet. Genom. 2017, 292, 397–405. [Google Scholar] [CrossRef]
- Fomeju, B.F.; Falentin, C.; Lassalle, G.; Manzanares-Dauleux, M.J.; Delourme, R. Homoeologous duplicated regions are involved in quantitative resistance of Brassica napus to stem canker. BMC Genom. 2014, 15, 498. [Google Scholar] [CrossRef] [Green Version]
- Fomeju, B.F.; Falentin, C.; Lassalle, G.; Manzanares-Dauleux, M.J.; Delourme, R. Comparative genomic analysis of duplicated homoeologous regions involved in the resistance of Brassica napus to stem canker. Front. Plant Sci. 2015, 6, 772. [Google Scholar] [CrossRef] [Green Version]
- Larkan, N.J.; Ma, L.; Haddadi, P.; Buchwaldt, M.; Parkin, I.A.P.; Djavaheri, M.; Borhan, M.H. The Brassica napus Wall-Associated Kinase-Like (WAKL) gene Rlm9 provides race-specific Blackleg resistance. Plant J. 2020, 104, 892–900. [Google Scholar] [CrossRef] [PubMed]
- Haddadi, P.; Larkan, N.J.; Van de Wouw, A.; Zhang, Y.; Neik, T.X.; Beynon, E.; Bayer, P.; Edwards, D.; Batley, J.; Borhan, M.H. Brassica napus genes Rlm4 and Rlm7 conferring resistance to Leptosphaeria maculans are alleles of the Rlm9 wall-associated kinase-like resistance locus. Plant Biotechnol. J. 2022. [Google Scholar] [CrossRef] [PubMed]
- Vollrath, P.; Chawla, H.S.; Alnajar, D.; Gabur, I.; Lee, H.; Weber, S.; Ehrig, L.; Koopmann, B.; Snowdon, R.J.; Obermeier, C. Dissection of quantitative blackleg resistance reveals novel variants of resistance gene Rlm9 in elite Brassica napus. Front. Plant Sci. 2021, 12, 749491. [Google Scholar] [CrossRef]
- Raman, H.; Raman, R.; Qiu, Y.; Zhang, Y.; Batley, J.; Liu, S. The Rlm13 gene, a new player of Brassica napus–Leptosphaeria maculans interaction maps on chromosome C03 in canola. Front. Plant Sci. 2021, 12, 675. [Google Scholar] [CrossRef] [PubMed]
- Chawla, H.S.; Lee, H.; Gabur, I.; Vollrath, P.; Tamilselvan-Nattar-Amutha, S.; Obermeier, C.; Schiessl, S.V.; Song, J.-M.; Liu, K.; Guo, L.; et al. Long-read sequencing reveals widespread intragenic structural variants in a recent allopolyploid crop plant. Plant Biotechnol. J. 2021, 19, 240–250. [Google Scholar] [CrossRef] [PubMed]
- Gabur, I.; Chawla, H.S.; Lopisso, D.T.; von Tiedemann, A.; Snowdon, R.J.; Obermeier, C. Gene presence-absence variation associates with quantitative Verticillium longisporum disease resistance in Brassica napus. Sci. Rep. 2020, 10, 4131. [Google Scholar] [CrossRef] [Green Version]
- Neik, T.X.; Ghanbarnia, K.; Ollivier, B.; Scheben, A.; Severn-Ellis, A.; Larkan, N.J.; Haddadi, P.; Fernando, D.W.G.; Rouxel, T.; Batley, J.; et al. Two independent approaches converge to the cloning of a new Leptosphaeria maculans avirulence effector gene, AvrLmS-Lep2. Mol. Plant Pathol. 2022, 23, 733–748. [Google Scholar] [CrossRef]
- Velásquez, A.C.; Castroverde, C.D.M.; He, S.Y. Plant–pathogen warfare under changing climate conditions. Curr. Biol. 2018, 28, R619–R634. [Google Scholar] [CrossRef] [Green Version]
- Raman, H.; Raman, R.; Diffey, S.; Qiu, Y.; McVittie, B.; Barbulescu, D.M.; Salisbury, P.A.; Marcroft, S.; Delourme, R. Stable quantitative resistance loci to Blackleg disease in canola (Brassica napus L.) over continents. Front. Plant Sci. 2018, 9, 1622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cantila, A.Y.; Saad, N.S.M.; Amas, J.C.; Edwards, D.; Batley, J. Recent findings unravel genes and genetic factors underlying Leptosphaeria maculans resistance in Brassica napus and its relatives. Int. J. Mol. Sci. 2020, 22, 313. [Google Scholar] [CrossRef]
- Amas, J.; Anderson, R.; Edwards, D.; Cowling, W.; Batley, J. Status and advances in mining for blackleg (Leptosphaeria maculans) quantitative resistance (QR) in oilseed rape (Brassica napus). Theor. Appl. Genet. 2021, 134, 3123–3145. [Google Scholar] [CrossRef]
- Zhai, C.; Liu, X.; Song, T.; Yu, F.; Peng, G. Genome-wide transcriptome reveals mechanisms underlying Rlm1-mediated blackleg resistance on canola. Sci. Rep. 2021, 11, 4407. [Google Scholar] [CrossRef]
- Larkan, N.J.; Ma, L.; Borhan, M.H. The Brassica napus receptor-like protein RLM2 is encoded by a second allele of the LepR3/Rlm2 Blackleg resistance locus. Plant Biotechnol. J. 2015, 13, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Larkan, N.J.; Lydiate, D.J.; Parkin, I.A.P.; Nelson, M.N.; Epp, D.J.; Cowling, W.A.; Rimmer, S.R.; Borhan, M.H. The Brassica napus blackleg resistance gene LepR3 encodes a receptor-like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phytol. 2013, 197, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Larkan, N.J.; Raman, H.; Lydiate, D.J.; Robinson, S.J.; Yu, F.; Barbulescu, D.M.; Raman, R.; Luckett, D.J.; Burton, W.; Wratten, N.; et al. Multi-environment QTL studies suggest a role for cysteine-rich protein kinase genes in quantitative resistance to blackleg disease in Brassica napus. BMC Plant Biol. 2016, 16, 183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karandeni Dewage, C.S.; Qi, A.; Stotz, H.U.; Huang, Y.-J.; Fitt, B.D.L. Interactions in the Brassica napus–Pyrenopeziza brassicae pathosystem and sources of resistance to P. brassicae (light leaf spot). Plant Pathol. 2021, 70, 2104–2114. [Google Scholar] [CrossRef]
- Bousset, L.; Sprague, S.J.; Thrall, P.H.; Barrett, L.G. Spatio-temporal connectivity and host resistance influence evolutionary and epidemiological dynamics of the canola pathogen Leptosphaeria maculans. Evol. Appl. 2018, 11, 1354–1370. [Google Scholar] [CrossRef]
- Hwang, S.F.; Strelkov, S.E.; Ahmed, H.U.; Manolii, V.P.; Zhou, Q.; Fu, H.; Turnbull, G.; Fredua-Agyeman, R.; Feindel, D. Virulence and inoculum density-dependent interactions between clubroot resistant canola (Brassica napus) and Plasmodiophora brassicae. Plant Pathol. 2017, 66, 1318–1328. [Google Scholar] [CrossRef]
- Crété, R.; Pires, R.N.; Barbetti, M.J.; Renton, M. Rotating and stacking genes can improve crop resistance durability while potentially selecting highly virulent pathogen strains. Sci. Rep. 2020, 10, 19752. [Google Scholar] [CrossRef] [PubMed]
- Naseem, M.; Kaltdorf, M.; Dandekar, T. The nexus between growth and defence signalling: Auxin and cytokinin modulate plant immune response pathways. J. Exp. Bot. 2015, 66, 4885–4896. [Google Scholar] [CrossRef] [Green Version]
- Zou, Z.; Liu, F.; Huang, S.; Fernando, W.G.D. Genome-wide identification and analysis of the valine-glutamine motif-containing gene family in Brassica napus and functional characterization of BnMKS1 in response to Leptosphaeria maculans. Phytopathology 2020, 111, 281–292. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, N.B.; Franks, S.J.; Kane, N.C.; Tittes, S.; Rest, J.S. Evolution of pathogen response genes associated with increased disease susceptibility during adaptation to an extreme drought in a Brassica rapa plant population. BMC Ecol. Evol. 2021, 21, 61. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Sun, X.; Wang, C.; Li, F.; Zhang, S.; Zhang, H.; Li, G.; Yuan, L.; Chen, G.; Sun, R.; et al. Gene co-expression network analysis reveals key pathways and hub genes in Chinese cabbage (Brassica rapa L.) during vernalization. BMC Genom. 2021, 22, 236. [Google Scholar] [CrossRef]
- Roy, J.; Shaikh, T.M.; del Río Mendoza, L.; Hosain, S.; Chapara, V.; Rahman, M. Genome-wide association mapping and genomic prediction for adult stage sclerotinia stem rot resistance in Brassica napus (L) under field environments. Sci. Rep. 2021, 11, 21773. [Google Scholar] [CrossRef] [PubMed]
- Jablonka, E.; Lamb, M.J. The changing concept of epigenetics. In From Epigenesis to Epigenetics: The Genome in Context; VanSpeybroeck, L., VandeVijver, G., DeWaele, D., Eds.; Annals of the New York Academy of Sciences: Medford, OR, USA, 2002; Volume 981, pp. 82–96. [Google Scholar]
- Tirnaz, S.; Batley, J. DNA methylation: Toward crop disease resistance improvement. Trends Plant Sci. 2019, 24, 1137–1150. [Google Scholar] [CrossRef]
- Zheng, H.X.; Sun, X.; Li, J.L.; Song, Y.S.; Song, J.; Wang, F.; Liu, L.N.; Zhang, X.S.; Sui, N. Analysis of N6-methyladenosine reveals a new important mechanism regulating the salt tolerance of sweet sorghum. Plant Sci. 2021, 304, 110801. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.; Lepere, G.; Jay, F.; Wang, J.Y.; Bapaume, L.; Wang, Y.; Abraham, A.L.; Penterman, J.; Fischer, R.L.; Voinnet, O.; et al. Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc. Natl. Acad. Sci. USA 2013, 110, 2389–2394. [Google Scholar] [CrossRef] [Green Version]
- Rambani, A.; Rice, J.H.; Liu, J.Y.; Lane, T.; Ranjan, P.; Mazarei, M.; Pantalone, V.; Stewart, C.N.; Staton, M.; Hewezi, T. The methylome of soybean roots during the compatible interaction with the soybean cyst nematode. Plant Physiol. 2015, 168, 1364–1377. [Google Scholar] [CrossRef]
- Wang, C.G.; Wang, C.N.; Xu, W.J.; Zou, J.Z.; Qiu, Y.H.; Kong, J.; Yang, Y.S.; Zhang, B.Y.; Zhu, S.F. Epigenetic changes in the regulation of Nicotiana tabacum response to Cucumber Mosaic Virus infection and symptom recovery through single-base resolution methylomes. Viruses 2018, 10, 402. [Google Scholar] [CrossRef] [Green Version]
- Geng, S.F.; Kong, X.C.; Song, G.Y.; Jia, M.L.; Guan, J.T.; Wang, F.; Qin, Z.R.; Wu, L.; Lan, X.J.; Li, A.L.; et al. DNA methylation dynamics during the interaction of wheat progenitor Aegilops tauschii with the obligate biotrophic fungus Blumeria graminis f. sp. tritici. New Phytol. 2019, 221, 1023–1035. [Google Scholar] [CrossRef] [Green Version]
- Hu, M.; Pei, B.L.; Zhang, L.F.; Li, Y.Z. Histone H2B monoubiquitination is involved in regulating the dynamics of microtubules during the defense response to Verticillium dahliae toxins in Arabidopsis (1 OPEN). Plant Physiol. 2014, 164, 1857–1865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, B.H.; Yang, D.L.; Shi, Z.Y.; Dong, H.S.; Hua, J. Monoubiquitination of histone 2B at the disease resistance gene locus regulates its expression and impacts immune responses in Arabidopsis. Plant Physiol. 2014, 165, 309–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.S.; Tong, M.X.Z.; Tian, L.; Zhu, C.P.; Liu, X.R.; Zhang, Y.L.; Li, X. Plant E3 ligases SNIPER1 and SNIPER2 broadly regulate the homeostasis of sensor NLR immune receptors. EMBO J. 2020, 39, e104915. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.R.; Shi, J.; Yu, L.; Zhao, X.Z.; Ran, L.L.; Hu, D.Y.; Song, B.A. N-6-methyl-adenosine level in Nicotiana tabacum is associated with tobacco mosaic virus. Virol. J. 2018, 15, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.Y.; Wang, Z.Q.; Hu, H.C.; Chen, Z.Q.; Liu, P.; Gao, S.Q.; Zhang, F.; He, L.; Jin, P.; Xu, M.Z.; et al. Transcriptome-wide N-6-Methyladenosine (m(6)A) profiling of susceptible and resistant wheat varieties reveals the involvement of variety-specific m(6)A modification involved in virus-host interaction pathways. Front. Microbiol. 2021, 12, 656302. [Google Scholar] [CrossRef]
- Ma, L.J.; Zhao, B.X.; Chen, K.; Thomas, A.; Tuteja, J.H.; He, X.; He, C.; White, K.P. Evolution of transcript modification by N-6-methyladenosine in primates. Genome Res. 2017, 27, 385–392. [Google Scholar] [CrossRef] [Green Version]
- Miao, Z.Y.; Zhang, T.; Xie, B.; Qi, Y.H.; Ma, C. Evolutionary implications of the RNA N-6-methyladenosine methylome in plants. Mol. Biol. Evol. 2022, 39, msab299. [Google Scholar] [CrossRef]
- Hashimoto, M.; Neriya, Y.; Yamaji, Y.; Namba, S. Recessive resistance to plant viruses: Potential resistance genes beyond translation initiation factors. Front. Microbiol. 2016, 7, 1695. [Google Scholar] [CrossRef] [Green Version]
- Fraser, R.S.S. The genetics of resistance to plant viruses. Annu. Rev. Phytopathol. 1990, 28, 179–200. [Google Scholar] [CrossRef]
- Agaoua, A.; Rittener, V.; Troadec, C.; Desbiez, C.; Bendahmane, A.; Moquet, F.; Dogimont, C. A single substitution in Vacuolar protein sorting 4 is responsible for resistance to watermelon mosaic virus in melon. J. Exp. Bot. 2022, 73, 4008–4021. [Google Scholar] [CrossRef]
- Diaz-Pendon, J.A.; Truniger, V.; Nieto, C.; Garcia-Mas, J.; Bendahmane, A.; Aranda, M.A. Advances in understanding recessive resistance to plant viruses. Mol. Plant Pathol. 2004, 5, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Piron, F.; Nicolai, M.; Minoia, S.; Piednoir, E.; Moretti, A.; Salgues, A.; Zamir, D.; Caranta, C.; Bendahmane, A. An induced mutation in tomato eIF4E leads to immunity to two potyviruses. PLoS ONE 2010, 5, e11313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Ruiz, H.; Szurek, B.; Van den Ackerveken, G. Stop helping pathogens: Engineering plant susceptibility genes for durable resistance. Curr. Opin. Biotechnol. 2021, 70, 187–195. [Google Scholar] [CrossRef]
- Lellis, A.D.; Kasschau, K.D.; Whitham, S.A.; Carrington, J.C. Loss-of-susceptibility mutants of arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr. Biol. 2002, 12, 1046–1051. [Google Scholar] [CrossRef] [Green Version]
- Yoshii, M.; Yoshioka, N.; Ishikawa, M.; Naito, S. Isolation of an Arabidopsis thaliana mutant in which the multiplication of both cucumber mosaic virus and turnip crinkle virus is affected. J. Virol. 1998, 72, 8731–8737. [Google Scholar] [CrossRef]
- Yoshii, M.; Nishikiori, M.; Tomita, K.; Yoshioka, N.; Kozuka, R.; Naito, S.; Ishikawa, M. The Arabidopsis cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J. Virol. 2004, 78, 6102–6111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruffel, S.; Gallois, J.L.; Lesage, M.L.; Caranta, C. The recessive potyvirus resistance gene pot-1 is the tomato orthologue of the pepper pvr2-eIF4E gene. Mol. Genet. Genom. 2005, 274, 346–353. [Google Scholar] [CrossRef]
- Nicaise, V.; German-Retana, S.; Sanjuan, R.; Dubrana, M.P.; Mazier, M.; Maisonneuve, B.; Candresse, T.; Caranta, C.; LeGall, O. The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the Potyvirus Lettuce mosaic virus. Plant Physiol. 2003, 132, 1272–1282. [Google Scholar] [CrossRef] [Green Version]
- Nieto, C.; Morales, M.; Orjeda, G.; Clepet, C.; Monfort, A.; Sturbois, B.; Puigdomenech, P.; Pitrat, M.; Caboche, M.; Dogimont, C.; et al. An eIF4E allele confers resistance to an uncapped and non-polyadenylated RNA virus in melon. Plant J. 2006, 48, 452–462. [Google Scholar] [CrossRef]
- Kanyuka, K.; Druka, A.; Caldwell, D.G.; Tymon, A.; McCallum, N.; Waugh, R.; Adams, M.J. Evidence that the recessive bymovirus resistance locus rym4 in barley corresponds to the eukaryotic translation initiation factor 4E gene. Mol. Plant Pathol. 2005, 6, 449–458. [Google Scholar] [CrossRef]
- Stein, N.; Perovic, D.; Kumlehn, J.; Pellio, B.; Stracke, S.; Streng, S.; Ordon, F.; Graner, A. The eukaryotic translation initiation factor 4E confers multiallelic recessive Bymovirus resistance in Hordeum vulgare (L.). Plant J. 2005, 42, 912–922. [Google Scholar] [CrossRef] [PubMed]
- Albar, L.; Bangratz-Reyser, M.; Hebrard, E.; Ndjiondjop, M.N.; Jones, M.; Ghesquiere, A. Mutations in the eIF(iso)4G translation initiation factor confer high resistance of rice to Rice yellow mottle virus. Plant J. 2006, 47, 417–426. [Google Scholar] [CrossRef]
- Rusholme, R.L.; Higgins, E.E.; Walsh, J.A.; Lydiate, D.J. Genetic control of broad-spectrum resistance to turnip mosaic virus in Brassica rapa (Chinese cabbage). J. Gen. Virol. 2007, 88, 3177–3186. [Google Scholar] [CrossRef]
- Kim, J.; Kang, W.-H.; Yang, H.-B.; Park, S.; Jang, C.-s.; Yu, H.-J.; Kang, B.-C. Identification of a broad-spectrum recessive gene in Brassica rapa and molecular analysis of the eIF4E gene family to develop molecular markers. Mol. Breed. 2013, 32, 385–398. [Google Scholar] [CrossRef]
- Qian, W.; Zhang, S.; Zhang, S.; Li, F.; Zhang, H.; Wu, J.; Wang, X.; Walsh, J.A.; Sun, R. Mapping and candidate-gene screening of the novel Turnip mosaic virus resistance gene retr02 in Chinese cabbage (Brassica rapa L.). Theor. Appl. Genet. 2013, 126, 179–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nellist, C.F.; Qian, W.; Jenner, C.E.; Moore, J.D.; Zhang, S.; Wang, X.; Briggs, W.H.; Barker, G.C.; Sun, R.; Walsh, J.A. Multiple copies of eukaryotic translation initiation factors in Brassica rapa facilitate redundancy, enabling diversification through variation in splicing and broad-spectrum virus resistance. Plant J. 2014, 77, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Shopan, J.; Mou, H.; Zhang, L.; Zhang, C.; Ma, W.; Walsh, J.A.; Hu, Z.; Yang, J.; Zhang, M. Eukaryotic translation initiation factor 2B-beta (eIF2Bbeta), a new class of plant virus resistance gene. Plant J. 2017, 90, 929–940. [Google Scholar] [CrossRef] [Green Version]
- Shopan, J.; Liu, C.; Hu, Z.; Zhang, M.; Yang, J. Identification of eukaryotic translation initiation factors and the temperature-dependent nature of Turnip mosaic virus epidemics in allopolyploid Brassica juncea. 3 Biotech 2020, 10, 75. [Google Scholar] [CrossRef]
- Büschges, R.; Hollricher, K.; Panstruga, R.; Simons, G.; Wolter, M.; Frijters, A.; van Daelen, R.; van der Lee, T.; Diergaarde, P.; Groenendijk, J.; et al. The barley Mlo Gene: A novel control element of plant pathogen resistance. Cell 1997, 88, 695–705. [Google Scholar] [CrossRef] [Green Version]
- Wretblad, S.; Bohman, S.; Dixelius, C. Overexpression of a Brassica nigra cDNA gives enhanced resistance to Leptosphaeria maculans in B. napus. Mol. Plant Microbe Interact. 2003, 16, 477–484. [Google Scholar] [CrossRef] [Green Version]
- Yan, P.; Zhou, S.; Li, X.; Zhao, S.; Zhou, H.; Zhou, Y.; Xu, S.; Ke, T. Genome-wide comparative analysis of MLO related genes in Brassica lineage. Chin. J. Oil Crop Sci. 2017, 39, 729–736. [Google Scholar] [CrossRef]
- Van de Wouw, A.P.; Sheedy, E.M.; Ware, A.H.; Marcroft, S.J.; Idnurm, A. Independent breakdown events of the Brassica napus Rlm7 resistance gene including via the off-target impact of a dual-specificity avirulence interaction. Mol. Plant Pathol. 2022, 23, 997–1010. [Google Scholar] [CrossRef] [PubMed]
- Zhan, J.; Thrall, P.H.; Papaïx, J.; Xie, L.; Burdon, J.J. Playing on a pathogen’s weakness: Using evolution to guide sustainable plant disease control strategies. Annu. Rev. Phytopathol. 2015, 53, 19–43. [Google Scholar] [CrossRef] [PubMed]
- Delmotte, F.; Bourguet, D.; Franck, P.; Guillemaud, T.; Reboud, X.; Vacher, C.; Walker, A.-S. Combining selective pressures to enhance the durability of disease resistance genes. Front. Plant Sci. 2016, 7, 1916. [Google Scholar]
- Van de Wouw, A.P.; Howlett, B.J. Advances in understanding the Leptosphaeria maculans—Brassica pathosystem and their impact on disease management. Can. J. Plant Pathol. 2020, 42, 149–163. [Google Scholar] [CrossRef]
- Zhang, X.; Peng, G.; Kutcher, H.R.; Balesdent, M.-H.; Delourme, R.; Fernando, W.G.D. Breakdown of Rlm3 resistance in the Brassica napus–Leptosphaeria maculans pathosystem in western Canada. Eur. J. Plant Pathol. 2016, 145, 659–674. [Google Scholar] [CrossRef]
- Van de Wouw, A.P.; Marcroft, S.J.; Ware, A.; Lindbeck, K.; Khangura, R.; Howlett, B.J. Breakdown of resistance to the fungal disease, blackleg, is averted in commercial canola (Brassica napus) crops in Australia. Field Crops Res. 2014, 166, 144–151. [Google Scholar] [CrossRef]
- Sprague, S.J.; Balesdent, M.-H.; Brun, H.; Hayden, H.L.; Marcroft, S.J.; Pinochet, X.; Rouxel, T.; Howlett, B.J. Major gene resistance in Brassica napus (oilseed rape) is overcome by changes in virulence of populations of Leptosphaeria maculans in France and Australia. Eur. J. Plant Pathol. 2006, 114, 33–40. [Google Scholar] [CrossRef]
- Mohd Saad, N.S.; Neik, T.X.; Thomas, W.J.W.; Amas, J.C.; Cantila, A.Y.; Craig, R.J.; Edwards, D.; Batley, J. Advancing designer crops for climate resilience through an integrated genomics approach. Curr. Opin. Plant Biol. 2022, 67, 102220. [Google Scholar] [CrossRef]
- Ton, L.B.; Neik, T.X.; Batley, J. The use of genetic and gene technologies in shaping modern rapeseed cultivars (Brassica napus L.). Genes 2020, 11, 1161. [Google Scholar] [CrossRef]
- Arora, S.; Steuernagel, B.; Gaurav, K.; Chandramohan, S.; Long, Y.; Matny, O.; Johnson, R.; Enk, J.; Periyannan, S.; Singh, N.; et al. Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat. Biotechnol. 2019, 37, 139–143. [Google Scholar] [CrossRef]
- Anjanappa, R.B.; Gruissem, W. Current progress and challenges in crop genetic transformation. J. Plant Physiol. 2021, 261, 153411. [Google Scholar] [CrossRef] [PubMed]
- Haddadi, P.; Larkan, N.J.; Borhan, M.H. Dissecting R gene and host genetic background effect on the Brassica napus defense response to Leptosphaeria maculans. Sci. Rep. 2019, 9, 6947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.-J.; Mitrousia, G.K.; Sidique, S.N.M.; Qi, A.; Fitt, B.D.L. Combining R gene and quantitative resistance increases effectiveness of cultivar resistance against Leptosphaeria maculans in Brassica napus in different environments. PLoS ONE 2018, 13, e0197752. [Google Scholar] [CrossRef] [PubMed]
- Pilet-Nayel, M.-L.; Moury, B.; Caffier, V.; Montarry, J.; Kerlan, M.-C.; Fournet, S.; Durel, C.-E.; Delourme, R. Quantitative resistance to plant pathogens in pyramiding strategies for durable crop protection. Front. Plant Sci. 2017, 8, 1838. [Google Scholar] [CrossRef] [Green Version]
- Dolatabadian, A.; Cornelsen, J.; Huang, S.; Zou, Z.; Fernando, W.G.D. Sustainability on the farm: Breeding for resistance and management of major canola diseases in Canada contributing towards an IPM approach. Can. J. Plant Pathol. 2022, 44, 157–190. [Google Scholar] [CrossRef]
- Mohd Saad, N.S.; Severn-Ellis, A.A.; Pradhan, A.; Edwards, D.; Batley, J. Genomics armed with diversity leads the way in Brassica improvement in a changing global environment. Front. Genet. 2021, 12, 600789. [Google Scholar] [CrossRef]
- Yuan, Y.; Bayer, P.E.; Batley, J.; Edwards, D. Current status of structural variation studies in plants. Plant Biotechnol. J. 2021, 19, 2153–2163. [Google Scholar] [CrossRef]
- Guo, N.; Wang, S.; Gao, L.; Liu, Y.; Wang, X.; Lai, E.; Duan, M.; Wang, G.; Li, J.; Yang, M.; et al. Genome sequencing sheds light on the contribution of structural variants to Brassica oleracea diversification. BMC Biol. 2021, 19, 93. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, J.; Fang, L.; Zhang, Z.; Ma, W.; Niu, Y.; Ju, L.; Deng, J.; Zhao, T.; Lian, J.; et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019, 51, 739–748. [Google Scholar] [CrossRef] [Green Version]
- Feng, Q.; Yu, J.; Yang, X.; Lv, X.; Lu, Y.; Yuan, J.; Du, X.; Zhu, B.; Li, Z. Development and characterization of an allooctaploid (AABBCCRR) incorporating Brassica and radish genomes via two rounds of interspecific hybridizations. Sci. Hortic. 2022, 293, 110730. [Google Scholar] [CrossRef]
- Chen, S.; Nelson, M.N.; Chèvre, A.-M.; Jenczewski, E.; Li, Z.; Mason, A.S.; Meng, J.; Plummer, J.A.; Pradhan, A.; Siddique, K.H.M.; et al. Trigenomic bridges for Brassica improvement. Crit. Rev. Plant Sci. 2011, 30, 524–547. [Google Scholar] [CrossRef]
- Mason, A.S.; Batley, J. Creating new interspecific hybrid and polyploid crops. Trends Biotechnol. 2015, 33, 436–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Li, R.; Chen, L.; Niu, S.; Chen, L.; Gao, J.; Wen, J.; Yi, B.; Ma, C.; Tu, J.; et al. Fine-mapping and candidate gene analysis of the Brassica juncea white-flowered mutant Bjpc2 using the whole-genome resequencing. Mol. Genet. Genom. 2018, 293, 359–370. [Google Scholar] [CrossRef]
- Itoh, N.; Segawa, T.; Tamiru, M.; Abe, A.; Sakamoto, S.; Uemura, A.; Oikawa, K.; Kutsuzawa, H.; Koga, H.; Imamura, T.; et al. Next-generation sequencing-based bulked segregant analysis for QTL mapping in the heterozygous species Brassica rapa. Theor. Appl. Genet. 2019, 132, 2913–2925. [Google Scholar] [CrossRef]
- Li, P.; Su, T.; Zhang, B.; Li, P.; Xin, X.; Yue, X.; Cao, Y.; Wang, W.; Zhao, X.; Yu, Y.; et al. Identification and fine mapping of qSB.A09, a major QTL that controls shoot branching in Brassica rapa ssp. chinensis Makino. Theor. Appl. Genet. 2020, 133, 1055–1068. [Google Scholar] [CrossRef]
- Dakouri, A.; Zhang, X.; Peng, G.; Falk, K.C.; Gossen, B.D.; Strelkov, S.E.; Yu, F. Analysis of genome-wide variants through bulked segregant RNA sequencing reveals a major gene for resistance to Plasmodiophora brassicae in Brassica oleracea. Sci. Rep. 2018, 8, 17657. [Google Scholar] [CrossRef]
- Lin, X.; Armstrong, M.; Baker, K.; Wouters, D.; Visser, R.G.F.; Wolters, P.J.; Hein, I.; Vleeshouwers, V.G.A.A. RLP/K enrichment sequencing; A novel method to identify receptor-like protein (RLP) and receptor-like kinase (RLK) genes. New Phytol. 2020, 227, 1264–1276. [Google Scholar] [CrossRef] [Green Version]
- Jupe, F.; Witek, K.; Verweij, W.; Śliwka, J.; Pritchard, L.; Etherington, G.J.; Maclean, D.; Cock, P.J.; Leggett, R.M.; Bryan, G.J.; et al. Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB-LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations. Plant J. 2013, 76, 530–544. [Google Scholar] [CrossRef] [Green Version]
- Bayer, P.E.; Petereit, J.; Danilevicz, M.F.; Anderson, R.; Batley, J.; Edwards, D. The application of pangenomics and machine learning in genomic selection in plants. Plant Genome 2021, 14, e20112. [Google Scholar] [CrossRef]
- Song, J.-M.; Liu, D.-X.; Xie, W.-Z.; Yang, Z.; Guo, L.; Liu, K.; Yang, Q.-Y.; Chen, L.-L. BnPIR: Brassica napus pan-genome information resource for 1689 accessions. Plant Biotechnol. J. 2021, 19, 412–414. [Google Scholar] [CrossRef] [PubMed]
- Miura, F.; Ito, T. Highly sensitive targeted methylome sequencing by post-bisulfite adaptor tagging. DNA Res. 2015, 22, 13–18. [Google Scholar] [CrossRef] [Green Version]
- Cokus, S.J.; Feng, S.; Zhang, X.; Chen, Z.; Merriman, B.; Haudenschild, C.D.; Pradhan, S.; Nelson, S.F.; Pellegrini, M.; Jacobsen, S.E. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 2008, 452, 215–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, Z.; Tang, B.; Xing, J.; Liu, C.; Cai, X.; Hendy, A.; Kamran, M.; Liu, H.; Zheng, L.; Huang, J.; et al. MTA1-mediated RNA m6 A modification regulates autophagy and is required for infection of the rice blast fungus. New Phytol. 2022, 235, 247–262. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Bai, X.; Zhang, C.; He, Y. Advanced high-throughput plant phenotyping techniques for genome-wide association studies: A review. J. Adv. Res. 2022, 35, 215–230. [Google Scholar] [CrossRef]
- Sun, D.; Robbins, K.; Morales, N.; Shu, Q.; Cen, H. Advances in optical phenotyping of cereal crops. Trends Plant Sci. 2022, 27, 191–208. [Google Scholar] [CrossRef]
- Bergsträsser, S.; Fanourakis, D.; Schmittgen, S.; Cendrero-Mateo, M.P.; Jansen, M.; Scharr, H.; Rascher, U. HyperART: Non-invasive quantification of leaf traits using hyperspectral absorption-reflectance-transmittance imaging. Plant Methods 2015, 11, 1. [Google Scholar] [CrossRef] [Green Version]
- Kuska, M.; Wahabzada, M.; Leucker, M.; Dehne, H.-W.; Kersting, K.; Oerke, E.-C.; Steiner, U.; Mahlein, A.-K. Hyperspectral phenotyping on the microscopic scale: Towards automated characterization of plant-pathogen interactions. Plant Methods 2015, 11, 28. [Google Scholar] [CrossRef] [Green Version]
- Yates, S.; Mikaberidze, A.; Krattinger, S.G.; Abrouk, M.; Hund, A.; Yu, K.; Studer, B.; Fouche, S.; Meile, L.; Pereira, D.; et al. Precision phenotyping reveals novel loci for quantitative resistance to septoria tritici blotch. Plant Phenomics 2019, 2019, 3285904. [Google Scholar] [CrossRef] [Green Version]
- Fordyce, R.F.; Soltis, N.E.; Caseys, C.; Gwinner, R.; Corwin, J.A.; Atwell, S.; Copeland, D.; Feusier, J.; Subedy, A.; Eshbaugh, R.; et al. Digital imaging combined with genome-wide association mapping links loci to plant-pathogen interaction traits. Plant Physiol. 2018, 178, 1406–1422. [Google Scholar] [CrossRef] [Green Version]
| Mechanism of R Gene Evolution | Main Findings | Impact on Disease Management and Crop Production |
|---|---|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, F.; Neik, T.X.; Wu, T.; Edwards, D.; Batley, J. Understanding R Gene Evolution in Brassica. Agronomy 2022, 12, 1591. https://doi.org/10.3390/agronomy12071591
Zhang F, Neik TX, Wu T, Edwards D, Batley J. Understanding R Gene Evolution in Brassica. Agronomy. 2022; 12(7):1591. https://doi.org/10.3390/agronomy12071591
Chicago/Turabian StyleZhang, Fangning, Ting Xiang Neik, Tingting Wu, David Edwards, and Jacqueline Batley. 2022. "Understanding R Gene Evolution in Brassica" Agronomy 12, no. 7: 1591. https://doi.org/10.3390/agronomy12071591
APA StyleZhang, F., Neik, T. X., Wu, T., Edwards, D., & Batley, J. (2022). Understanding R Gene Evolution in Brassica. Agronomy, 12(7), 1591. https://doi.org/10.3390/agronomy12071591








