A Dated Phylogeny of the Pantropical Genus Dalbergia L.f. (Leguminosae: Papilionoideae) and Its Implications for Historical Biogeography
Abstract
:1. Introduction
2. Materials and Methods
2.1. Taxon Sampling and Sequence Alignment
2.2. Phylogenetic Reconstruction of Dalbergia
2.3. Divergence Time Estimation
2.4. Ancestral Area Estimations
3. Results
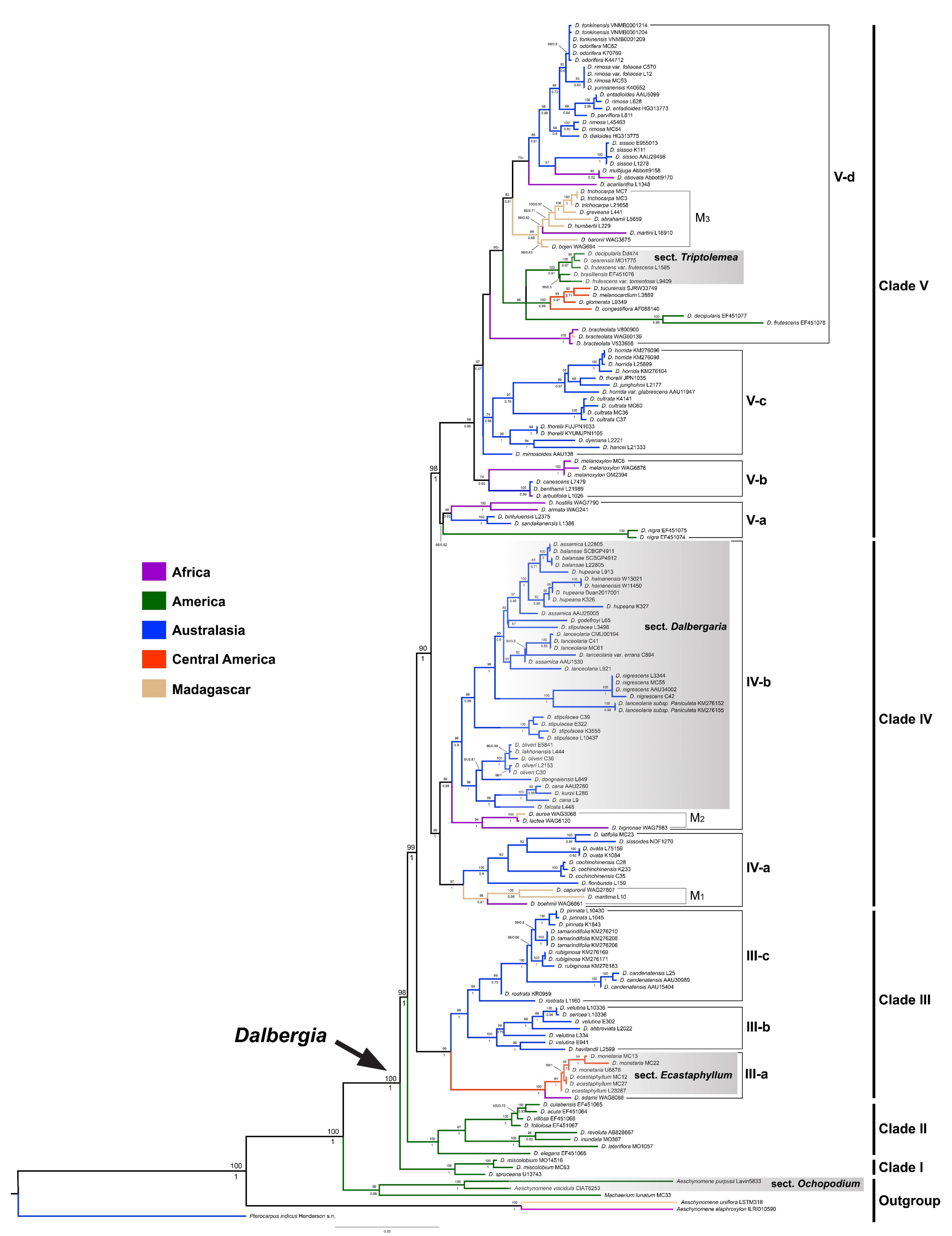
3.1. Phylogenetic Analyses
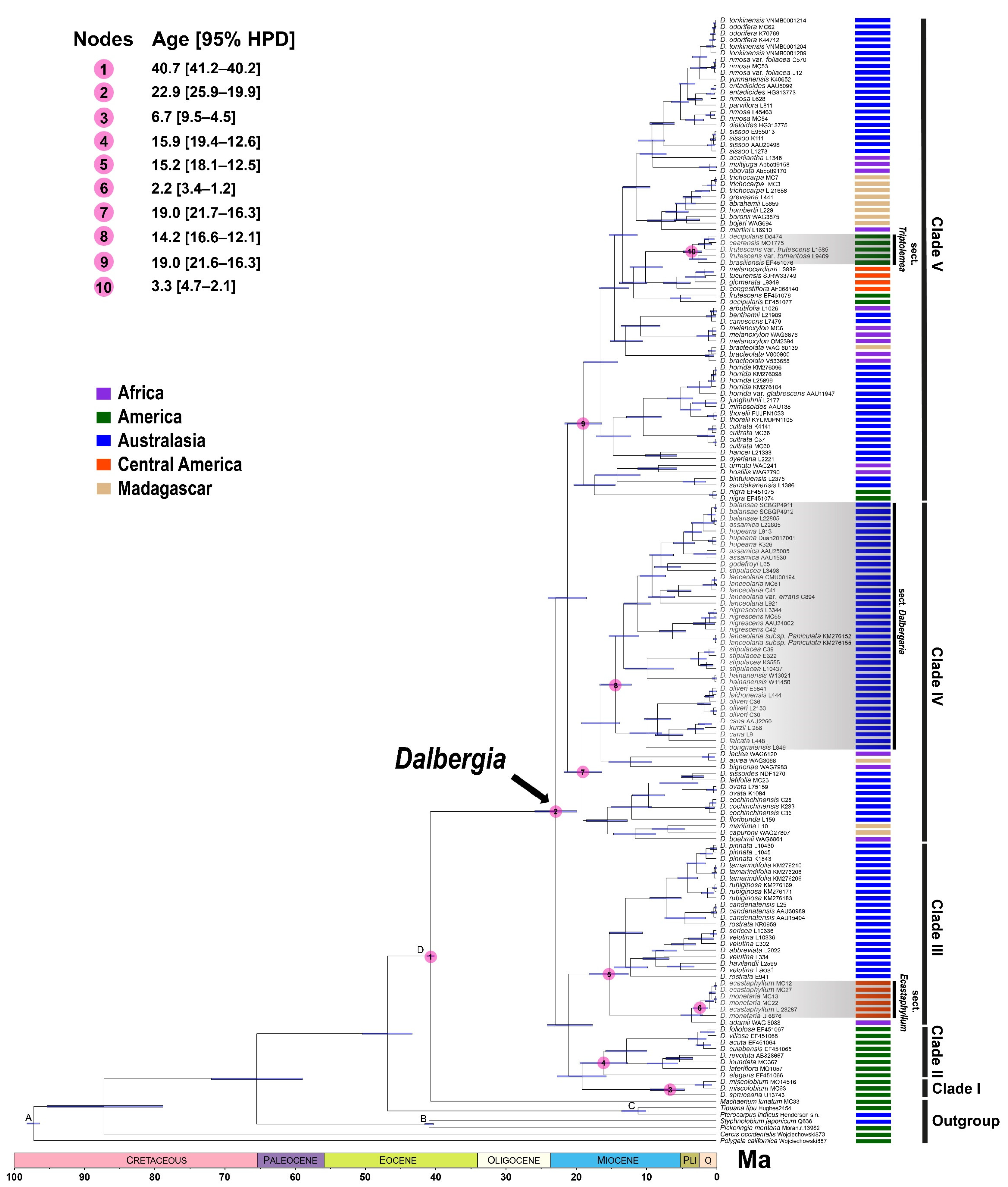
3.2. Divergence Times and Biogeographical Analyses
4. Discussion
4.1. Phylogenetic Relationships and Taxonomic Implications
4.2. Origin and Biogeographical Diversification of Dalbergia
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klitgaard, B.; Lavin, M. Tribe Dalbergieae sens. lat. In Legumes of the World; Royal Botanic Gardens Kew: Richmond, UK, 2005; pp. 307–335. [Google Scholar]
- de Carvalho, A. A Synopsis of the Genus Dalbergia (Fabaceae: Dalbergieae) in Brazil. Brittonia 1997, 49, 87–109. [Google Scholar] [CrossRef]
- Du Puy, D.J.; Labat, J.; Rabevohitra, R.; Villiers, J.; Bosser, J.; Moat, J. The Leguminosae of Madagascar; Royal Botanic Gardens Kew: Richmond, UK, 2002; p. 737. [Google Scholar]
- Mabberley, D. Mabberley’s Plant-Book: A Portable Dictionary of Plants, Their Classifications, and Uses; Cambridge University Press: Cambridge, UK, 2008; p. 1021. [Google Scholar]
- Lewis, G.P.; Schrire, B.; Mackinder, B.; Lock, M. Legumes of the World; Royal Botanic Gardens Kew: Richmond, UK, 2005; p. 577. [Google Scholar]
- Saha, S.; Shilpi, J.A.; Mondal, H.; Hossain, F.; Anisuzzman, M.; Hasan, M.M.; Cordell, G.A. Ethnomedicinal, Phytochemical, and Pharmacological Profile of the Genus Dalbergia L.f. (Fabaceae). Phytopharmacology 2013, 2, 291–346. [Google Scholar]
- Bentham, G. Synopsis of Dalbergieae, a Tribe of Leguminosae. Bot. J. Linn. Soc. 1860, 4, 1–128. [Google Scholar] [CrossRef]
- Prain, D. The Species of Dalbergia of Southeastern Asia. Ann. Roy. Bot. Gard. 1904, 10, 1–114. [Google Scholar]
- Thothathri, K. Taxonomic Revision of the Tribe Dalbergieae. In The Indian Subcontinent; Botanical Survey of India: Culcutta, India, 1987; pp. 231–239. [Google Scholar]
- Lavin, M.; Pennington, R.T.; Klitgaard, B.B.; Sprent, J.I.; de Lima, H.C.; Gasson, P.E. The Dalbergioid Legumes (Fabaceae): Delimitation of a Pantropical Monophyletic Clade. Am. J. Bot. 2001, 88, 503–533. [Google Scholar] [CrossRef]
- Cardoso, D.B.; Ramos, G.; Barbosa São-Mateus, W.M.; Paganucci de Queiroz, L. Aeschynomene chicocesariana, a Striking New Unifoliolate Legume Species from the Brazilian Chapada Diamantina and its Phylogenetic Placement in the Dalbergioid Clade. Syst. Bot. 2019, 44, 810–817. [Google Scholar] [CrossRef]
- Ribeiro, R.A.; Lavin, M.; Lemos-Filho, J.P.; Santos, F.R.d.; Lovato, M.B. The Genus Machaerium (Leguminosae) is More Closely Related to Aeschynomene sect. Ochopodium than to Dalbergia: Inferences from Combined Sequence Data. Syst. Bot. 2007, 32, 762–771. [Google Scholar]
- Vatanparast, M.; Klitgård, B.B.; Adema, F.A.; Pennington, R.T.; Yahara, T.; Kajita, T. First Molecular Phylogeny of the Pantropical Genus Dalbergia: Implications for Infrageneric Circumscription and Biogeography. S. Afr. J. Bot. 2013, 89, 143–149. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Farris, J.S.; Kallersjo, M.; Kluge, A.G.; Bult, C. Testing Significance of Incongruence. Cladistics 1994, 10, 315–319. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0b10; Sinauer: Sunderland, MA, USA, 2002. [Google Scholar]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More Models, New Heuristics and Parallel Computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A.; Drummond, A.J. Tracer Version 1.6. 2013. Available online: http://tree.bio.ed.ac.uk/software (accessed on 11 December 2013).
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A. FigTree Version 1.4.2. 2014. Available online: http://tree.bio.ed.ac.uk/software/Figtree (accessed on 9 July 2014).
- Kress, W.J.; Prince, L.M.; Williams, K.J. The Phylogeny and a New Classification of the Gingers (Zingiberaceae): Evidence from Molecular Data. Am. J. Bot. 2002, 89, 1682–1696. [Google Scholar] [CrossRef]
- Magallón, S.; Gómez-Acevedo, S.; Sánchez-Reyes, L.L.; Hernández-Hernández, T. A Metacalibrated Time-Tree Documents the Early Rise of Flowering Plant Phylogenetic Diversity. New Phytol. 2015, 207, 437–453. [Google Scholar] [CrossRef]
- Matzke, N.J. BioGeoBEARS: BioGeography with Bayesian (and Likelihood) Evolutionary Analysis in R Scripts. R Package, Version 0.2.1. 2013. Available online: http://phylo.wikidot.com/biogeobears (accessed on 2 January 2013).
- Yu, Y.; Blair, C.; He, X. RASP 4: Ancestral State Reconstruction Tool for Multiple Genes and Characters. Mol. Biol. Evol. 2020, 37, 604–606. [Google Scholar] [CrossRef]
- Ree, R.H.; Smith, S.A. Maximum Likelihood Inference of Geographic Range Evolution by Dispersal, Local Extinction, and Cladogenesis. Syst. Biol. 2008, 57, 4–14. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F. Dispersal-Vicariance Analysis: A New Approach to the Quantification of Historical Biogeography. Syst. Biol. 1997, 46, 195–203. [Google Scholar] [CrossRef]
- Landis, M.J.; Matzke, N.J.; Moore, B.R.; Huelsenbeck, J.P. Bayesian Analysis of Biogeography When the Number of Areas is Large. Syst. Biol. 2013, 62, 789–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ree, R.H.; Sanmartín, I. Conceptual and Statistical Problems with the DEC+J Model of Founder-Event Speciation and its Comparison with DEC via Model Selection. J. Biogeogr. 2018, 45, 741–749. [Google Scholar] [CrossRef]
- Murthy, K.; Rani, S.S.; Pullaiah, T. Genus Dalbergia L.f. (Leguminosae: Faboideae) in Eastern Ghats. J. Econ. Taxon. Bot. 2000, 24, 133–139. [Google Scholar]
- Niyomdham, C. An Account of Dalbergia (Leguminosae-Papilionoideae) in Thailand. Thai For. Bull. 2002, 30, 124–166. [Google Scholar]
- Sunarno, B.; Ohashi, H. Dalbergia (Leguminosae) of Sulawesi, Indonesia. J. Jpn. Bot. 1996, 71, 241–248. [Google Scholar]
- Sunarno, B.; Ohashi, H. Dalbergia (Leguminosae) of Borneo. J. Jpn. Bot. 1997, 72, 198–220. [Google Scholar]
- Carvalho, d.A.e.M. Systematic Studies of the Genus Dalbergia L.f. in Brazil. Ph.D. Thesis, University of Reading, Reading, UK, 1989. [Google Scholar]
- Lavin, M.; Herendeen, P.S.; Wojciechowski, M.F. Evolutionary Rates Analysis of Leguminosae Implicates a Rapid Diversification of Lineages During the Tertiary. Syst. Biol. 2005, 54, 575–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hung, T.H.; So, T.; Sreng, S.; Thammavong, B.; Boounithiphonh, C.; Boshier, D.H.; MacKay, J.J. Reference Transcriptomes and Comparative Analyses of Six Species in the Threatened Rosewood Genus Dalbergia. Sci. Rep. 2020, 10, 17749. [Google Scholar] [CrossRef]
- Saporta, d.G. Dalbergia phleboptera Saporta. Muséum National d’Histoire Naturelle (MNHN), Paris, France. 2015. Available online: http://coldb.mnhn.fr/catalognumber/mnhn/f/14084 (accessed on 14 December 2015).
- Kučerová, J. Miocénna Flóra z Lokalít Kalonda a Mučín. Acta Geol. Slovaca 2009, 1, 65–70. [Google Scholar]
- Guo, S.; Zhou, Z. The Megafossil Legumes from China. In Advances in Legume Systematics Part 4; Royal Botanic Gardens Kew: Richmond, UK, 1994; pp. 207–223. [Google Scholar]
- Lomolino, M.V. Four Darwinian Themes on the Origin, Evolution and Preservation of Island Life. J. Biogeogr. 2010, 37, 985–994. [Google Scholar] [CrossRef]
- Jin, J.J.; Yang, M.Q.; Fritsch, P.W.; van Velzen, R.; Li, D.Z.; Yi, T.S. Born Migrators: Historical Biogeography of the Cosmopolitan Family Cannabaceae. J. Syst. Evol. 2020, 58, 461–473. [Google Scholar] [CrossRef]
- Li, J.; Jiang, J.; Stel, H.V.; Homkes, A.; Corajod, J.; Brown, K.; Chen, Z. Phylogenetics and Biogeography of Apios (Fabaceae) Inferred from Sequences of Nuclear and Plastid Genes. Int. J. Plant Sci. 2014, 175, 764–780. [Google Scholar] [CrossRef]
- Vatanparast, M.; Takayama, K.; Sousa, M.S.; Tateishi, Y.; Kajita, T. Origin of Hawaiian Endemic Species of Canavalia (Fabaceae) from Sea-Dispersed Species Revealed by Chloroplast and Nuclear DNA Sequences. J. Jpn. Bot. 2011, 86, 15–25. [Google Scholar]
- Fortuna-Perez, A.P.; da Silva, M.J.; de Queiroz, L.P.; Lewis, G.P.; Simões, A.O.; de Azevedo Tozzi, A.M.G.; Sarkinen, T.; de Souza, A.P. Phylogeny and Biogeography of the Genus Zornia (Leguminosae: Papilionoideae: Dalbergieae). Taxon 2013, 62, 723–732. [Google Scholar] [CrossRef]
- Schaefer, H.; Hechenleitner, P.; Santos-Guerra, A.; de Sequeira, M.M.; Pennington, R.T.; Kenicer, G.; Carine, M.A. Systematics, Biogeography, and Character Evolution of the Legume Tribe Fabeae with Special Focus on the Middle-Atlantic Island Lineages. BMC Evol. Biol. 2012, 12, 250. [Google Scholar] [CrossRef] [Green Version]
- Lavin, M.; Schrire, B.P.; Lewis, G.; Pennington, R.T.; Delgado–Salinas, A.; Thulin, M.; Hughes, C.E.; Matos, A.B.; Wojciechowski, M.F. Metacommunity Process Rather than Continental Tectonic History Better Explains Geographically Structured Phylogenies in Legumes. Philos. Trans. R. Soc. London Ser. B Biol. Sci. 2004, 359, 1509–1522. [Google Scholar] [CrossRef] [Green Version]
- Pennington, R.T.; Richardson, J.E.; Lavin, M. Insights into the Historical Construction of Species-Rich Biomes from Dated Plant Phylogenies, Neutral Ecological Theory and Phylogenetic Community Structure. New Phytol. 2006, 172, 605–616. [Google Scholar] [CrossRef]
- Croat, T.B. Flora of Barro Colorado Island, Vol. VIII; Stanford University Press: Stanford, CA, USA, 1978; p. 943. [Google Scholar]
- Bacon, C.D.; Silvestro, D.; Jaramillo, C.; Smith, B.T.; Chakrabarty, P.; Antonelli, A. Biological Evidence Supports an Early and Complex Emergence of the Isthmus of Panama. Proc. Natl. Acad. Sci. USA 2015, 112, 6110–6115. [Google Scholar] [CrossRef] [Green Version]
- Hoorn, C.; Flantua, S. An Early Start for the Panama Land Bridge. Science 2015, 348, 186–187. [Google Scholar] [CrossRef]
- Montes, C.; Cardona, A.; Jaramillo, C.; Pardo, A.; Silva, J.; Valencia, V.; Ayala, C.; Pérez-Angel, L.; Rodriguez-Parra, L.; Ramirez, V. Middle Miocene Closure of the Central American Seaway. Science 2015, 348, 226–229. [Google Scholar] [CrossRef] [Green Version]
- Cody, S.; Richardson, J.E.; Rull, V.; Ellis, C.; Pennington, R.T. The Great American Biotic Interchange Revisited. Ecography 2010, 33, 326–332. [Google Scholar] [CrossRef]
- Dupin, J.; Matzke, N.J.; Särkinen, T.; Knapp, S.; Olmstead, R.G.; Bohs, L.; Smith, S.D. Bayesian Estimation of the Global Biogeographical History of the Solanaceae. J. Biogeogr. 2017, 44, 887–899. [Google Scholar] [CrossRef]
- Grose, S.O.; Olmstead, R. Evolution of a Charismatic Neotropical Clade: Molecular Phylogeny of Tabebuia s. l., Crescentieae, and Allied Genera (Bignoniaceae). Syst. Bot. 2007, 32, 650–659. [Google Scholar] [CrossRef]
- Olmstead, R.G. Phylogeny and Biogeography in Solanaceae, Verbenaceae and Bignoniaceae: A Comparison of Continental and Intercontinental Diversification Patterns. Bot. J. Linn. Soc. 2013, 171, 80–102. [Google Scholar] [CrossRef] [Green Version]
- Davis, C.C.; Bell, C.D.; Fritsch, P.W.; Mathews, S. Phylogeny of Acridocarpus-Brachylophon (Malpighiaceae): Implications for Tertiary Tropical Floras and Afroasian Biogeography. Evolution 2002, 56, 2395–2405. [Google Scholar] [CrossRef]
- Hoorn, C.; Wesselingh, F.; Ter Steege, H.; Bermudez, M.; Mora, A.; Sevink, J.; Sanmartín, I.; Sanchez-Meseguer, A.; Anderson, C.; Figueiredo, J. Amazonia Through Time: Andean Uplift, Climate Change, Landscape Evolution, and Biodiversity. Science 2010, 330, 927–931. [Google Scholar] [CrossRef] [Green Version]
- Davis, C.C.; Bell, C.D.; Mathews, S.; Donoghue, M.J. Laurasian Migration Explains Gondwanan Disjunctions: Evidence from Malpighiaceae. Proc. Natl. Acad. Sci. USA 2002, 99, 6833–6837. [Google Scholar] [CrossRef] [Green Version]
- Conti, E.; Eriksson, T.; Schönenberger, J.; Sytsma, K.J.; Baum, D.A. Early Tertiary Out-of-India Dispersal of Crypteroniaceae: Evidence from Phylogeny and Molecular Dating. Evolution 2002, 56, 1931–1942. [Google Scholar] [CrossRef]
- Samonds, K.E.; Godfrey, L.R.; Ali, J.R.; Goodman, S.M.; Vences, M.; Sutherland, M.R.; Irwin, M.T.; Krause, D.W. Spatial and Temporal Arrival Patterns of Madagascar’s Vertebrate Fauna Explained by Distance, Ocean Currents, and Ancestor Type. Proc. Natl. Acad. Sci. USA 2012, 109, 5352–5357. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.-M.; Wohlhauser, S.; Möller, M.; Klackenberg, J.; Callmander, M.W.; Küpfer, P. Phylogeny and Biogeography of Exacum (Gentianaceae): A Disjunctive Distribution in the Indian Ocean Basin Resulted from Long Distance Dispersal and Extensive Radiation. Syst. Biol. 2005, 54, 21–34. [Google Scholar] [CrossRef] [Green Version]
- Kosuch, J.; Vences, M.; Dubois, A.; Ohler, A.; Böhme, W. Out of Asia: Mitochondrial DNA Evidence for an Oriental Origin of Tiger Frogs, Genus Hoplobatrachus. Mol. Phylogenet. Evol. 2001, 21, 398–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.-Q.; Maki, M.; Drew, B.T.; Paton, A.J.; Li, H.-W.; Zhao, J.-L.; Conran, J.G.; Li, J. Phylogeny and historical biogeography of Isodon (Lamiaceae): Rapid Radiation in South-West China and Miocene Overland Dispersal into Africa. Mol. Phylogenet. Evol. 2014, 77, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Su, Y.C.; Thomas, D.C.; Saunders, R.M. “Out-of-Africa” Dispersal of Tropical Floras During the Miocene Climatic Optimum: Evidence from Uvaria (Annonaceae). J. Biogeogr. 2012, 39, 322–335. [Google Scholar] [CrossRef]
- Popov, S.; Rögl, F.; Rozanov, A.; Steininger, F.F.; Shcherba, I.; Kovac, M. Lithological-Paleogeographic Maps of Paratethys (Maps 1–10). Cour. Forschungsinst. Senckenberg 2004, 250, 46. [Google Scholar]
- Rögl, F. Palaeogeographic Considerations for Mediterranean and Paratethys Seaways (Oligocene to Miocene). Ann. Naturhist. Mus. Wien A 1997, 99, 279–310. [Google Scholar]
- Zachos, J.; Pagani, M.; Sloan, L.; Thomas, E.; Billups, K. Trends, Rhythms, and Aberrations in Global Climate 65 Ma to Present. Science 2001, 292, 686–693. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Deng, T.; Moore, M.J.; Wang, H.; Li, Z.; Lin, N.; Yusupov, Z.; Tojibaev, K.S.; Wang, Y.; Sun, H. Tropical Asian Origin, Boreotropical Migration and Long-Distance Dispersal in Nettles (Urticeae, Urticaceae). Mol. Phylogenet. Evol. 2019, 137, 190–199. [Google Scholar] [CrossRef]
- Van der Pijl, L. Principles of Dispersal in Higher Plants; Springer: New York, NY, USA, 1982; p. 153. [Google Scholar]
- McLoughlin, S. The Breakup History of Gondwana and its Impact on Pre-Cenozoic Floristic Provincialism. Aust. J. Bot. 2001, 49, 271–300. [Google Scholar] [CrossRef]
- Yoder, A.D.; Nowak, M.D. Has Vicariance or Dispersal Been the Predominant Biogeographic Force in Madagascar? Only Time Will Tell. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 405–431. [Google Scholar] [CrossRef] [Green Version]
- Baum, D.A.; Small, R.L.; Wendel, J.F. Biogeography and Floral Evolution of Baobabs Adansonia, Bombacaceae as Inferred from Multiple Data Sets. Syst. Biol. 1998, 47, 181–207. [Google Scholar] [CrossRef]
- Clayton, J.W.; Soltis, P.S.; Soltis, D.E. Recent Long-Distance Dispersal Overshadows Ancient Biogeographical Patterns in a Pantropical Angiosperm Family (Simaroubaceae, Sapindales). Syst. Biol. 2009, 58, 395–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meimberg, H.; Wistuba, A.; Dittrich, P.; Heubl, G. Molecular Phylogeny of Nepenthaceae Based on Cladistic Analysis of Plastid trnK Intron Sequence Data. Plant Biol. 2001, 3, 164–175. [Google Scholar] [CrossRef]
- Renner, S.S. Multiple Miocene Melastomataceae Dispersal between Madagascar, Africa and India. Philos. Trans. R. Soc. London Ser. B Biol. Sci. 2004, 359, 1485–1494. [Google Scholar] [CrossRef] [PubMed]


| Label | Node Constrained (MRCA) | Species | Morphology | Age | References |
|---|---|---|---|---|---|
| A | Legume stem | Polygala californica–Cercis occidentalis | 96.33 | Magallon et al. (2015) | |
| B | Styphnolobium stem | Styphnolobium japonicum–Pickeringia montana | Leaf and fruit | 40 | Lavin et al. (2005) |
| C | Tipuana stem | Tipuana tipu–Pterocarpus indicus | Fruit | 10 | Lavin et al. (2005) |
| D | Dalbergia stem | Dalbergia hupeana–Machaerium lunatum | Leaf | 40 | Lavin et al. (2005) |
| DNA Region | Alignment Length (bp) | Number of Variable Sites | Number of Potentially Informative Sites | ||
|---|---|---|---|---|---|
| rbcL | 695 | 17 | (2.44%) | 24 | (3.45%) |
| matK | 1070 | 74 | (6.91%) | 119 | (11.12%) |
| ITS | 1041 | 105 | (10.08%) | 338 | (32.46%) |
| Concatenated dataset | 2806 | 196 | (6.98%) | 481 | (17.14%) |
| Node | Estimated Divergence Time in Ma with [95% HPD] | Estimated Ancestral Area (DEC) |
|---|---|---|
| 1 | 22.9 [25.9–19.9] | E |
| 2 | 6.7 [9.5–4.5] | E |
| 3 | 15.9 [19.4–12.6] | E |
| 4 | 15.2 [18.1–12.5] | AE |
| 5 | 3.4 [5.1–2.0] | B |
| 6 | 2.2 [3.4–1.2] | E 78; DE 22 |
| 7 | 0.9 [1.7–0.3] | E 68; DE 21; D 11 |
| 8 | 21.1 [24.0–18.5] | A 77; AE 12; AB 11 |
| 9 | 18.9 [21.7–16.3] | A 75; AB 25 |
| 10 | 11.5 [14.6–8.6] | BC 55; AC 20; C 17; B 8 |
| 11 | 16.3 [19.2–13.8] | AB 79; A 21.62 |
| 12 | 1.0 [2.1–0.3] | BC |
| 13 | 18.9 [21.6–16.3] | A 63; AB 20; AE 17 |
| 14 | 17.3 [20.3–14.4] | A 52; AE 26; B 11; AB 11 |
| 15 | 14.1 [17.3–10.9] | AB 76; A 24 |
| 16 | 1.0 [1.7–0.4] | BC |
| 17 | 10.7 [13.6–8.0] | B 75 AB 25 |
| 18 | 11.7 [13.8–9.0] | E 72; DE 15; BE 14 |
| 19 | 10.0 [12.3–7.7] | DE 74; E 26 |
| 20 | 5.6 [7.3–4] | C |
| 21 | 7.5 [9.4–5.9] | A 84; AB 16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahaingoson, F.R.; Oyebanji, O.; Stull, G.W.; Zhang, R.; Yi, T.-S. A Dated Phylogeny of the Pantropical Genus Dalbergia L.f. (Leguminosae: Papilionoideae) and Its Implications for Historical Biogeography. Agronomy 2022, 12, 1612. https://doi.org/10.3390/agronomy12071612
Rahaingoson FR, Oyebanji O, Stull GW, Zhang R, Yi T-S. A Dated Phylogeny of the Pantropical Genus Dalbergia L.f. (Leguminosae: Papilionoideae) and Its Implications for Historical Biogeography. Agronomy. 2022; 12(7):1612. https://doi.org/10.3390/agronomy12071612
Chicago/Turabian StyleRahaingoson, Fabien Robert, Oyetola Oyebanji, Gregory W. Stull, Rong Zhang, and Ting-Shuang Yi. 2022. "A Dated Phylogeny of the Pantropical Genus Dalbergia L.f. (Leguminosae: Papilionoideae) and Its Implications for Historical Biogeography" Agronomy 12, no. 7: 1612. https://doi.org/10.3390/agronomy12071612
APA StyleRahaingoson, F. R., Oyebanji, O., Stull, G. W., Zhang, R., & Yi, T.-S. (2022). A Dated Phylogeny of the Pantropical Genus Dalbergia L.f. (Leguminosae: Papilionoideae) and Its Implications for Historical Biogeography. Agronomy, 12(7), 1612. https://doi.org/10.3390/agronomy12071612






