Genome-Wide Analysis of the Rice Gibberellin Dioxygenases Family Genes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Sequence Retrieval and Identification and Analysis of GAox Genes in Rice
2.3. Analysis of Phylogenetic Relationship and Sequences
2.4. Analysis of GA Oxidase Family Gene Expression Patterns
2.5. RNA Extraction, qRT-PCR and RNA seq
2.6. Statistical Analysis
3. Results
3.1. Identification and Analysis of the GAox Family Genes in the Rice
3.2. Phylogenetic Analysis of the GAox Gene Family
3.3. Gene Structure and Conserved Motif Analysis of GAox Gene Family
3.4. Expression Patterns of GAox Genes
3.5. GAox Gene Expression Profiles under GA3 Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Peng, J.; Harberd, N.P. The role of GA-mediated signalling in the control of seed germination. Curr. Opin. Plant Biol. 2002, 5, 376–381. [Google Scholar] [CrossRef]
- Zhang, N.; Xie, Y.D.; Guo, H.J.; Zhao, L.S.; Xiong, H.C.; Gu, J.Y.; Li, J.H.; Kong, F.Q.; Sui, L.; Zhao, Z.W.; et al. Gibberellins regulate the stem elongation rate without affecting the mature plant height of a quick development mutant of winter wheat (Triticum aestivum L.). Plant Physiol. Biochem. 2016, 1072, 28–236. [Google Scholar] [CrossRef] [PubMed]
- Lo, S.F.; Yang, S.Y.; Chen, K.T.; Hsing, Y.I.; Zeevaart, J.A.; Chen, L.J.; Yu, S.M. A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 2008, 20, 2603–2618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakata, T.; Oda, S.; Tsunaga, Y.; Shomura, H.; Kawagishi-Kobayashi, M.; Aya, K.; Saeki, K.; Endo, T.; Nagano, K.; Kojima, M.; et al. Reduction of gibberellin by low temperature disrupts pollen development in rice. Plant Physiol. 2014, 164, 2011–2019. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, E.; Atsmon, D.; Halevy, A.H. Adventitious staminate flower formation in gibberellin treated gynoecious cucumber plants. Plant Cell Physiol. 1977, 18, 1193–1201. [Google Scholar] [CrossRef]
- MacMillan, J. Occurrence of Gibberellins in Vascular Plants, Fungi, and Bacteria. J. Plant Growth Regul. 2001, 20, 387–442. [Google Scholar] [CrossRef]
- Hedden, P.; Thomas, S.G. Gibberellin biosynthesis and its regulation. Biochem. J. 2012, 444, 11–25. [Google Scholar] [CrossRef] [Green Version]
- Olszewski, N.; Sun, T.P.; Gubler, F. Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 2002, 14, S61–S80. [Google Scholar] [CrossRef] [Green Version]
- Hedden, P. Gibberellin Metabolism and Its Regulation. J. Plant Growth Regul. 2001, 20, 317–318. [Google Scholar] [CrossRef]
- Arabidopsis Genome, I. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef] [Green Version]
- Hedden, P.; Kamiya, Y. GIBBERELLIN BIOSYNTHESIS: Enzymes, Genes and Their Regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 484, 31–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Carolis, E.; De Luca, V. 2-oxoglutarate-dependent dioxygenase and related enzymes: Biochemical characterization. Phytochemistry 1994, 36, 1093–1107. [Google Scholar] [CrossRef]
- Monna, L.; Kitazawa, N.; Yoshino, R.; Suzuki, J.; Masuda, H.; Maehara, Y.; Tanji, M.; Sato, M.; Nasu, S.; Minobe, Y. Positional cloning of rice semidwarfing gene, sd-1: Rice “green revolution gene” encodes a mutant enzyme involved in gibberellin synthesis. Dna Res. Int. J. Rapid Publ. Rep. Genes Genomes 2002, 9, 11–17. [Google Scholar] [CrossRef] [Green Version]
- Khush, G.S. Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 1997, 35, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Tang, D.; Shen, Y.; Qin, B.; Hong, L.; You, A.; Li, M.; Wang, X.; Yu, H.; Gu, M.; et al. Activation of gibberellin 2-oxidase 6 decreases active gibberellin levels and creates a dominant semi-dwarf phenotype in rice (Oryza sativa L.). J. Genet. Genom. 2010, 37, 23–36. [Google Scholar] [CrossRef]
- Oikawa, T.; Koshioka, M.; Kojima, K.; Yoshida, H.; Kawata, M. A role of OsGA20ox1, encoding an isoform of gibberellin 20-oxidase, for regulation of plant stature in rice. Plant Mol. Biol. 2004, 55, 687–700. [Google Scholar] [CrossRef]
- Itoh, H.; Ueguchi-Tanaka, M.; Sentoku, N.; Kitano, H.; Matsuoka, M.; Kobayashi, M. Cloning and functional analysis of two gibberellin 3 beta -hydroxylase genes that are differently expressed during the growth of rice. Proc. Natl. Acad. Sci. USA 2001, 98, 8909–8914. [Google Scholar] [CrossRef] [Green Version]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucl. Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucl. Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [Green Version]
- Vilella, A.J.; Severin, J.; Ureta-Vidal, A.; Heng, L.; Durbin, R.; Birney, E. Ensembl Compara Gene Trees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009, 19, 327–335. [Google Scholar] [CrossRef] [Green Version]
- Guo, A.Y.; Zhu, Q.H.; Chen, X.; Luo, J.C. GSDS: A gene structure display server. Yi Chuan = Hered. 2007, 29, 1023–1026. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W. Noble WS: MEME SUITE: Tools for motif discovery and searching. Nucl. Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Wang, Y.; Liu, Z.; Cheng, H.; Xue, Y. HemI: A toolkit for illustrating heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Zhu, B. Evolutionary analysis of three gibberellin oxidase genes in rice, Arabidopsis, and soybean. Gene 2011, 473, 23–35. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucl. Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef] [PubMed]
- Shan, C.; Mei, Z.; Duan, J.; Chen, H.; Feng, H.; Cai, W. OsGA2ox5, a gibberellin metabolism enzyme, is involved in plant growth, the root gravity response and salt stress. PLoS ONE 2014, 9, e87110. [Google Scholar] [CrossRef]
- Long, M.; Rosenberg, C.; Gilbert, W. Intron phase correlations and the evolution of the intron/exon structure of genes. Proc. Natl. Acad. Sci. USA 1995, 92, 12495–12499. [Google Scholar] [CrossRef] [Green Version]
- Kolkman, J.A.; Stemmer, W.P. Directed evolution of proteins by exon shuffling. Nat. Biotechnol. 2001, 19, 423–428. [Google Scholar] [CrossRef]
- Morgante, M.; Brunner, S.; Pea, G.; Fengler, K.; Zuccolo, A.; Rafalski, A. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat. Genet. 2005, 37, 997–1002. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.; Abraham, Z.; Carbonero, P.; Diaz, I. Comparative phylogenetic analysis of cystatin gene families from arabidopsis, rice and barley. Mol. Genet. Genom. Mgg 2005, 273, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Pimenta Lange, M.J.; Liebrandt, A.; Arnold, L.; Chmielewska, S.M.; Felsberger, A.; Freier, E.; Heuer, M.; Zur, D.; Lange, T. Functional characterization of gibberellin oxidases from cucumber, Cucumis sativus L. Phytochemistry 2013, 906, 2–69. [Google Scholar] [CrossRef] [PubMed]
- Lange, T. Cloning gibberellin dioxygenase genes from pumpkin endosperm by heterologous expression of enzyme activities in Escherichia coli. Proc. Natl. Acad. Sci. USA 1997, 94, 6553–6558. [Google Scholar] [CrossRef] [Green Version]
- Salazar-Cerezo, S.; Martinez-Montiel, N.; Garcia-Sanchez, J.; Perez, Y.T.R.; Martinez-Contreras, R.D. Gibberellin biosynthesis and metabolism: A convergent route for plants, fungi and bacteria. Microbiol. Res. 2018, 2088, 5–98. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, Y.; Liu, X.; Zhang, X.; Liu, S.; Yu, X.; Ren, Y.; Zheng, X.; Zhou, K.; Jiang, L.; et al. A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev. Cell 2013, 27, 113–122. [Google Scholar] [CrossRef] [Green Version]
- Hedden, P.; Phillips, A.L. Gibberellin metabolism: New insights revealed by the genes. Trends Plant Sci. 2000, 5, 523–530. [Google Scholar] [CrossRef]
- Sakamoto, T.; Morinaka, Y.; Ishiyama, K.; Kobayashi, M.; Itoh, H.; Kayano, T.; Iwahori, S.; Matsuoka, M.; Tanaka, H. Genetic manipulation of gibberellin metabolism in transgenic rice. Nat. Biotechnol. 2003, 21, 909–913. [Google Scholar] [CrossRef]

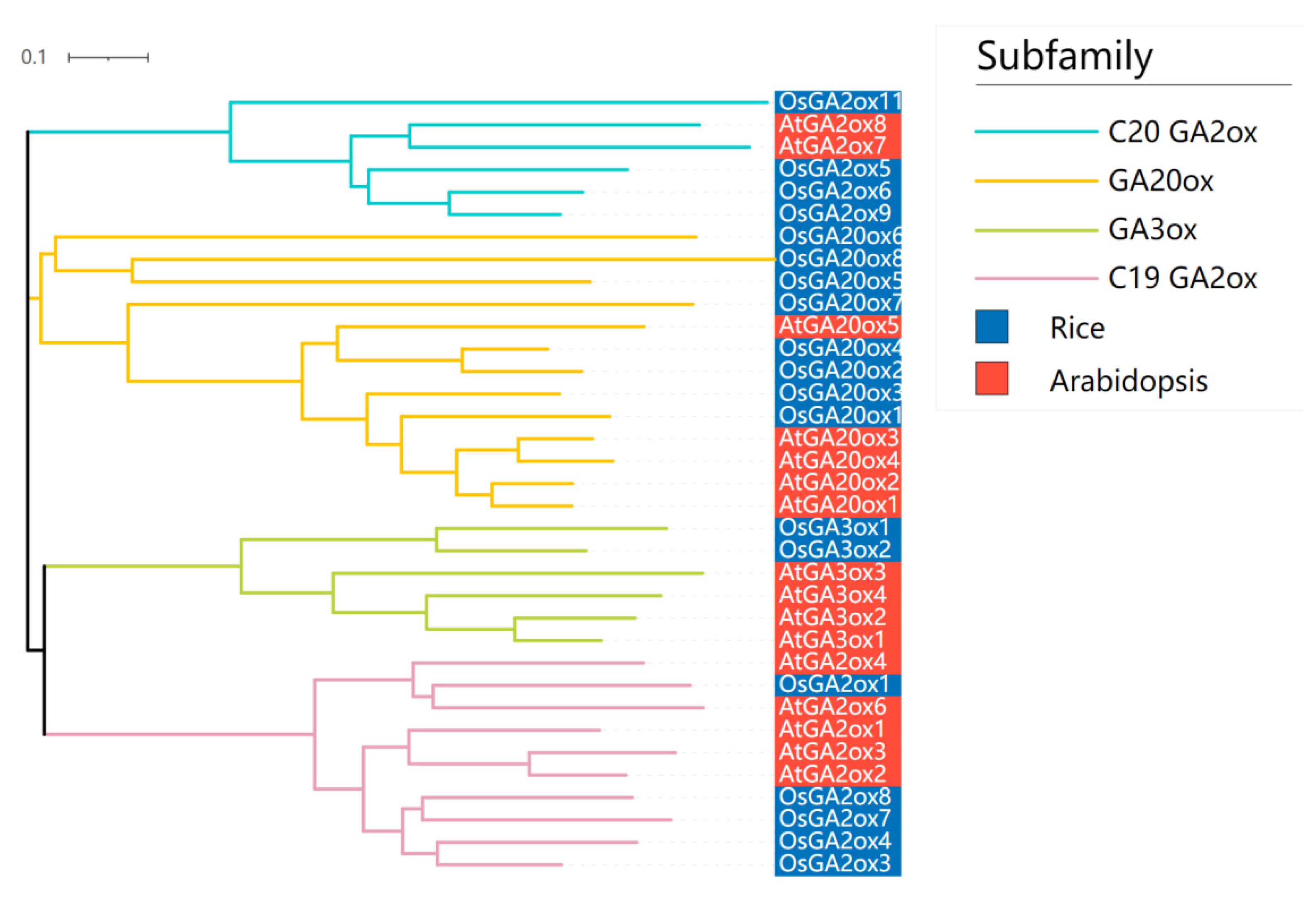
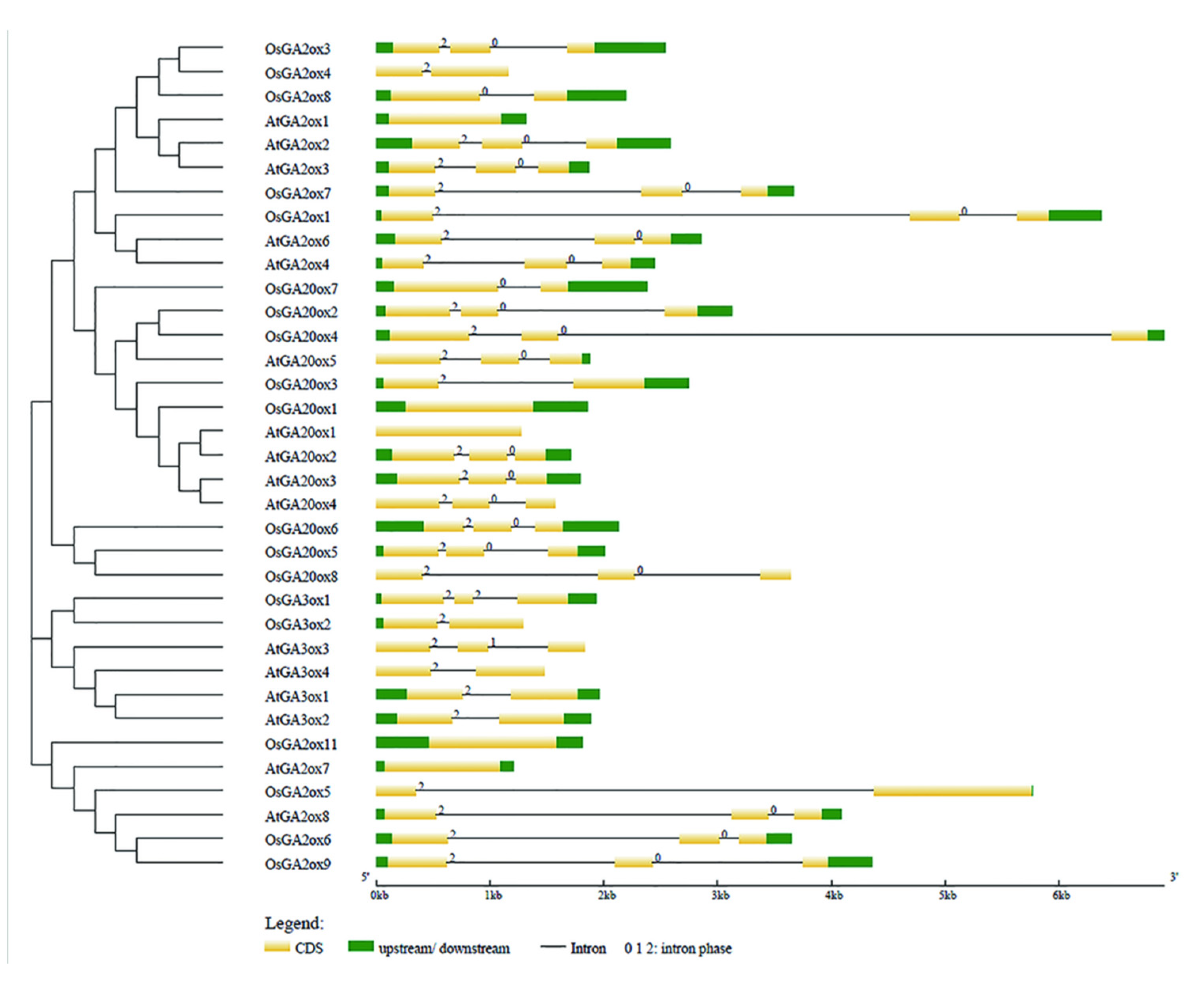
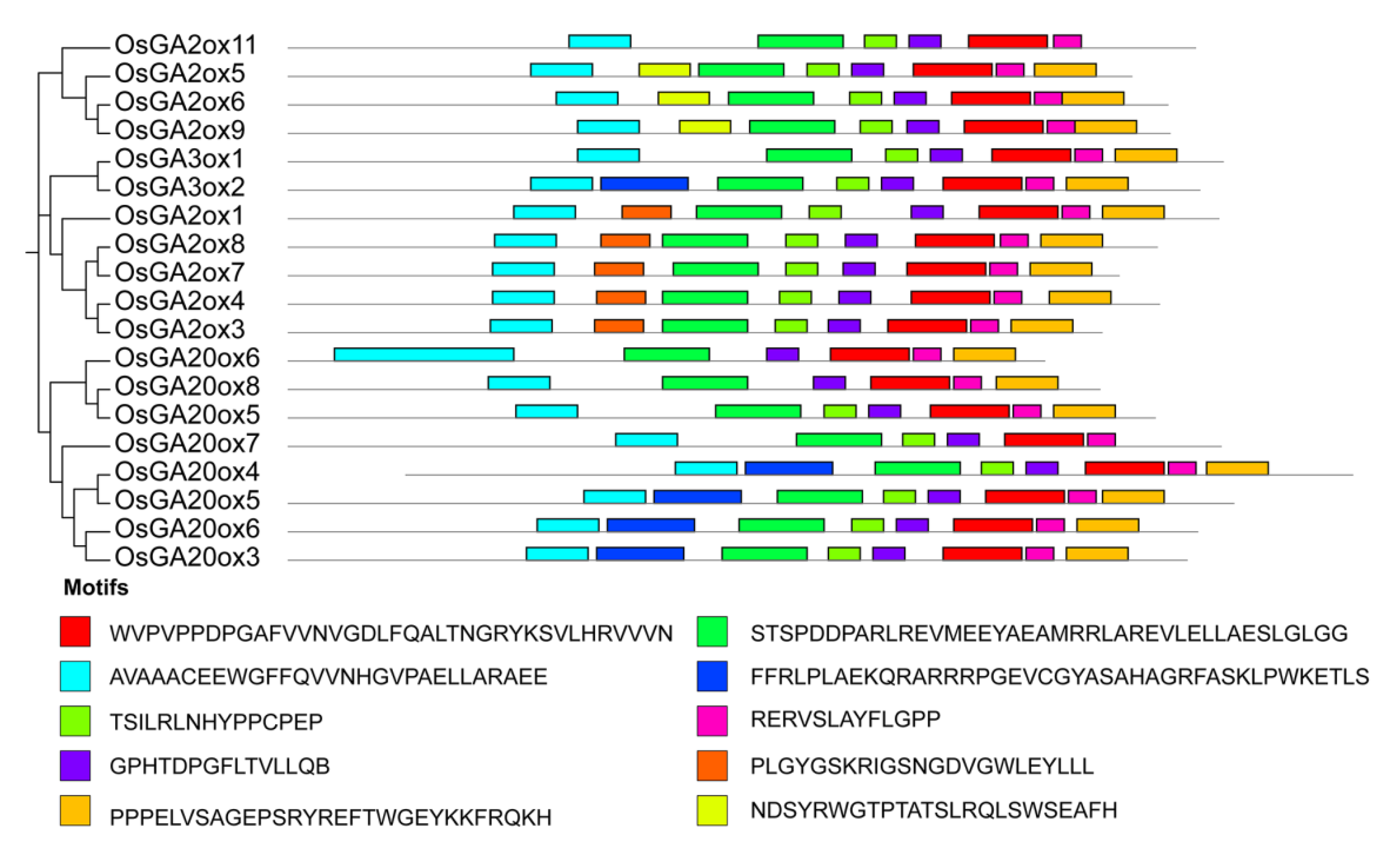

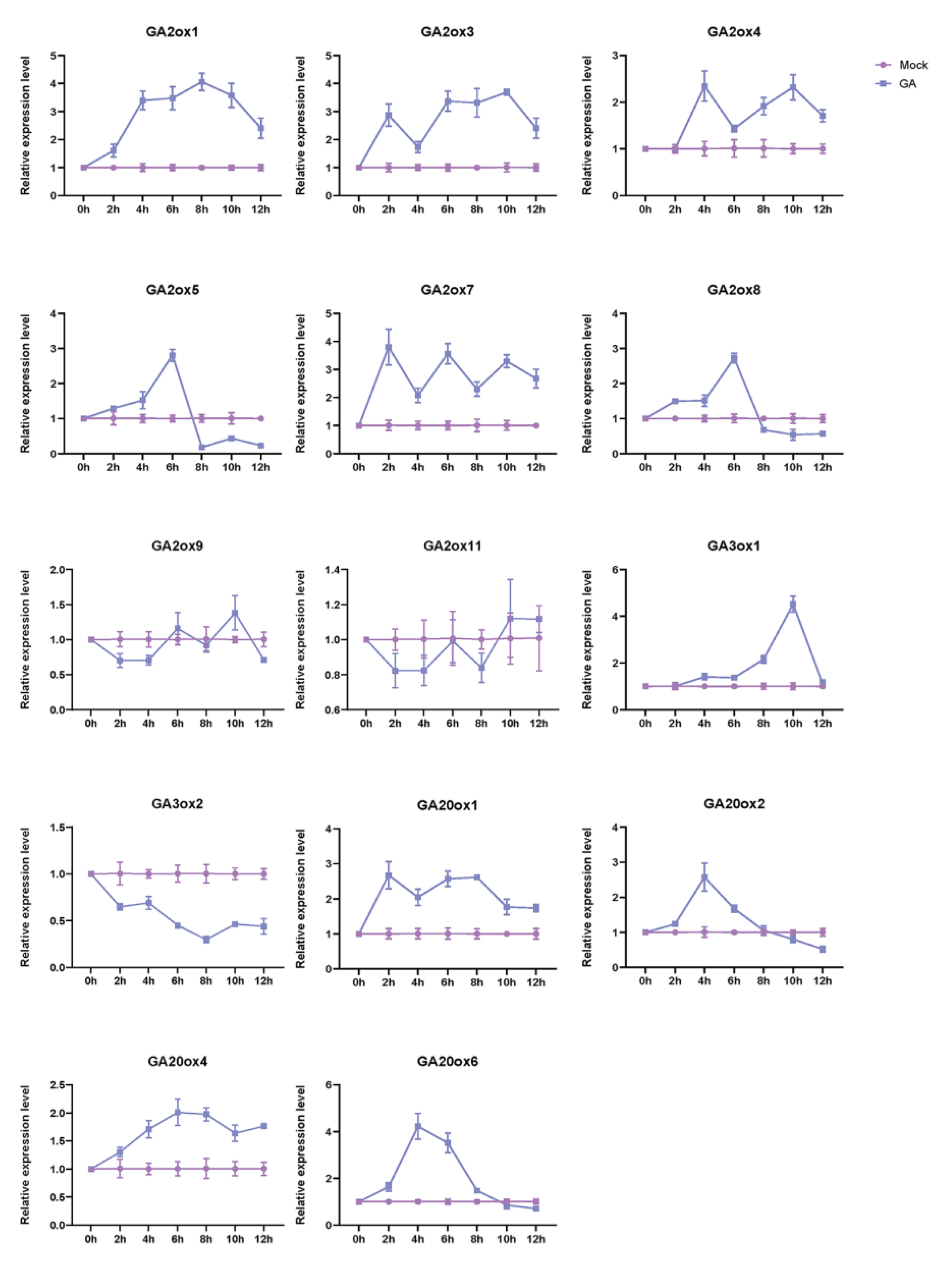
| Gene Names | Entry ID | Number of Deduced Amino Acid | Molecular Weight(kDa) | Subcellular Location (TargetP 2.0) | Subcellular Location (WoLFPSORT) | Groups | Theoretical pI |
|---|---|---|---|---|---|---|---|
| OsGA2ox1 | Os05g06670 | 383 | 40.62 | other/sp | pero/cyto/nucl | C19GA2ox | 7.11 |
| OsGA2ox3 | Os01g55240 | 328 | 35.33 | other/sp | cyto/chlo/pero | C20GA2ox | 6.67 |
| OsGA2ox4 | Os05g43880 | 355 | 37.79 | other/sp | chlo/cyto/nucl | C19GA2ox | 6.86 |
| OsGA2ox5 | Os07g01340 | 342 | 38.69 | other | nucl/cyto/chlo | C19GA2ox | 6.27 |
| OsGA2ox6 | Os04g44150 | 359 | 39.04 | other/sp | cyto/chlo/pero | C20GA2ox | 7.44 |
| OsGA2ox7 | Os01g11150 | 336 | 35.14 | other/sp/mito/chlo | chlo/plas/cyto | C19GA2ox | 7.08 |
| OsGA2ox8 | Os05g48700 | 354 | 37.31 | other/chlo | cyto/chlo/pero | C19GA2ox | 6.4 |
| OsGA2ox9 | Os02g41954 | 360 | 39.02 | other/sp | cyto/nucl/mito | C20GA2ox | 5.66 |
| OsGA2ox11 | Os04g33360 | 372 | 40.23 | other/mito | chlo/nucl/cyto | C20GA2ox | 6.71 |
| OsGA3ox1 | Os05g08540 | 385 | 41.56 | other/chlo | chlo/mito | GA3ox | 6.36 |
| OsGA3ox2 | Os01g08220 | 374 | 40.57 | other/sp | cyto/chlo/nucl | GA3ox | 6.96 |
| OsGA20ox1 | Os03g63970 | 373 | 42.26 | other/sp | cyto/chlo/nucl | GA20ox | 6.41 |
| OsGA20ox2 | Os01g66100 | 390 | 42.51 | other/sp | cyto/nucl/plas | GA20ox | 6.01 |
| OsGA20ox3 | Os07g07420 | 368 | 40.49 | other/sp | chlo/plas | GA20ox | 6.09 |
| OsGA20ox4 | Os05g34854 | 446 | 47.63 | other/chlo | chlo/cyto/nucl | GA20ox | 7.18 |
| OsGA20ox5 | Os03g42130 | 353 | 39.29 | other/sp | cyto/chlo/mito | GA20ox | 4.98 |
| OsGA20ox6 | Os04g39980 | 301 | 32.1 | other/sp | cyto/mito | GA20ox | 5.61 |
| OsGA20ox7 | Os08g44590 | 384 | 41.83 | other/sp | cyto | GA20ox | 6.35 |
| OsGA20ox8 | Os04g55070 | 327 | 35.79 | other/sp | cyto/chlo/nucl | GA20ox | 5.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Liu, W.; Huang, Z.; Huang, J.; Xu, Y.; Zhang, Q.; Hu, J. Genome-Wide Analysis of the Rice Gibberellin Dioxygenases Family Genes. Agronomy 2022, 12, 1627. https://doi.org/10.3390/agronomy12071627
He Y, Liu W, Huang Z, Huang J, Xu Y, Zhang Q, Hu J. Genome-Wide Analysis of the Rice Gibberellin Dioxygenases Family Genes. Agronomy. 2022; 12(7):1627. https://doi.org/10.3390/agronomy12071627
Chicago/Turabian StyleHe, Yurong, Wei Liu, Zhihao Huang, Jishuai Huang, Yanghong Xu, Qiannan Zhang, and Jun Hu. 2022. "Genome-Wide Analysis of the Rice Gibberellin Dioxygenases Family Genes" Agronomy 12, no. 7: 1627. https://doi.org/10.3390/agronomy12071627
APA StyleHe, Y., Liu, W., Huang, Z., Huang, J., Xu, Y., Zhang, Q., & Hu, J. (2022). Genome-Wide Analysis of the Rice Gibberellin Dioxygenases Family Genes. Agronomy, 12(7), 1627. https://doi.org/10.3390/agronomy12071627





