Influence of Kosakonia sp. on the Growth of Arachis hypogaea L. on Arid Soil
Abstract
1. Introduction
2. Materials and Methods
2.1. Rhizosphere Sample Collection and Processing
2.2. Chemicals Used
2.3. Enumeration of Nitrogen-Fixing Bacterial Populations
2.4. Plant Growth-Promoting (PGP) Activities of Test Isolates
2.4.1. Analysis of Ammonia-Producing Potential
2.4.2. Hydrogen Cyanide (HCN)-Producing Efficiency of Isolates
2.4.3. Siderophore Synthesis
2.4.4. Quantitative Examination of IAA Production
2.4.5. Phosphate-Solubilizing Competence of Test Isolates
2.4.6. Characterization and Identification of Nitrogen-Fixing Isolates
2.4.7. Morphological and Biochemical Characterization
2.4.8. 16S rRNA Sequencing
2.4.9. PCR Amplification of 16S rRNA
2.4.10. Sequencing of Amplified 16SrRNA
2.4.11. Multiple Sequence Alignment and Phylogenetic Analysis
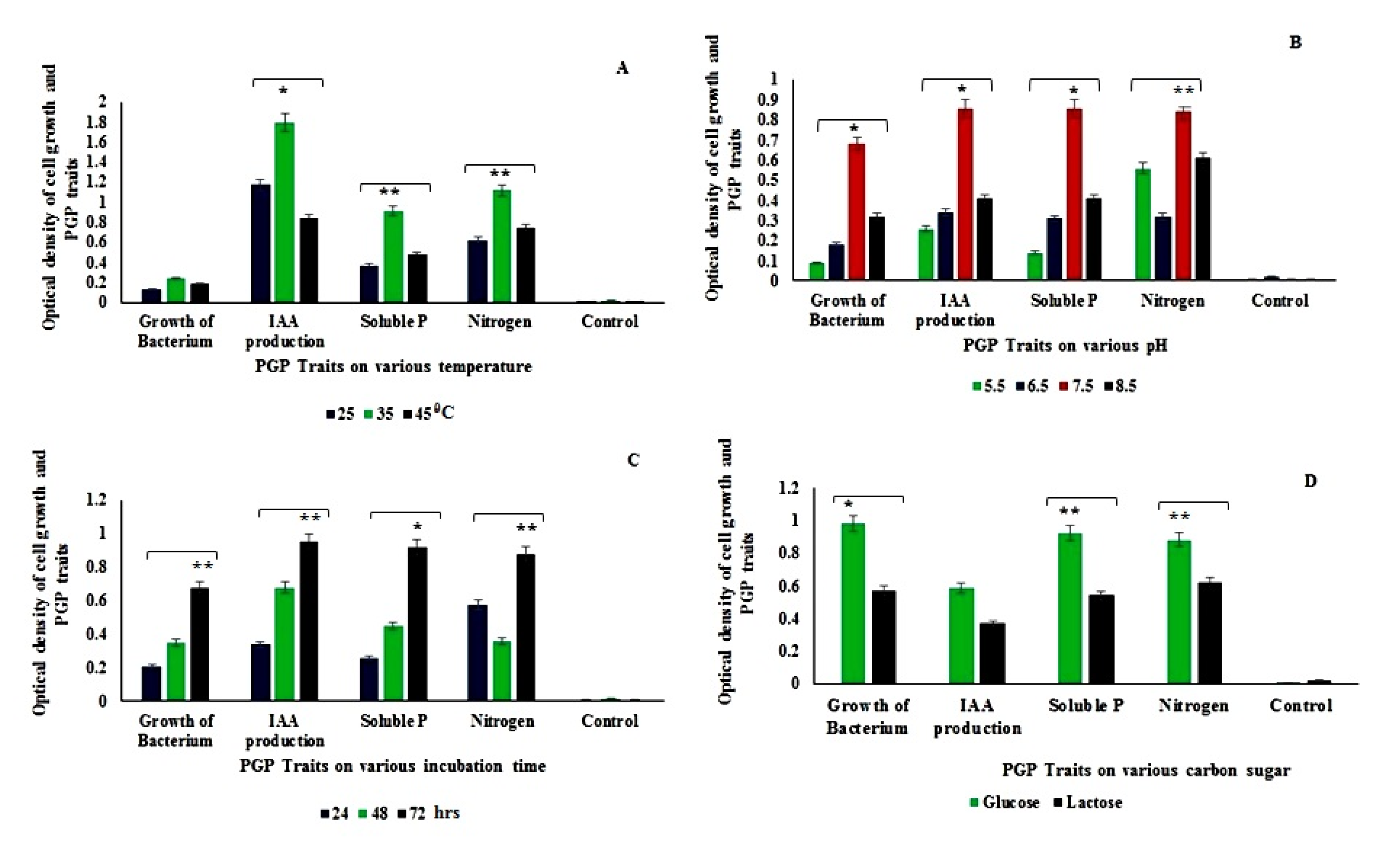
2.4.12. Optimization of Growth Parameters for Better PGP Trait Activities of Kosakonia sp. MGR1
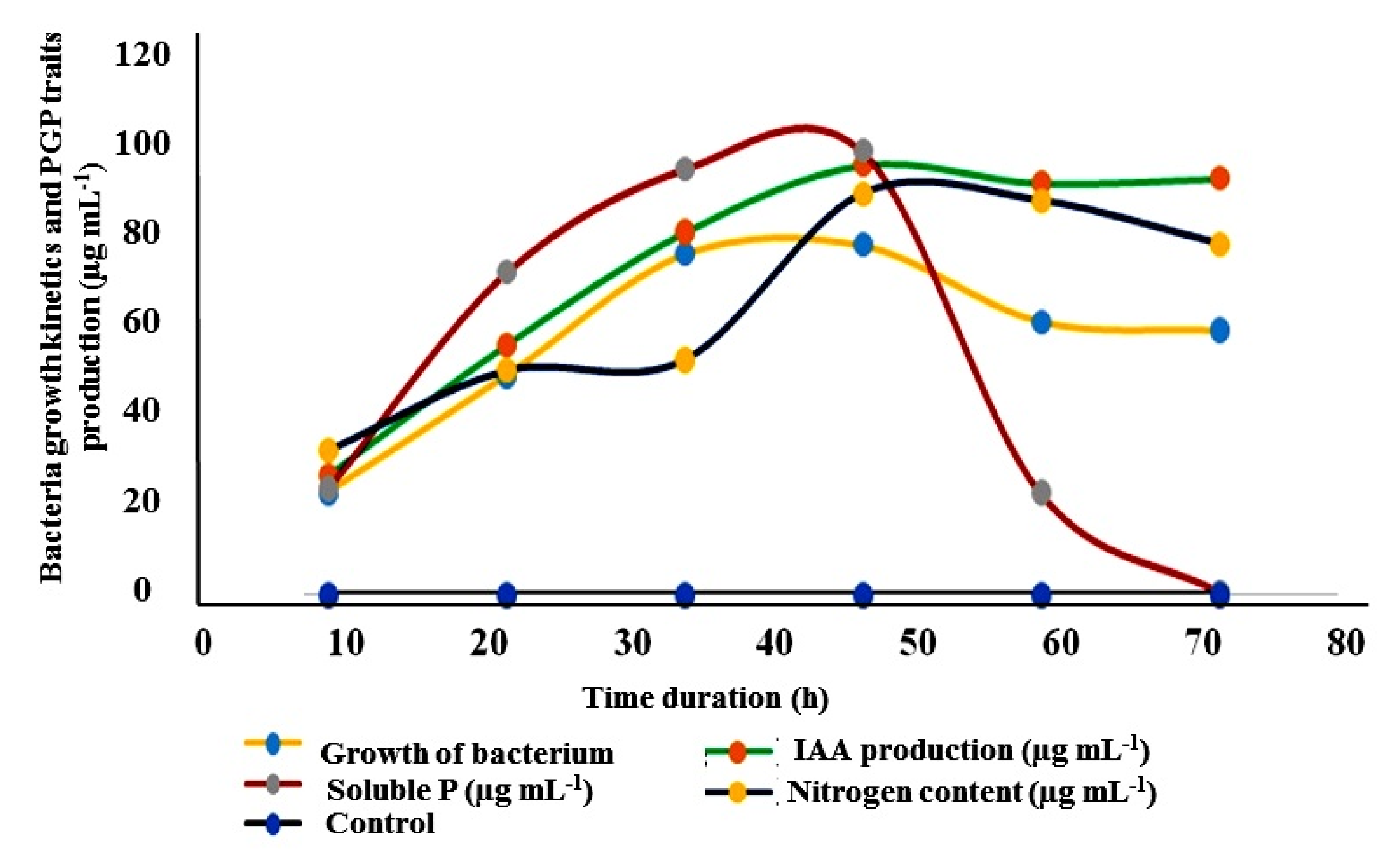
2.4.13. IAA Production, Nitrogen, and Phosphate Solubilization Potential of Kosakonia sp. MGR1
2.4.14. Stimulus Effect of Kosakonia sp. MGR1 on Physiology and Biomolecule (Protein, Carbohydrate, and Chlorophyll) Contents of A. hypogaea L.
2.5. Statistical Analysis
3. Results and Discussion
3.1. PGP Activities of Isolates
3.2. Quantitative Analyses of IAA and P-Solubilizing Potential of Isolates
3.3. Preliminary Identification of Potential PGP Isolate Ah4
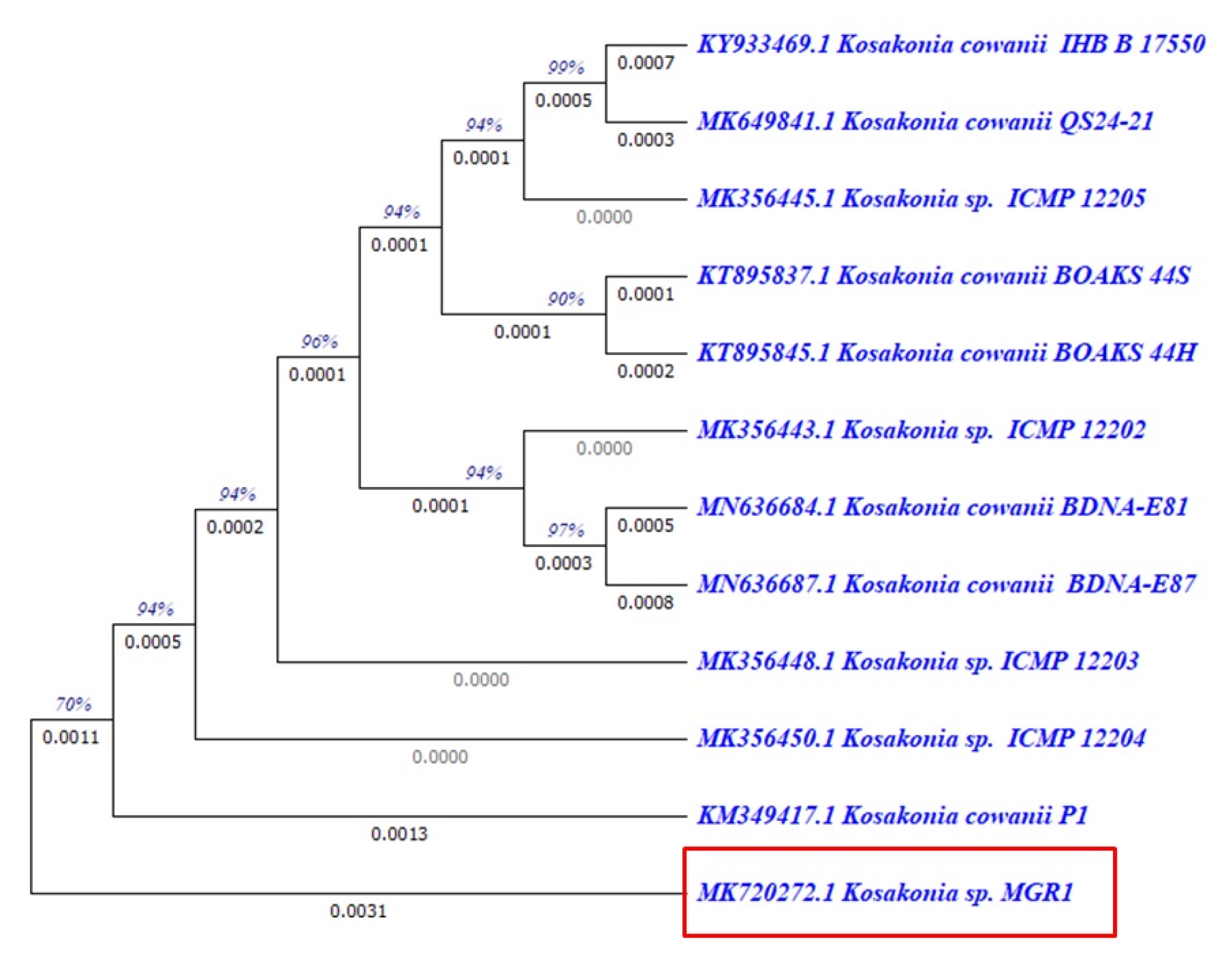
3.4. 16S rRNA Sequencing Analysis
3.4.1. Similarity and Phylogenetic Tree Analysis
3.4.2. Optimization, Production of IAA, Nitrogen, and Phosphate Solubilization
3.4.3. Influence of Kosakonia sp. MGR1 on Physiology and Biomolecule Contents of A. hypogaea L.
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Narayanan, M.; Kumarasamy, S.; Ranganathan, M.; Kandasamy, S.; Kandasamy, G.; Gnanavel, K. Enzyme and metabolites attained in degradation of chemical pesticides ß Cypermethrin by Bacillus cereus. Mater. Today Proc. 2020, 33, 3640–3645. [Google Scholar] [CrossRef]
- Obele, I.I.; Danladi, M.M.; Akwashiki, O.; Owuna, G.; Peter, O.E.; Obiekezie, S.; Paul, T.; Kenneth, E.I.; Olokunle, A.-A. Isolation, Identification and Screening for Nitrogen Fixing Activities by Azotobacter chroococcum Isolated from Soil of Keffi, Nigeria as Agent for Bio-fertilizer Production. Front. Environ. Microbiol. 2019, 5, 70. [Google Scholar] [CrossRef][Green Version]
- Leghari, S.J.; Wahocho, N.A.; Laghari, G.M.; HafeezLaghari, A.; MustafaBhabhan, G.; HussainTalpur, K.; Bhutto, T.A.; Wahocho, S.A.; Lashari, A.A. Role of nitrogen for plant growth and development: A review. Adv. Environ. Biol. 2016, 10, 209–219. [Google Scholar]
- Mathiyazhagan, N.; Natarajan, D. Phytoremediation efficiency of edible and economical crops on waste dumps of bauxite mines, Salem district, Tamil Nadu, India. In On a Sustainable Future of the Earth’s Natural Resources; Springer: Berlin, Heidelberg, 2013; pp. 493–508. [Google Scholar]
- Sun, S.; Chen, Y.; Cheng, J.; Li, Q.; Zhang, Z.; Lan, Z. Isolation, characterization, genomic sequencing, and GFP-marked insertional mutagenesis of a high-performance nitrogen-fixing bacterium, Kosakonia radicincitans GXGL-4A and visualization of bacterial colonization on cucumber roots. Folia Microbiol. 2018, 63, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Ambreen, A.; Robab, M.I.; Rushda, S. Plant growth promoting rhizobacteria: An overview. J. Nat. Prod. Plant Resour. 2012, 2, 19–31. [Google Scholar]
- Chen, Y.; Huang, Z.; Li, J.; Su, G.; Feng, B. Complete Genome Sequence of Kosakonia radicincitans GXGL-4A, a Nitrogen-Fixing Bacterium with Capability to Degrade TEX. Curr. Microbiol. 2020, 77, 1848–1857. [Google Scholar] [CrossRef]
- Ahmad, F.; Ahmad, I.; Khan, M. Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol. Res. 2008, 163, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Kleingesinds, C.K.; de Santi Ferrara, F.I.; Floh, E.I.S.; Aidar, M.P.M.; Barbosa, H.R. Sugarcane growth promotion by Kosakonia sp. ICB117 an endophytic and diazotrophic bacterium. Afr. J. Microbiol. Res. 2018, 12, 105–114. [Google Scholar]
- Inui, R.; Takeshi, L.; Picchi, S.; Barbosa, J.; Lemos, M.; Marcondes, J.; Macedo Eg Lemos, D. Phosphorus solubilizing and IAA production activities in plant growth promoting rhizobacteria from brazilian soils under sugarcane cultivation. ARPN J. Eng. Appl. Sci. 2012, 7, 1446–1454. [Google Scholar]
- Narayanan, M.; Kumarasamy, S.; Ranganathan, M.; Kandasamy, S.; Kandasamy, G.; Gnanavel, K.; Mamtha, K. Production and characterization of polyhydroxyalkanoates synthesized by E. coli Isolated from sludge soil. Mater. Today Proc. 2020, 33, 3646–3653. [Google Scholar] [CrossRef]
- Mohite, B. Isolation and characterization of indole acetic acid (IAA) producing bacteria from rhizospheric soil and its effect on plant growth. J. Soil Sci. Plant Nutr. 2013, 13, 638–649. [Google Scholar] [CrossRef]
- Sadeghi, A.; Karimi, E.; Dahaji, P.A.; Javid, M.G.; Dalvand, Y.; Askari, H. Plant growth promoting activity of an auxin and siderophore producing isolate of Streptomyces under saline soil conditions. World J. Microbiol. Biotechnol. 2012, 28, 1503–1509. [Google Scholar] [CrossRef] [PubMed]
- Khamna, S.; Yokota, A.; Peberdy, J.F.; Lumyong, S. Indole-3-acetic acid production by Streptomyces sp. isolated from some Thai medicinal plant rhizosphere soils. EurAsian J. BioSci. 2010, 4, 23–32. [Google Scholar] [CrossRef]
- Fang, Q.; Fan, Z.; Xie, Y.; Wang, X.; Li, K.; Liu, Y. Screening and Evaluation of the Bioremediation Potential of Cu/Zn-Resistant, Autochthonous Acinetobacter sp. FQ-44 from Sonchus oleraceus L. Front. Plant Sci. 2016, 7, 1487. [Google Scholar] [CrossRef] [PubMed]
- Wahyudi, A.T.; Astuti, R.P.; Widyawati, A.; Mery, A.; Nawangsih, A.A. Characterization of Bacillus sp. strains isolated from rhizosphere of soybean plants for their use as potential plant growth for promoting rhizobacteria. J. Microbiol. Antimicrob. 2011, 3, 34–40. [Google Scholar]
- Rodrigues, A.A.; Araújo, M.V.F.; Soares, R.S.; De Oliveira, B.F.; Ribeiro, I.D.; Sibov, S.T.; Vieira, J.D.G. Isolation and prospection of diazotrophic rhizobacteria associated with sugarcane under organic management. An. Acad. Bras. Ciências 2018, 90, 3813–3829. [Google Scholar] [CrossRef]
- Da Silva, M.A.C.; Cavalett, A.; Spinner, A.; Rosa, D.C.; Jasper, R.B.; Quecine, M.C.; Bonatelli, M.L.; Pizzirani-Kleiner, A.; Corção, G.; Lima, A.O.D.S. Phylogenetic identification of marine bacteria isolated from deep-sea sediments of the eastern South Atlantic Ocean. SpringerPlus 2013, 2, 127. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Tamura, Y.; Harada, Y.; Nishikawa, S.-I.; Yamano, K.; Kamiya, M.; Shiota, T.; Kuroda, T.; Kuge, O.; Sesaki, H.; Imai, K.; et al. Tam41 is a CDP-diacylglycerol synthase required for cardiolipin biosynthesis in mitochondria. Cell Metab. 2013, 17, 709–718. [Google Scholar] [CrossRef]
- Celloto, V.R.; Oliveira, A.J.; Gonçalves, J.E.; Watanabe, C.S.; Matioli, G.; Gonçalves, R.A. Biosynthesis of indole-3-acetic acid by new Klebsiella oxytoca free and immobilized cells on inorganic matrices. Sci. World J. 2012, 2012, 495970. [Google Scholar] [CrossRef]
- Singh, A.K.; Singh, G.; Bhatt, R.P. Effects of salt stress on cell surface properties and symbiotic performance of root nodulating bacteria. Pharm. Biosci. J. 2015, 3, 23–29. [Google Scholar] [CrossRef]
- Mathiyazhagan, N.; Natarajan, D. Impact of mine waste dumps on growth and biomass of economically important crops. J. Environ. Biol. 2012, 33, 1069. [Google Scholar] [PubMed]
- Talreja, T. Biochemical estimation of three primary metabolites from medicinally important plant Moringa oleifera. Int. J. Pharm. Sci. Rev. Res. 2011, 7, 186–188. [Google Scholar]
- Chandra, S.; Askari, K.; Kumari, M. Optimization of indole acetic acid production by isolated bacteria from Stevia rebaudiana rhizosphere and its effects on plant growth. J. Genet. Eng. Biotechnol. 2018, 16, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Sudha, M.; Gowri, R.S.; Prabhavath, P.; Astapriya, P.; Devi, S.Y.; Saranya, A. Production and optimization of indole acetic acid by indigenous micro flora using agro waste as substrate. Pak. J. Biol. Sci. 2012, 15, 39–43. [Google Scholar] [CrossRef]
- Gopalakrishnan, S.; Srinivas, V.; Alekhya, G.; Prakash, B. Effect of plant growth-promoting Streptomyces sp. on growth promotion and grain yield in chickpea (Cicer arietinum L). 3 Biotech 2015, 5, 799–806. [Google Scholar] [CrossRef]
- Castro-González, R.; Martínez-Aguilar, L.; Ramírez-Trujillo, A.; Santos, P.E.-D.L.; Caballero-Mellado, J. High diversity of culturable Burkholderia species associated with sugarcane. Plant Soil 2011, 345, 155–169. [Google Scholar] [CrossRef]
- Brady, C.; Cleenwerck, I.; Venter, S.; Coutinho, T.; De Vos, P. Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): Proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov. and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter. Syst. Appl. Microbiol. 2013, 36, 309–319. [Google Scholar]
- Yang, X.-J.; Wang, S.; Cao, J.-M.; Hou, J.-H. Complete genome sequence of human pathogen Kosakonia cowanii type strain 888-76T. Braz. J. Microbiol. 2018, 49, 16–17. [Google Scholar] [CrossRef] [PubMed]
- Yamanouchi, K.; Sakamoto, Y.; Fujioka, M.; Kasai, K.; Tsujiguchi, T.; Kimura, T.; Shimazu, M.; Kato, A.; Ito, K. The effects of Pantoea and Kosakonia isolated from buckwheat sprouts on obese mice. Appl. Microbiol. Open Access 2018, 4, 23–35. [Google Scholar] [CrossRef]
- Berger, B.; Baldermann, S.; Ruppel, S. The plant growth-promoting bacterium Kosakonia radicincitans improves fruit yield and quality of Solanum lycopersicum. J. Sci. Food Agric. 2017, 97, 4865–4871. [Google Scholar] [CrossRef] [PubMed]
- Witzel, K.; Üstün, S.; Schreiner, M.; Grosch, R.; Börnke, F.; Ruppel, S. A proteomic approach suggests unbalanced proteasome functioning induced by the growth-promoting bacterium Kosakonia radicincitans in Arabidopsis. Front. Plant Sci. 2017, 8, 661. [Google Scholar] [CrossRef] [PubMed]
- Khattab, E.A.; Afifi, M.H.; Amin, G.A. Significance of nitrogen, phosphorus, and boron foliar spray on jojoba plants. Bull. Natl. Res. Cent. 2019, 43, 66. [Google Scholar] [CrossRef]
- Gendy, A.S.; Said-Al Ahl, H.A.; Mahmoud, A.A.; Mohamed, H.F. Effect of nitrogen sources, bio-fertilizers and their interaction on the growth, seed yield and chemical composition of guar plants. Life Sci. J. 2013, 10, 389–402. [Google Scholar]
- Khan, A. Phosphorus and compost management influence maize (Zea mays) productivity under semiarid condition with and without phosphate solubilizing bacteria. Front. Plant Sci. 2015, 6, 1083. [Google Scholar]

| Name of Isolates | Qualitative Analysis of Plant Growth-Promoting Activities of Isolates | |||||
|---|---|---|---|---|---|---|
| N2 Fixing | NH3 | IAA | HCN | Siderophore | Phosphate Solubilizing | |
| Ah1 | ++ | ++ | ++ | + | ++ | ++ |
| Ah2 | +++ | ++ | ++ | ++ | ++ | ++ |
| Ah3 | ++ | +++ | +++ | ++ | + | + |
| Ah4 | +++ | +++ | +++ | +++ | +++ | +++ |
| Ah5 | ++ | ++ | ++ | ++ | ++ | ++ |
| Ah6 | +++ | +++ | +++ | + | ++ | + |
| Ah7 | ++ | ++ | +++ | ++ | ++ | ++ |
| Isolates | Various Concentrations of Tryptophan (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 4 | 6 | 8 | 10 | ||||||
| 1 h | 2 h | 1 h | 2 h | 1 h | 2 h | 1 h | 2 h | 1 h | 2 h | |
| Ah1 | 3.2 ± 0.24 | 3.8 ± 0.37 | 8.12 ± 1.04 | 8.19 ± 1.54 | 13.64 ± 1.45 | 13.75 ± 1.64 | 12.18 ± 1.64 | 12.22 ± 1.07 | 12.42 ± 0.69 | 12.24 ± 0.98 |
| Ah2 | 5.3 ± 0.49 | 5.8 ± 0.39 | 9.32 ± 0.96 | 9.25 ± 1.3 | 12.45 ± 1.07 | 12.52 ± 0.56 | 13.21 ± 1.84 | 13.68 ± 1.64 | 13.1 ± 1.04 | 13.85 ± 1.32 |
| Ah3 | 2.6 ± 0.05 | 3.6 ± 0.05 | 5.7 ± 0.67 | 5.9 ± 0.34 | 6.8 ± 0.96 | 7.2 ± 0.69 | 6.9 ± 0.68 | 7.5 ± 0.69 | 6.1 ± 0.79 | 6.6 ± 0.68 |
| Ah4 | 10.5 ± 1.3 | 10.8 ± 0.94 | 18.24 ± 2.94 | 18.24 ± 2.31 | 26.17 ± 3.47 | 26.69 ± 2.67 | 25.32 ± 2.74 | 25.59 ± 2.49 | 25.14 ± 1.34 | 25.56 ± 3.04 |
| Ah5 | 6.4 ± 0.67 | 6.8 ± 0.64 | 8.6 ± 0.68 | 8.9 ± 0.57 | 11.46 ± 1.62 | 11.54 ± 1.37 | 10.34 ± 0.97 | 10.78 ± 0.67 | 10.42 ± 1.54 | 10.57 ± 0.67 |
| Ah6 | 5.4 ± 0.62 | 5.5 ± 0.67 | 9.4 ± 0.39 | 9.9 ± 0.59 | 10.64 ± 1.47 | 10.44 ± 0.96 | 11.37 ± 1.48 | 11.21 ± 1.64 | 10.95 ± 1.05 | 10.81 ± 1.56 |
| Ah7 | 2.3 ± 0.04 | 2.8 ± 0.37 | 5.21 ± 0.47 | 5.65 ± 0.62 | 8.4 ± 0.38 | 8.9 ± 0.68 | 8.6 ± 0.35 | 8.8 ± 0.67 | 8.1 ± 0.63 | 8.8 ± 0.67 |
| Isolate | Morphological Characterization | Biochemical Characterization | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I | MR | VP | C | CA | O | H2S | U | S.H | Carbohydrate Test | |||||||||
| C.M | G.S | E.S | C.S | Shape | M | GL | SU | LA | ||||||||||
| Ah4 | Mucoid | − | − | + | rod | + | − | − | + | + | + | − | − | + | + | + | − | + |
| Growth Parameters of A. hypogaea | Experimental Groups (45th Day Analysis) | |
|---|---|---|
| Test | Control | |
| Percentage germination | 100 | 100 |
| Shoot length (cm) (average of twigs) | 59.16 ± 0.55 | 41.75 ± 20.75 |
| Root length (cm) | 17.24 ± 0.50 | 14.5 ± 0.40 |
| Height of plant (cm) | 36.28 ± 4.19 | 55.88 ± 1.41 |
| Width of stem (cm) | 0.8 ± 0.1 | 0.6 ± 0.1 |
| Total wet biomass (root and shoot) (g) | 24.54 ± 2.36 | 15.46 ± 1.34 |
| Total dry biomass (root and shoot) (g) | 6.38 ± 1.34 | 5.55 ± 1.28 |
| Total chlorophyll (mg·g−1) | 25.42 ± 0.54 | 20.32 ± 0.89 |
| Total carbohydrate (mg·g−1) | 129.15 ± 2.03 | 101.96 ± 1.25 |
| Total protein (mg·g−1) | 189.35 ± 1.76 | 174.79 ± 1.44 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Narayanan, M.; Pugazhendhi, A.; David, S.; Chi, N.T.L.; Nasif, O.; Alharbi, S.A.; Ma, Y. Influence of Kosakonia sp. on the Growth of Arachis hypogaea L. on Arid Soil. Agronomy 2022, 12, 1801. https://doi.org/10.3390/agronomy12081801
Narayanan M, Pugazhendhi A, David S, Chi NTL, Nasif O, Alharbi SA, Ma Y. Influence of Kosakonia sp. on the Growth of Arachis hypogaea L. on Arid Soil. Agronomy. 2022; 12(8):1801. https://doi.org/10.3390/agronomy12081801
Chicago/Turabian StyleNarayanan, Mathiyazhagan, Arivalagan Pugazhendhi, Selvaraj David, Nguyen Thuy Lan Chi, Omaima Nasif, Sulaiman Ali Alharbi, and Ying Ma. 2022. "Influence of Kosakonia sp. on the Growth of Arachis hypogaea L. on Arid Soil" Agronomy 12, no. 8: 1801. https://doi.org/10.3390/agronomy12081801
APA StyleNarayanan, M., Pugazhendhi, A., David, S., Chi, N. T. L., Nasif, O., Alharbi, S. A., & Ma, Y. (2022). Influence of Kosakonia sp. on the Growth of Arachis hypogaea L. on Arid Soil. Agronomy, 12(8), 1801. https://doi.org/10.3390/agronomy12081801






