Genetic Improvement in Sesame (Sesamum indicum L.): Progress and Outlook: A Review
Abstract
1. Introduction
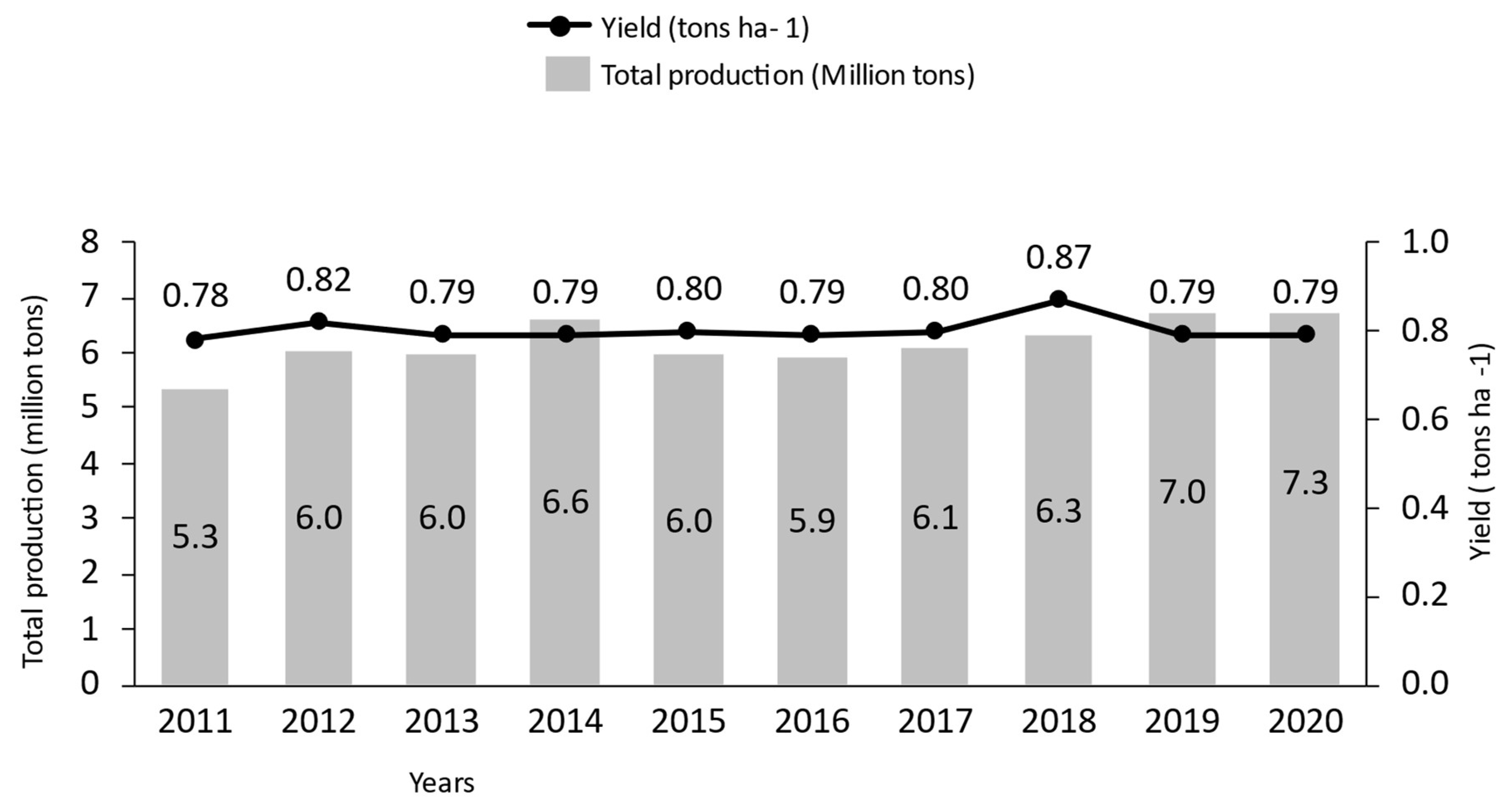
2. Global Sesame Production
3. Constraints to Sesame Production
4. Sesame Breeding
Progress and Achievements in Sesame Genetic Improvement
5. Sesame Genetic Resources and Gene Banks
6. Landraces and Improved Sesame Varieties
7. Breeding Methods and Associated Technologies for Sesame Improvement
7.1. Conventional Breeding
7.2. Mutation Breeding
7.3. Genomics-Assisted Breeding
7.3.1. Genetic Diversity Analysis
7.3.2. Quantitative Trait Locus (QTL) Analysis
7.3.3. Next-Generation Sequencing
7.3.4. Genetic Engineering and Genome Editing
8. Waterlogging and Drought Tolerance
9. Insect Pest Resistance in Sesame
10. Market-Driven Breeding in Sesame
11. Conclusion and Outlook
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ashri, A. Sesame breeding. In Plant Breeding Reviews; Janick, J., Ed.; John Wiley: Oxford, UK, 2010; Volume 16. [Google Scholar]
- Bedigian, D.; Harlan, J. Evidence for cultivation of sesame in the ancient world. Econ. Bot. 1986, 40, 137–154. [Google Scholar] [CrossRef]
- De la Vega, A.J.; Hall, A.J. Effect of planting date, genotype, and their interactions on sunflower yield: II. Components of oil yield. Crop Sci. 2002, 42, 1202–1210. [Google Scholar] [CrossRef]
- Zheljazkov, V.D.; Vick, B.A.; Ebelhar, M.W.; Buehring, N.; Baldwin, B.S.; Astatkie, T.; Mille, J.F. Yield, oil content, and composition of sunflower grown at multiple locations in Mississippi. Agron. J. 2008, 100, 635–639. [Google Scholar] [CrossRef]
- Wei, W.; Zhang, Y.; Lv, H.; Li, D.; Wang, L.; Zhang, X. Association analysis for quality traits in a diverse panel of Chinese sesame (Sesamum indicum L.) germplasm. J. Integr. Plant Biol. 2013, 55, 745–758. [Google Scholar] [CrossRef] [PubMed]
- Dossa, K.; Wei, X.; Niang, M.; Liu, P.; Zhang, Y.; Wang, L.; Liao, B.; Cissé, N.; Zhang, X.; Diouf, D. Near-infrared reflectance spectroscopy reveals wide variation in major components of sesame seeds from Africa and Asia. Crop J. 2018, 6, 202–206. [Google Scholar] [CrossRef]
- Gulluoglu, L.; Arioglu, H.; Bakal, H.; Onat, B.; Kurt, C. The Effect of Harvesting on Some Agronomic and Quality Characteristics of Peanut Grown in the Mediterranean Region of Turkey. Turk. J. Field Crop 2016, 21, 224–232. [Google Scholar] [CrossRef]
- Were, B.A.; Onkware, A.O.; Gudu, S.; Welander, M.; Carlsson, A.S. Seed oil content and fatty acid composition in East African sesame (Sesamum indicum L.) accessions evaluated over 3 years. Field Crops Res. 2006, 97, 254–260. [Google Scholar] [CrossRef]
- Anastasi, U.; Sortino, O.; Tuttobene, R. Agronomic performance and grain quality of sesame (Sesamum indicum L.) landraces and improved varieties grown in a Mediterranean environment. Genet. Resour. Crop Evol. 2017, 64, 127–137. [Google Scholar]
- Yen, G.C. Influence of seed roasting process on the changes in composition and quality of sesame (Sesame indicum L.) oil. J. Sci. Food Agric. 1990, 50, 563–570. [Google Scholar] [CrossRef]
- Yol, E.; Toker, R.; Golukcu, M.; Uzun, B. Oil Content and fatty acid Characteristics in mediterranean sesame core collection. Crop Sci. 2015, 55, 2177–2185. [Google Scholar] [CrossRef]
- Myint, D.; Gilani, S.A.; Kawase, M.; Watanabe, K.N. Sustainable Sesame (Sesamum indicum L.) Production through Improved Technology: An Overview of Production, Challenges, and Opportunities in Myanmar. Sustainability 2020, 12, 3515. [Google Scholar] [CrossRef]
- FAOSTAT. The Food and Agriculture Organization Corporate Statistical Database (FAOSTAT), Rome, Italy. 2020. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 16 March 2022).
- Morris, J.B. Food, industrial, nutraceutical, and pharmaceutical uses of sesame genetic resources. In Trends in News Crops and New Uses; Janick, J., Whipkey, A., Eds.; ASHS Press: Alexandria, Egypt, 2002; pp. 153–156. [Google Scholar]
- Abdellatef, E.; Sirelkhatem, R.; Ahmed, M.M.; Radwan, K.H.; Khalafalla, M.M. Study of genetic diversity in Sudanese sesame (Sesamum indicum L.) germplasm using random amplified polymorphic DNA (RAPD) markers. Afr. J. Biotechnol. 2008, 24, 4423–4427. [Google Scholar]
- Uzun, B.; Çagirgan, M.I. Identification of molecular markers linked to determinate growth habit in sesame. Euphytica 2009, 166, 379–384. [Google Scholar] [CrossRef]
- Teklu, D.H.; Shimelis, H.; Tesfaye, A.; Abady, S. Appraisal of the sesame production opportunities and constraints, and farmer-preferred varieties and traits, in Eastern and Southwestern Ethiopia. Sustainability 2021, 13, 11202. [Google Scholar] [CrossRef]
- Langham, D.R.; Wiemers, T. Progress in mechanizing sesame in the US through breeding. In Trends in New Crops and New Uses; Janick, J., Whipkey, A., Eds.; ASHS Press: Alexandria, Egypt, 2002; pp. 157–173. [Google Scholar]
- Khidir, M.O. Natural cross-fertilization in sesame under Sundan conditions. Exp. Agric. 1972, 8, 55–59. [Google Scholar] [CrossRef]
- Bedigian, D. Origin, diversity, exploration and collection of sesame. Sesame: Status and Improvement. In Proceedings of the Expert Consultation, Rome, Italy, 8–12 December 1980; FAO Plant Production and Protection Paper: Rome, Italy, 1981; Volume 29, pp. 164–169. [Google Scholar]
- Seegeler, C.J. Oil Seeds in Ethiopia: Their Taxonomy and Agricultural Significance; Centre for Agricultural Publication and Documentation: Wageningen, The Netherlands, 1983. [Google Scholar]
- Teklu, D.H.; Shimelis, H.; Tesfaye, A.; Mashilo, J.; Zhang, X.; Zhang, Y.; Dossa, K.; Shayanowako, A.I.T. Genetic variability and population structure of Ethiopian sesame (Sesamum indicum L.) germplasm assessed through phenotypic traits and simple sequence repeats markers. Plants 2021, 10, 1129. [Google Scholar] [CrossRef]
- Were, B.A.; Lee, M.; Stymne, S. Variation in seed oil content and fatty acid composition of Sesamum indicum L. and its wild relatives in Kenya. Swed. Seed Assoc. 2001, 4, 178–183. [Google Scholar]
- Dossa, K.; Konteye, M.; Niang, M.; Doumbia, Y.; Cissé, N. Enhancing sesame production in West Africa’s Sahel: A comprehensive insight into the cultivation of this untapped crop in Senegal and Mali. Agric. Food Secur. 2017, 6, 68. [Google Scholar] [CrossRef]
- Nyongesa, B.O.; Were, B.A.; Gudu, S.; Dangasuk, O.G.; Onkware, A.O. Genetic diversity in cultivated sesame (Sesamum indicum L.) and related wild species in East Africa. J. Crop Sci. Biotechnol. 2013, 16, 9–15. [Google Scholar] [CrossRef]
- Woldesenbet, D.T.; Tesfaye, K.; Bekele, E. Genetic diversity of sesame germplasm collection (Sesamum indicum L.): Implication for conservation, improvement and use. Int. J. Biotechnol. Mol. Biol. Res. 2015, 6, 7–18. [Google Scholar]
- Anyanga, W.O.; Hohl, K.H.; Burg, A.; Gaubitzer, S.; Rubaihayo, P.R.; Vollmann, J.; Gibson, P.T.; Fluch, S.; Sehr, E.M. Genetic variability and population structure of global collection of sesame (Sesamum indicum L.) germplasm assessed through phenotypic traits and simple sequence repeats markers for Uganda. J. Agric. Sci. 2017, 9, 13–14. [Google Scholar]
- Tripathy, S.K.; Kar, J.; Sahu, D. Advances in Sesame (Sesamum indicum L.) Breeding. In Advances in Plant Breeding Strategies: Industrial and Food Crops; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer: Cham, Switzerland, 2019; pp. 577–635. [Google Scholar]
- Yol, E.; Uzun, B. Inheritance of indehiscent capsule character, heritability and genetic advance analyses in the segregation generations of dehiscent x indehiscent capsules in sesame. Tarim Bilim. Derg. 2019, 25, 79–85. [Google Scholar] [CrossRef]
- Weiss, E.A. Sesame. Oilseed Crops, 2nd ed.; Blackwell Science: London, UK, 2000. [Google Scholar]
- Gebregergis, Z.; Dereje, A.; Fitwy, I. Assessment of incidence of sesame webworm (Antigastra catalaunalis (Duponchel)) in Western Tigray, North Ethiopia. J. Agric. Ecol. Res. Int. 2016, 9, 1–9. [Google Scholar] [CrossRef]
- Ministry of Agriculture (MoA). Crop extension Package and Manual; MOA: Addis Ababa, Ethiopia, 2018; pp. 126–132. [Google Scholar]
- Thiyagu, K.; Kandasamy, G.; Manivannan, N.; Muralidharan, V.; Manoranjitham, S.K. Identification of resistant genotypes to root rot disease (Macrophomina phaseolina) of sesame (Sesamum indicum L). Agric. Sci. Digest. 2007, 27, 34–37. [Google Scholar]
- Li, D.; Wang, L.; Zhang, Y.; Lv, H.; Qi, X.; Wei, W.; Zhang, X. Pathogenic variation and molecular characterization of Fusarium species isolated from wilted sesame in China. Afr. J. Microbiol. Res. 2012, 6, 149–154. [Google Scholar] [CrossRef]
- Wei, X.; Zhu, X.; Yu, J.; Wang, L.; Zhang, Y.; Li, D. Identification of sesame genomic variations from genome comparison of landrace and variety. Front. Plant Sci. 2016, 7, 1169. [Google Scholar] [CrossRef] [PubMed]
- El-Barougy, E.S. Pathological Studies on Sesame (Sesamum indicum L.) Plant in Egypt. Master’s Thesis, Agriculture and Botany Department, Faculty of Agriculture, Seuz Canal University, Ismailia, Egypt, 1990; p. 145. [Google Scholar]
- El-Shakhess, S.A.M. Inheritance of some economic characters and disease reaction in some sesame (Sesamum indicum L.). Ph. D. Thesis, Agronomy Department, Faculty of Agriculture, Cario University, Cario, Egypt, 1998; p. 135. [Google Scholar]
- Boureima, S.; Oukarroum, A.; Diouf, M.; Cisse, N.; Van Damme, P. Screening for drought tolerance in mutant germplasm of sesame (Sesamum indicum) probing by chlorophyll A inflorescence. Environ. Exp. Bot. 2012, 81, 37–43. [Google Scholar] [CrossRef]
- Kadkhodaie, A.; Zahedi, M.; Razmjoo, J.; Pessarakli, M. Changes in some anti-oxidative enzymes and physiological indices among sesame genotypes (Sesamum indicum L.) in response to soil water deficits under field conditions. Acta Physiol. Plant 2014, 36, 641–650. [Google Scholar] [CrossRef]
- Aye, M.; Khaing, T.T.; Hom, N.H. Morphological characterization and genetic divergence in myanmar sesame (Sesamum indicum L.) germplasm. Int. J. Curr. Adv. Res. 2018, 6, 297–307. [Google Scholar] [CrossRef]
- Yousif, H.Y.; Bingham, F.T.; Yermason, D.M. Growth, mineral composition, and seed oil of sesame (Sesamum indicum L.) as affected by NaCl. Soil Sci. Soc. Am. J. 1972, 36, 450–453. [Google Scholar] [CrossRef]
- Uçan, K.; Kıllı, F.; Gençoglan, C.; Merdun, H. Effect of irrigation frequency and amount on water use efficiency and yield of sesame (Sesamum indicum L.) under field conditions. Field Crops Res. 2007, 101, 249–258. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, X.R.; Zhang, Y.X.; Wang, L.H.; Huang, B. Effects of waterlogging on leaf protective enzyme activities and seed yield of sesame at different growth stages. Chin. J. Appl. Environ. 2009, 15, 790–795. [Google Scholar] [CrossRef]
- Gebretsadik, D.; Haji, J.; Tegegne, B. Sesame post-harvest loss from small-scale producers in Kafta Humera District, Ethiopia. J. Dev. Agric. Econ. 2019, 11, 33–42. [Google Scholar]
- Dossa, K.; Diouf, D.; Cissé, N. Genome-Wide investigation of Hsf genes in Sesame reveals their segmental duplication expansion and their active role in drought stress response. Front. Plant Sci. 2016, 7, 1522. [Google Scholar] [CrossRef]
- Dixit, A.; Jin, M.H.; Chung, J.W.; Yu, J.W.; Chung, H.K.; Ma, K.H. Development of polymorphic microsatellite markers in sesame (Sesamum indicum L.). Mol. Ecol. Resour. 2005, 5, 736–738. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Y.; Qi, X.; Gao, Y.; Zhang, X. Development and characterization of 59 polymorphic cDNA-SSR markers for the edible oil crop Sesamum indicum L. (Pedaliaceae). Am. J. Bot. 2012, 99, 394–398. [Google Scholar] [CrossRef]
- Hodgkin, T.; Qingyuan, G.; Xiurong, Z.; Ying-zhong, Z.; Xiang-yun, F.; Gautam, P.L.; Mahajan, R.; Bisht, I.S.; Loknathan, T.R.; Mathur, P.N.; et al. Developing sesame core collections in China and India. In Core Collections for Today and Tomorrow; International Plant Genetic Resources Institute: Rome, Italy, 1999. [Google Scholar]
- Ministry of Agriculture (MoA). Plant variety release. Protection and seed quality control directorate. In Crop Variety Register; MoA: Addis Ababa, Ethiopia, 2019; Volume 22, p. 330. [Google Scholar]
- Stamatov, S.; Velcheva, N.; Deshev, M. Introduced sesame accessions as donors of useful qualities for breeding of Introduced sesame accessions as donors of useful qualities for breeding of mechanized harvesting cultivars. Bulg. J. Agric. Sci. 2018, 24, 820–824. [Google Scholar]
- Nyanapah, J.O.; Ayiecho, P.O.; Nyabundi, J.O. Evaluation of sesame cultivars for resistance to Cercospora leaf spot. East Afr. Agric. J. 1995, 60, 115–121. [Google Scholar] [CrossRef]
- Ayiecho, P.O.; Nyabundi, J.O. Yield Improvement of Kenyan Sesame Varieties Using Induced Mutations; 1st year report to IAEA; Department of Crop Science, University of Nairobi: Nairobi, Kenya, 1994. [Google Scholar]
- Pandey, S.K.; Das, A.; Rai, P.; Dasgupta, T. Morphological and genetic diversity assessment of sesame (Sesamum indicum L.) accessions differing in origin. Physiol. Mol. Biol. Plants. 2015, 21, 519–529. [Google Scholar] [CrossRef]
- Chellamuthu, M.; Subramanian, S.; Swaminathan, M. Genetic potential and possible improvement of Sesamum indicum L. IntechOpen 2020, 11, 1–18. [Google Scholar] [CrossRef]
- NBPGR. National Bureau of Plant Genetic Resources, India. 2021. Available online: www.nbpgr.ernet.in (accessed on 5 December 2021).
- OCRI. Oil Crops Research Institute, China. 2021. Available online: http://www.sesame-bioinfo.org/phenotype/index.html (accessed on 5 December 2021).
- NACRDA. National Agrobiodiversity Center, Rural Development Administration, South Korea. 2021. Available online: http://www.rda.go.kr/foreign/ten/ (accessed on 5 December 2021).
- USDA-ARS-PGRU. United States Department of Agriculture-Agricultural Research Service-Plant Genetic Resource Unit, USA. 2021. Available online: https://www.ars.usda.gov/ (accessed on 5 December 2021).
- Zhang, Y.; Zhang, X.; Che, Z.; Wang, L.; Wei, W.; Li, D. Genetic diversity assessment of sesame core collection in China by phenotype and molecular markers and extraction of a mini-core collection. BMC Genet. 2012, 13, 102. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Suresh, S.; Raveendar, S.; Baek, H.; Kim, C.; Lee, S. Development and evaluation of core collection using qualitative and quantitative trait descriptor in sesame (Sesamum indicum L.) germplasm. Korean J. 2015, 60, 75–84. [Google Scholar] [CrossRef]
- Wei, X.; Liu, K.; Zhang, Y.; Feng, Q.; Wang, L.; Zhao, Y. Genetic discovery for oil production and quality in sesame. Nat. Commun 2015, 6, 8609. [Google Scholar] [CrossRef]
- Hodgkin, T.; Brown, A.H.D.; Hintum, T.J.L.V.; Morales, E.A.V. Core Collections of Plant Genetic Resources; A co-publication with the International Plant Genetic Resources Institute (IPGRI) and Sayce publishing: London, UK, 1995. [Google Scholar]
- Bisht, I.S.; Mahajan, R.K.; Loknathan, T.R.; Agarwal, R.C. Diversity in Indian sesame collection and stratification of germplasm accessions in different diversity groups. Genet. Resour. Crop Evol. 1998, 45, 325–335. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, Y.; Cheng, Y.; Feng, X.; Guo, Q.; Zhou, M. Establishment of sesame germplasm core collection in China. Genet. Resour. Crop Evol. 2000, 47, 273–279. [Google Scholar] [CrossRef]
- Dossa, K.; Wei, X.; Zhang, Y.; Fonceka, D.; Yang, W.; Diouf, D. Analysis of genetic diversity and population structure of sesame accessions from Africa and Asia as major centers of its cultivation. Genes 2016, 7, 14. [Google Scholar] [CrossRef]
- Lopes, M.S.; El-Basyonim, I.; Baenziger, P.S.; Singh, S.; Royo, C.; Ozbek, K.; Aktas, H.; Ozer, E.; Ozdemir, F.; Manickavelu, A. Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. J. Exp. Bot. 2015, 66, 3477–3486. [Google Scholar] [CrossRef]
- Pícha, K.; Navrátil, J.; Švec, R. Preference to local food vs. Preference to “national” and regional food. J. Food Prod. Mark. 2018, 24, 125–145. [Google Scholar] [CrossRef]
- Sehr, E.M.; Okello-Anyanga, W.; Hasel-Hohl, K.; Burg, A.; Gaubitzer, S.; Rubaihayo, P.R. Assessment of genetic diversity amongst Ugandan sesame (Sesamum indicum L.) landraces based on agro-morphological traits and genetic markers. J. Crop Sci. Biotechnol. 2016, 19, 117–129. [Google Scholar] [CrossRef]
- Ashri, A. Report on FAO/IAEA. Expert consultation on breeding improved sesame cultivars; Hebrew University: Jerusalem, Israel, 1987. [Google Scholar]
- Furat, S.; Uzun, B. The use of agro-morphological characters for the assessment of genetic diversity in sesame (Sesamum indicum L.). Plant Omics 2010, 3, 85–91. [Google Scholar]
- Akbar, F.; Rabbani, M.A.; Shinwari, Z.K.; Khan, S.J. Genetic divergence in sesame (Sesamum Indicum L.) landraces based on qualitative and quantitative traits. Pak. J. Bot. 2011, 43, 2737–2744. [Google Scholar]
- Gidey, Y.T.; Kebede, S.A.; Gashawbeza, G.T. Extent and pattern of the genetic diversity for morpho-agronomic traits in Ethiopian sesame landraces (Sesamum indicum L.). Asian J. Agric. Res. 2012, 6, 118–128. [Google Scholar] [CrossRef][Green Version]
- Teklu, D.H.; Kebede, S.A.; Gebremichael, D.E. Assessment of genetic variability, genetic advance, correlation, and path analysis for morphological traits in sesame genotypes. Asian J. Agric. Res. 2014, 7, 118–128. [Google Scholar] [CrossRef]
- Hika, G.; Geleta, N.; Jaleta, Z. Correlation and divergence analysis for phenotypic traits in sesame (Sesamum indicum L.) Genotypes. Sci. Technol. Arts Res. J. 2014, 3, 1–9. [Google Scholar] [CrossRef]
- Hika, G.; Geleta, N.; Jaleta, Z. Genetic variability, heritability and genetic advance for the phenotypic sesame (Sesamum indicum L.) Populations from Ethiopia. Sci. Technol. Arts Res. J. 2015, 4, 20–26. [Google Scholar] [CrossRef]
- Abdou, R.I.Y.; Moutari, A.; Ali, B.; Basso, Y.; Djibo, M. Variability study in sesame (Sesamum indicum L.) cultivars based on agro-morphological characters. Int. J. Agric. Fish. 2015, 6, 237–242. [Google Scholar]
- Harfi, M.E.; Jbilou, M.; Hanine, H.; Rizki, H.; Fechtali, M.; Nabloussi, A. Genetic Diversity Assessment of Moroccan Sesame (Sesamum indicum L.) Populations Using Agro-morphological Traits. J. Agric. Sci. Technol. 2018, 8, 296–305. [Google Scholar] [CrossRef]
- Teklu, D.H.; Shimelis, H.; Tesfaye, A.; Mashilo, J. Genetic diversity and association of yield-related traits in sesame. Plant Breed. 2021, 40, 331–341. [Google Scholar] [CrossRef]
- Johnson, M.W.; Robinson, H.F.; Comstock, R.E. Genotypic and phenotypic correlations in soybeans and their implication in selection. Agron. J. 1955, 47, 477–483. [Google Scholar] [CrossRef]
- Panse, V.G. Genetics of quantitative characters in relation to plant breeding. Indian J. Genet. Plant Br. 1957, 17, 318–346. [Google Scholar]
- Divya, K.; Rani, T.S.; Babu, T.K.; Padmaja, D. Assessment of genetic variability, heritability and genetic gain in advanced mutant breeding lines of sesame (Sesamum indicum L.). Int. J. Curr. Microbiol. Ap. 2018, 71, 565–1574. [Google Scholar] [CrossRef]
- Aye, M.; Htwe, N.M. Trait association and path coefficient analysis for yield traits in myanmar sesame (Sesamum indicum L.). Germplasm. J. Exp. Agric. Int. 2019, 4, 1–10. [Google Scholar] [CrossRef]
- Abraha, M.; Shimelis, H.; Laing, M.; Assefa, K. Performance of tef [Eragrostis tef (Zucc.) Trotter] genotypes for yield and yield components under drought-stressed and non-stressed conditions. Crop Sci. 2016, 56, 1799–1806. [Google Scholar] [CrossRef]
- Teklu, D.H.; Shimelis, H.; Tesfaye, A.; Shayanowako, A.I.T. Analyses of genetic diversity and population structure of sesame (Sesamum indicum L.) germplasm collections through seed oil and fatty acid compositions and SSR markers. J. Food Compos. Anal. 2022, 110, 104545. [Google Scholar] [CrossRef]
- Maluszynski, M.; Ahloowalia, B.S.; Sigurbjornsson, B. Application of in vivo and in vitro mutation techniques for crop improvement. Euphytica 1995, 85, 303–315. [Google Scholar] [CrossRef]
- Muduli, K.C.; Mishra, R.C. Efficacy of mutagenic treatments in producing useful mutants in finger millet (Eleusine coracana Gaertn.). Indian J. Genet. Plant Breed. 2007, 67, 232–237. [Google Scholar]
- Ashri, A. Induced Mutations in Sesame Breeding. Proceedings of 2nd FAO/IAEA, Co-Ordinated Research Project Organized by the Joint FAO/IAEA Division of Nuclear Techniques in Food and Agriculture. Sesame Improvement by Induced Mutations. IAEA, Vienna, Austria, 1997, 13–20. Available online: https://inis.iaea.org/collection/NCLCollectionStore/_Public/32/007/32007909.pdf (accessed on 9 August 2022).
- Mutant Variety Database (MVD). The Joint FAO/IAEA Mutant Variety Database. 2022. Available online: https://mvd.iaea.org/ (accessed on 23 March 2022).
- Lee, J.I.; Choi, B.H. Progress and prospects of sesame breeding in Korea. In Sesame and Safflower: Status and Potential; Ashri, A., Ed.; FAO Plant Production and Protection Paper 66: Rome, Italy, 1985; pp. 137–144. [Google Scholar]
- Kang, C.W. Breeding for diseases and shatter resistant high yielding varieties using induced mutations in sesame. In Proceedings of the 2nd FAO/IAEA Res. Coord. Mtg, Induced Mutations for Sesame Improvement; IAEA: Vienna, Austria, 1997; pp. 48–57. [Google Scholar]
- Pathirana, R. Gamma ray-induced field tolerance to Phytophthora blight in sesame. Plant Breed. 1992, 108, 314–319. [Google Scholar] [CrossRef]
- Khan, A.W.; Garg, V.; Roorkiwal, M.; Golicz, A.A.; Edwards, D.; Varshney, R.K. Super-Pangenome by integrating the wild side of a species for accelerated crop improvement. Trends Plant Sci. 2020, 25, 148–158. [Google Scholar] [CrossRef]
- Yu, J.; Golicz, A.A.; Lu, K.; Dossa, K.; Zhang, Y.; Chen, J.; Wang, L.; You, J.; Fan, D.; Edwards, D.; et al. Insight into the evolution and functional characteristics of the pan-genome assembly from sesame landraces and modern cultivars. Plant Biotechnol. J. 2019, 17, 881–892. [Google Scholar] [CrossRef]
- Wang, L.; Yu, S.; Tong, C.; Zhao, Y.; Liu, Y.; Song, C. Genome sequencing of the high oil crop sesame. Genome Biol. 2014, 15, R39. [Google Scholar] [CrossRef]
- Wei, X.; Gong, H.; Yu, J.; Liu, P.; Wang, L.; Zhang, Y. Sesame FG: An integrated database for the functional genomics of sesame. Sci. Rep. 2017, 7, 2342. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, L.; Xin, H.; Li, D.; Ma, C.; Ding, X.; Hong, W.; Zhang, X. Construction of a high-density genetic map for sesame based on large scale marker development by specific length amplified fragment (SLAF) sequencing. BMC Plant Biol. 2013, 13, 141. [Google Scholar] [CrossRef] [PubMed]
- Ke, T.; Yu, J.; Dong, C.; Mao, H.; Hua, W.; Liu, S. OcsESTdb: A database of oil crop seed EST sequences for comparative analysis and investigation of a global metabolic network and oil accumulation metabolism. BMC Plant Biol. 2015, 15, 19. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Ke, T.; Tehrim, S.; Sun, F.; Liao, B.; Hua, W. PTGBase: An integrated database to study tandem duplicated genes in plants. Database 2015, 2015, bav017. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Dossa, K.; Wang, L.; Zhang, Y.; Wei, X.; Liao, B. PMDBase: A database for studying microsatellite DNA and marker development in plants. Nucleic Acids Res. 2016, 45, D1046–D1053. [Google Scholar] [CrossRef] [PubMed]
- Jones, N.; Ougham, H.; Thomas, H.; Pasakinskiene, I. Markers and mapping revisited: Finding your gene. New Phytol. 2009, 183, 935–966. [Google Scholar] [CrossRef]
- Thomson, M.J.; Septiningsih, E.M.; Suwardjo, F.; Santoso, T.J.; Silitonga, T.S.; McCouch, S.R. Genetic diversity analysis of traditional and improved Indonesian rice (Oryza sativa L.) germplasm using microsatellite markers. Theor. Appl. Genet. 2007, 114, 559–568. [Google Scholar] [CrossRef]
- Laurentin, E.H.; Karlovsky, P. Genetic relationship and diversity in a sesame (Sesamum indicum L.) germplasm collection using amplified fragment length polymorphism (AFLP). BMC Genet. 2006, 7, 10. [Google Scholar] [CrossRef]
- Laurentin, H.; Karlovsky, P. AFLP fingerprinting of sesame (Sesamum indicum L.) cultivars: Identification, genetic relationship and comparison of AFLP informativeness parameters. Genet. Resour. Crop Evol. 2007, 54, 1437–1446. [Google Scholar] [CrossRef]
- Bhat, K.V.; Babrekar, P.P.; Lakhanpaul, S. Study of genetic diversity in Indian and exotic sesame (Sesamum indicum L.) germplasm using random amplified polymorphic DNA (RAPD) markers. Euphytica 1999, 110, 21–33. [Google Scholar] [CrossRef]
- Ercan, A.G.; Taskin, M.; Turgut, K. Analysis of genetic diversity in Turkish sesame (Sesamum indicum L.) populations using RAPD markers. Genet. Resour. Crop Evol. 2004, 51, 599–607. [Google Scholar] [CrossRef]
- Kim, D.H.; Zur, G.; Danin-Poleg, Y.; Lee, S.W.; Shim, K.B.; Kang, C.W.; Kashi, Y. Genetic relationships of sesame germplasm collection as revealed by inter-simple sequence repeats. Plant Breed. 2002, 121, 259–262. [Google Scholar] [CrossRef]
- Gebremichael, D.E.; Parzies, H.K. Genetic variability among landraces of sesame in Ethiopia. Afr. Crop Sci. J. 2011, 19, 1–13. [Google Scholar] [CrossRef][Green Version]
- Wei, X.; Wang, L.; Zhang, Y.; Qi, X.; Wang, X.; Ding, X.; Zhang, J.; Zhang, X. Development of Simple Sequence Repeat (SSR) Markers of Sesame (Sesamum indicum L.) from a Genome Survey. Molecules 2014, 19, 5150–5162. [Google Scholar] [CrossRef] [PubMed]
- Asekova, S.; Kulkarni, K.P.; Oh, K.W.; Lee, M.H.; Oh, E.; Kim, J.I.; Yeo, U.; Pae, U.S.; Ha, T.J.; Kim, S.U. Analysis of molecular variance and population structure of sesame (Sesamum indicum L.) genotypes using SSR markers. Plant Breed. Biotechnol. 2018, 6, 321–336. [Google Scholar] [CrossRef]
- Araújo, E.D.S.; Arriel, N.H.C.; Santos, R.C.D.; Lima, L.M.D. Assessment of genetic variability in sesame accessions using SSR markers and morpho agronomic traits. Aust. J. Crop Sci. 2019, 13, 45–54. [Google Scholar] [CrossRef]
- Basak, M.; Uzun, B.; Yol, E. Genetic diversity and population structure of the Mediterranean sesame core collection with use of genome-wide SNPs developed by double digest RAD-Seq. PLoS ONE 2019, 14, e0223757. [Google Scholar] [CrossRef]
- Tesfaye, T.; Tesfaye, K.; Keneni, G.; Ziyomo, C.; Alemu, T. Genetic diversity of Sesame (Sesamum indicum L.) using high throughput diversity array technology. J. Crop Sci. Biotechnol. 2022, 25, 359–371. [Google Scholar] [CrossRef]
- Wei, W.; Qi, X.; Wang, L.; Zhang, Y.; Hua, W.; Li, D.; Lv, H.; Zhang, X. Characterization of the sesame (Sesamum indicum L.) global transcriptome using Illumina paired-end sequencing and development of EST-SSR markers. BMC Genom. 2011, 19, 12–451. [Google Scholar] [CrossRef]
- Frary, A.; Tekin, P.; Celik, I.; Furat, S.; Uzun, B.; Doganlar, S. Morphological and molecular diversity in Turkish sesame germplasm and selection of a core set for inclusion in the national collection. Crop Sci. 2014, 54, 702–711. [Google Scholar] [CrossRef]
- Pham, T.D.; Geleta, M.; Bui, T.M.; Bui, T.C.; Merker, A.; Carlsson, A.S. Comparative analysis of genetic diversity of sesame (Sesamum indicum L.) from Vietnam and Cambodia using agro-morphological and molecular markers. Hereditas 2011, 148, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Tabatabaei, I.; Pazouki, L.; Bihamta, M.R.; Mansoori, S.; Javaran, M.J.; Niinemets, Ü. Genetic variation among Iranian sesame (Sesamum indicum L.) accessions vis-à-vis exotic genotypes on the basis of morpho-physiological traits and RAPD markers. Aust. J. Crop Sci. 2011, 11, 1396–1407. [Google Scholar]
- Uncu, A.O.; Gultekin, V.; Allmer, J.; Frary, A.; Doganlar, S. Genomic simple sequence repeat markers reveal patterns of genetic relatedness and diversity in sesame. Plant Genome. 2015, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Joshi, A.B. Sesamum; Indian Central Oilseed Committee: Hyderabad, India, 1961. [Google Scholar]
- Tiwari, S.; Kumar, S.; Gontia, I. Biotechnological approaches for sesame (Sesamum indicum L.) and Niger (Guizotia abyssinica L.f. Cass.). Asia Pac. J. Mol. Biol. Biotechnol. 2011, 19, 2–9. [Google Scholar]
- Zhang, X.; Zhang, K.; Wu, J.; Guo, N.; Liang, J.; Wang, X. QTL-Seq and sequence assembly rapidly mapped the gene BrMYBL2.1 for the purple trait in Brassica rapa. Sci. Rep. 2020, 10, 2328. [Google Scholar] [CrossRef] [PubMed]
- Tanksley, S.D.; Ganal, M.W.; Prince, J.P.; de Vicente, M.C.; Bonierbale, M.W.; Broun, P. High density molecular linkage maps of the tomato and potato genomes. Genetics 1992, 132, 1141–1160. [Google Scholar] [CrossRef]
- Wang, L.; Xia, Q.; Zhang, Y.; Zhu, X.; Zhu, X.; Li, D.; Ni, X.; Gao, Y.; Xiang, H.; Wei, X.; et al. Updated sesame genome assembly and fine mapping of plant height and seed coat color QTLs using a new high-density genetic map. BMC Genom. 2016, 17, 31. [Google Scholar] [CrossRef]
- Mei, H.; Liu, Y.; Du, Z.; Wu, K.; Cui, C.; Jiang, X. High-density Genetic map construction and gene Mapping of basal branching habit and flowers per leaf axil in Sesame plant materials and trait investigation. Front. Plant Sci. 2017, 8, 636. [Google Scholar] [CrossRef]
- Wang, L.; Li, D.; Zhang, Y.; Gao, Y.; Yu, J.; Wei, X.; Zhang, X. Tolerant and susceptible sesame genotypes reveal waterlogging stress response patterns. PLoS ONE 2016, 11, e0149912. [Google Scholar] [CrossRef]
- Wu, K.; Liu, H.; Yang, M.; Tao, Y.; Ma, H.; Wu, W.; Zuo, Y.; Zhao, Y. High-density genetic map construction and QTLs analysis of grain yield-related traits in Sesame (Sesamum indicum L.) based on RAD-Seq technology. BMC Plant Biol. 2014, 14, 1–14. [Google Scholar] [CrossRef]
- Zhang, H.; Miao, H.; Li, C.; Wei, L.; Duan, Y.; Ma, Q.; Kong, J.; Xu, F.; Chang, S. Ultra-dense SNP genetic map construction and identification of SiDt gene controlling the determinate growth habit in Sesamum indicum L. Sci. Rep. 2016, 6, 31556. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yang, M.; Wu, K.; Liu, H.; Wu, J.; Liu, K. Characterization and genetic mapping of a novel recessive genic male sterile gene in sesame (Sesamum indicum L.). Mol. Breed. 2013, 32, 901–908. [Google Scholar] [CrossRef]
- Li, C.; Miao, H.; Wei, L.; Zhang, T.; Han, X.; Zhang, H. Association Mapping of Seed Oil and Protein Content in Sesamum indicum L. Using SSR Markers. PLoS ONE 2014, 9, e105757. [Google Scholar] [CrossRef] [PubMed]
- Yan-xin, Z.; Lin-hai, W.; Dong-hua, L.I.; Yuan, G.A.O.; Hai-xia, L.Ü.; Xiu-rong, Z. Mapping of Sesame Waterlogging Tolerance QTL and Identification of Excellent Waterlogging Tolerant Germplasm. Sci. Agric. Sin. 2014, 47, 422–430. [Google Scholar]
- Dossa, K.; Niang, M.; Assogbadjo, A.E.; Cisse, N.; Diouf, D. Whole genome homology-based identification of candidate genes for drought tolerance in sesame (Sesamum indicum L.). Afr. J. Biotechnol. 2016, 15, 1464–1475. [Google Scholar]
- Dossa, K.; Diouf, D.; Wang, L.; Wei, X.; Zhang, Y.; Niang, M.; Fonceka, D.; Yu, J.; Mmadi, M.A.; Yehouessi, L.W.; et al. The Emerging Oilseed Crop Sesamum indicum Enters the “Omics” Era. Front. Plant Sci. 2017, 8, 1154. [Google Scholar] [CrossRef]
- Chowdhury, S.; Basu, A.; Kundu, S. Overexpression of a new osmotinlike protein gene (SindOLP) confers tolerance against biotic and abiotic stresses in sesame. Front. Plant Sci. 2017, 8, 410. [Google Scholar] [CrossRef]
- Davey, J.W.; Hohenlohe, P.A.; Etter, P.D.; Boone, J.Q.; Catchen, J.M.; Blaxter, M.L. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 2011, 17, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Ganal, M.W.; Durstewitz, G.; Polley, A.; Berard, A.; Buckler, E.S.; Charcosset, A.; Clarke, J.D.; Graner, E.M.; Hansen, M.; Joets, J.; et al. A large maize (Zea mays L.) SNP genotyping array: Development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS ONE 2011, 6, e28334. [Google Scholar] [CrossRef]
- Sim, S.C.; Durstewitzm, G.; Plieske, J.; Wieseke, R.; Ganal, M.W.; Deynze, A.V.; Hamilton, J.P.; Buell, C.R.; Causse, M.; Wijeratne, S.; et al. Development of a large SNP genotyping array and generation of high-density genetic maps in tomato. PLoS ONE 2012, 7, e40563. [Google Scholar]
- Wang, S.; Chen, J.; Zhang, W.; Hu, Y.; Chang, L.; Fang, L.; Wang, Q.; Lv, F.; Wu, H.; Si, Z.; et al. Sequence-based ultra-dense genetic and physical maps reveal structural variations of allopolyploid cotton genomes. Genome Biol. 2015, 16, 108. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Han, Y.; Li, Y.; Liu, D.; Sun, M.; Zhao, Y.; Lv, C.; Li, D.; Yang, Z.; Huang, L.; et al. Loci and candidate gene identification for resistance to Sclerotinia sclerotiorum in soybean (Glycine max L. Merr.) via association and linkage maps. Plant J. 2015, 82, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Miao, H.; Wei, L.; Li, C.; Zhao, R.; Wang, C. Genetic analysis and QTL mapping of seed coat color in sesame (Sesamum indicum L.). PLoS ONE 2013, 8, e63898. [Google Scholar]
- Miao, H. The genome of Sesamum indicum L. In Proceedings of the Plant and Animal Genome XXII Conference. Plant and Animal Genome, San Diego, CA, USA, 7–13 January 2016. [Google Scholar]
- Zhang, H.; Miao, H.; Wei, L.; Li, C.; Duan, Y.; Xu, F.; Qu, W.; Zhao, R.; Ju, M.; Chang, S. Identification of a SiCL1 gene controlling leaf curling and capsule indehiscence in sesame via cross-population association mapping and genomic variants screening. BMC Plant Biol. 2018, 18, 296. [Google Scholar] [CrossRef] [PubMed]
- Yol, E.; Uzun, B. Geographical patterns of sesame (Sesamum indicum L.) accessions grown under Mediterranean environmental conditions, and establishment of a core collection. Crop Sci. 2012, 52, 2206–2214. [Google Scholar] [CrossRef]
- Yadav, M.; Sainger, D.C.M.; Jaiwal, P.K. Agrobacterium tumefaciensmediated genetic transformation of sesame (Sesamum indicum L.). Plant Cell Tissue Organ Cult. 2010, 103, 377–386. [Google Scholar] [CrossRef]
- Al-Shafeay, A.F.; Ibrahim, A.S.; Nesiem, M.R.; Tawfik, M.S. Establishment of regeneration and transformation system in Egyptian sesame (Sesamum indicum L.) cv Sohag1. GM Crops. 2011, 2, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Basu, A.; Kundu, S. A new high-frequency Agrobacterium-mediated transformation technique for Sesamum indicum L. using de-embryonated cotyledon as explant. Protoplasma 2014, 251, 1175–1190. [Google Scholar] [CrossRef]
- David, V.; Řepková, J. Application of next-generation sequencing in plant breeding. Czech J. Genet. Plant Breed. 2017, 53, 89–96. [Google Scholar] [CrossRef]
- Zhu, H.; Li, C.; Gao, C. Applications of CRISPR–Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 2020, 21, 661–677. [Google Scholar] [CrossRef]
- Zhang, K.; Nie, L.; Cheng, Q.; Yin, Y.; Chen, K.; Qi, F.; Zou, D.; Liu, H.; Zhao, W.; Wang, B. Effective editing for lysophosphatidic acid acyltransferase 2/5 in allotetraploid rapeseed (Brassica napus L.) using CRISPR-Cas9 system. Biotechnol. Biofuels. 2019, 12, 225. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhang, K.; Tang, M.; Zhou, W.; Chen, L.; Chen, Z.; Li, m. CRISPR-based genome editing technology and its applications in oil crops. Oil Crop Sci. 2021, 6, 105–113. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Y.; Qi, X.; Li, D.; Wei, W.; Zhang, X. Global gene expression responses to waterlogging in roots of sesame (Sesamum indicum L.). Acta Physiol. Plant. 2012, 34, 2241–2249. [Google Scholar] [CrossRef]
- Ashri, A. Sesame (Sesamum indium L.). In Genetic Resources, Chromosome Engineering, and Crop Improvement; Singh, R.J., Ed.; CRC Press: Boca Raton, FL, USA, 2007; Volume 4, pp. 231–289. [Google Scholar]
- Dossa, K.; Wei, X.; Li, D.; Zhang, Y.; Wang, L.; Fonceka, D.; Zhang, Y.; Wang, L.; Yu, J.; Boshuo, L.; et al. Insights into the AP2/ERF transcription factor superfamily in sesame (Sesamum indicum) and expression profiling of the DREB subfamily under drought stress. BMC Plant Biol. 2016, 16, 171. [Google Scholar] [CrossRef]
- Dossa, K.; Li, D.; Wang, L.; Zheng, X.; Yu, J.; Wei, X.; Fonceka, D.; Zhang, Y.; Wang, L.; Yu, J.; et al. Dynamic transcriptome landscape of sesame (Sesamum indicum L.) under progressive drought and after rewatering. Genomic Data 2017, 11, 122–124. [Google Scholar] [CrossRef]
- Simoglou, K.B.; Anastasiades, A.I.; Baixeras, J.; Roditakis, E. First report of Antigastra catalaunalis on sesame in Greece. Entomol. Hell. 2017, 26, 6–12. [Google Scholar] [CrossRef]
- Ramanujam, S. The cytogenetics of an amphidiploid Sesamum orientale and S. prostratum. Curr. Sci. 1944, 13, 40–41. [Google Scholar]
- Rao, G.P.; Un, N.S.; MadhuPriya. Overview on a century progress in research on sesame phyllody disease. Phytopathogenic Mollicutes 2015, 5, 74–83. [Google Scholar] [CrossRef]
- Rao, G.P.; Kumar, A.; Baranwal, V.K. Classification of sesame phytoplasma strain in India at 16Sr subgroup level. J. Plant Pathol. 2015, 3, 523–528. [Google Scholar] [CrossRef]
- Win, N.K.K.; Back, C.; Jung, H.Y. Phyllody phytoplasma infecting sesame in Myanmar belongs to group 16SrI and subgroup 16SrI-B. Tropical. Plant Pathol. 2010, 35, 310–313. [Google Scholar] [CrossRef]
- Tseng, Y.W.; Deng, W.L.; Chang, C.J.; Huang, J.W.; Jan, F.J. First report on the association of a 16SrII-A phytoplasma with sesame (Sesamum indicum) exhibiting abnormal stem curling and phyllody in Taiwan. Plant Dis. 2014, 98, 990. [Google Scholar] [CrossRef] [PubMed]
- Catal, M.; Ikten, C.; Yol, E.; Ustun, R.; Uzun, B. First report of a 16SrIX group (pigeon pea witches’-broom) phytoplasma associated with sesame phyllody in Turkey. Plant Dis. 2013, 97, 835. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Akram, M.; Vajpeyi, M.; Srivastava, R.L.; Kumar, K.; Naresh, R. Screening and development of resistant sesame varieties against phytoplasma. Bull. Insectol. 2007, 60, 303–304. [Google Scholar]
- Ahirwar, R.M.; Banerjee, S.; Gupta, M.P. Insect pest management in sesame crop by intercropping system. Ann. Plant Sci. 2009, 17, 225–226. [Google Scholar]
- Altieri, M.A.; Koohafkan, P. Enduring Farms: Climate Change, Smallholders and Traditional Farming Communities. In Environment and Development Series 6; Third World Network: Pulau Pinang, Malaysia, 2008. [Google Scholar]
- Chambers, R. Rapid Appraisal: Rapid, Relaxed and Participatory IDS Discussion Paper; Institute of Development Studies: Brighton, UK, 1992; Volume 311, p. 90. [Google Scholar]
| Region/Continent | Area (× 103 ha) | Production (× 103 tons) | % of World Production | Average Yield (tons·ha−1) |
|---|---|---|---|---|
| Africa | 9692.17 | 4282.99 | 59.05 | 0.54 |
| Asia | 4064.40 | 2645.23 | 36.47 | 1.13 |
| Latin America | 462.37 | 306.37 | 4.22 | 0.71 |
| Europe | 25.56 | 18.69 | 0.26 | 0.88 |
| Australia | NA | NA | NA | NA |
| World | 14,244.50 | 7253.28 | NA | 0.79 |
| Country | Area (× 103 ha) | Production (× 103 tons) | % of World Production | Yield (tons·ha−1) |
|---|---|---|---|---|
| Sudan | 5173.52 | 1525.10 | 21.02 | 0.29 |
| China | 554.97 | 896.63 | 12.36 | 1.62 |
| Myanmar | 1500.00 | 740.00 | 10.20 | 0.49 |
| The United Republic of Tanzania | 960.00 | 710.00 | 9.78 | 0.74 |
| India | 1520.00 | 658.00 | 9.07 | 0.43 |
| Nigerian | 621.41 | 490.00 | 6.76 | 0.79 |
| Burkina Faso | 450.00 | 270.00 | 3.72 | 0.60 |
| Ethiopia | 369.90 | 260.26 | 3.59 | 0.70 |
| Chad | 392.24 | 202.07 | 2.79 | 0.52 |
| South Sudan | 608.16 | 189.72 | 2.61 | 0.31 |
| World | 14,244.50 | 7253.28 | NA | 0.79 |
| Variety | Pedigree | Trait | Country | Year of Release | References |
|---|---|---|---|---|---|
| Sin-Yadana 4 | - | Good export quality | China | 1994 | [12] |
| Ju-Ni-Poke | - | Stable yield and high oil content | Myanmar | 1994 | [12] |
| Me-Daw-Let-The | - | Stable yield and high oil content | 1994 | ||
| Gwa-Taya | - | Stable yield | 1994 | ||
| Gwa-Kyaw-Net | - | Stable yield | 1994 | ||
| Humera-1 | ACC.038 sel.1 | Early maturity, better yield and broad adaptability | Ethiopia | 2010 | [17] |
| Setit-1 | Col sel p#1 | Early maturity, better yield and oil content and broad adaptability | 2010 | ||
| Dangur | E.W.013.(8) | High oil content | Ethiopia | 2015 | [49] |
| BaHaNecho | W-109/WSS/ (Acc-EW-012(5) | Better yield and oil content | 2016 | ||
| BaHaZeyit | W- 119/WSM/ (Acc-EW-023(1) | Better yield and oil content | 2016 | ||
| Setit-2 | J-03 | Early maturity, better yield, and broad adaptability | 2016 | ||
| Setit-3 | HuARC-4 | Early maturity, better yield and oil content, and broad adaptability | 2017 | ||
| Waliin | BG-004-1 | Better yield and oil content | 2017 | ||
| Gida Ayana | Ass-acc-29 | Late maturity, better yield and oil content, and broad adaptability | 2018 | ||
| Hagalo | EW002 × Obsa22-1 | Late maturity, better yield, resistance to bacterial blight, and broad adaptability | 2019 | ||
| Yale | EW002 × Dicho 5-3 | Late maturity, better yield and oil content, resistance to bacterial blight, and broad adaptability | 2019 | ||
| RAMA | ‘Khosla’ local | Medium seed size and brown seed color | India | 1989 | [53] |
| OSC-593 | - | White seed color | 1995 | ||
| TKG-352 | - | White seed color | 1995 | ||
| TMV 1 | - | Erect, fairly bushy with moderate branching, 4-loculed, red brown to black seeds, and better oil content | 1939 | [54] | |
| TMV 2 | Nagpur white × Sattur | Open, moderate branching, 6–8-loculed, cylindrical big sized capsules, and dark brown to black seeds Suitable for cold weather conditions and better oil content | 1942 | ||
| TMV 3 | South Arcot variety × Malabar Variety | Bushy with profuse branching, 4-loculed, dark brown to black seeds, and better oil content | 1943 | ||
| KRR 1 | - | Bushy with profuse branching, 4-loculed, brown seeds, and better oil content | 1967 | ||
| KRR 2 | Karur local × Bombay white | Bushy with profuse branching, 4-loculed, better oil content, and white seeds | 1970 | ||
| TMV 4 | - | Bushy with profuse branching, 4-loculed, brown seeds, and better oil content | 1977 | ||
| TMV 5 | - | Erect with moderate branching, 4-loculed, brown seeds, and better oil content | 1978 | ||
| TMV 6 | - | Erect with moderate branching, 4-loculed, brown seeds, and better oil content | 1980 | ||
| CO 1 | (TMV 3 × Si 1878) × Si 1878 | Bushy plant, 4-loculed, black warty seeds, and better oil content | 1983 | ||
| Paiyur 1 | Si2511 × Si 2314 | Resistance to powdery mildew, 4-loculed, bushy, suitable for irrigated condition, black seeds, and better oil content | 1990 | ||
| SVPR 1 | - | White seeds, 4-loculed, high yield, suitable for irrigated conditions, and better oil content | 1992 | ||
| VRI 1 | - | Early maturity, 4-loculed, and better oil content | 1995 | ||
| VRISV2 | US9003 × TMV6 | Moderate resistance to shoot webber, 4-loculed, and higher oil content | 2005 | ||
| TMV (Sv) 7 | High yield, 4-loculed, tolerance to root rot disease, lustrous brown testa, and higher oil content | 2009 | |||
| VRI 3 | SVPR 1 × TKG 87 | Moderate resistance to phyllody and root rot diseases, white seed, and higher oil content | 2017 |
| Country | Institution | Total Number of Accessions | Website | Reference |
|---|---|---|---|---|
| India | National Bureau of Plant Genetic Resources | 10,359 | www.nbpgr.ernet.in (accessed on 5 December 2021) | [55] |
| China | Oil Crops Research Institute | >8000 | http://www.sesame-bioinfo.org/phenotype/index.html (accessed on 5 December 2021) | [56] |
| South Korea | National Agrobiodiversity Center, Rural Development Administration | 7698 | http://www.rda.go.kr/foreign/ten/ (accessed on 5 December 2021) | [57] |
| United States of America | USDA-ARS-PGRU | 1226 | www.ars.usda.gov (accessed on 5 December 2021) | [58] |
| Variety Name | Trait | Country | Year of Release | Reference |
|---|---|---|---|---|
| NIAB-Pearl | Higher capsules per plant | Pakistan | 2017 | [88] |
| NIAB-Sesame 2016 | High oil content | 2016 | ||
| Binatil-3 | High yield | Bangladesh | 2013 | [88] |
| Cairo white 8 | Nonbranching | Egypt | 1992 | [87] |
| Senai white 48 | Seed color | 1992 | ||
| Kalika | Short stature | India | 1980 | [87] |
| UMA | Uniform maturity | 1990 | ||
| USHA | Higher yield | 1990 | ||
| Babil | Earliness | Iraq | 1992 | [87] |
| Rafiden | Earliness | 1992 | ||
| Eshtar | Capsule size | 1992 | ||
| Ahnsan | Disease resistance | South Korea | 1985 | [87] |
| Suweon | Lodging and disease resistance | 1991 | ||
| Yangbaek | Higher oil content | 1995 | ||
| Pungsan | Determinate growth habit and high seed retention | 1996 | ||
| Seodun | Higher oleic acid content and phytophthora blight tolerance | 1997 | ||
| ANK-2 | Disease resistance | Sri Lanka | 1995 | [87] |
| Database | Website | Utility | Reference |
|---|---|---|---|
| Sinbase | http://www.ocri-genomics.org/Sinbase/index.html. (accessed on 9 August 2022) | Genomics, comparative genomics, genetics, phenotypes, etc. | [94] |
| SesameHapMap | http://202.127.18.228/SesameHapMap/ (accessed on 9 August 2022) | Genome-wide SNP | [61] |
| SesameFG | http://www.ncgr.ac.cn/SesameFG (accessed on 9 August 2022) | Genomics, evolution, breeding, comparative genomics, molecular markers, phenotypes, transcriptomics | [95] |
| SisatBase | http://www.sesame-bioinfo.org/SisatBase/ (accessed on 9 August 2022) | Genome-wide SSR | - |
| The Sesame Genome Project | http://www.sesamegenome.org (accessed on 9 August 2022) | Genomics | [96] |
| Sesame Germplasm Resource Information Database | http://www.sesame-bioinfo.org/phenotype/index.html (accessed on 9 August 2022) | Plant phenotype | - |
| NCBI∗ | http://www.ncbi.nlm.nih.gov/genome/?term=sesame (accessed on 9 August 2022) | Versatile | - |
| ocsESTdb∗ | http://www.ocri-genomics.org/ocsESTdb/index.html (accessed on 9 August 2022) | Seed expression sequence tags, comparative genomics | [97] |
| PTGBase∗ | http://www.ocri-genomics.org/PTGBase/index.html (accessed on 9 August 2022) | Tandem duplication, evolution | [98] |
| PMDBase∗ | http://www.sesame-bioinfo.org/PMDBase (accessed on 9 August 2022) | SSR information | [99] |
| Marker Sequence | |||
|---|---|---|---|
| Primers | Forward Primer Sequence | Reverse Primer Sequence | References |
| GBssr-sa-05 | TCATATATAAAAGGAGCCCAAC | GTCATCGCTTCTCTCTTCTTC | [46] |
| GBssr-sa-08 | GGAGAAATTTTCAGAGAGAAAAA | ATTGCTCTGCCTACAAATAAAA | |
| Sesame-09 | CCCAACTCTTCGTCTATCTC | TAGAGGTAATTGTGGGGGA | |
| GBssr-sa-33 | TTTTCCTGAATGGCATAGTT | GCCCAATTTGTCTATCTCCT | |
| GBssr-sa-123 | GCAAACACATGCATCCCT | GCCCTGATGATAAAGCCA | |
| GBssr-sa-182 | CCATTGAAAACTGCACACAA | TCCACACACAGAGAGCCC | |
| GBssr-sa-184 | TCTTGCAATGGGGATCAG | CGAACTATAGATAATCACTTGGAA | |
| SSR-ES-12 | GCTGAGGAGTCTTGAAGCAGA | CAAAATCCCCCAACTCGATA | [53] |
| SSR-ES-15 | TGCAGGAATGAACTCAAGGA | ACCTTATTCCCAGCCCACTT | |
| ZM_2 | CTTCTTGAAGTTCTGGTGTTG | ATTCTTGGAGAAAGAGTGAGG | [111] |
| ZM_3 | ATCACCACACACTGACACAG | CGTGTCTGAGAATCCAATATC | |
| ZM_6 | GGTGTGTTCTCTCTCTCACAC | GGGCTGCTCAATAAATGTAG | |
| ZM_7 | ATCCTCTGCTCCTAACTTCAT | TCTGGTACTATCCTCAAGCAA | |
| ZM_10 | ATGCCCATCTCCATATACTCT | AATTCTTGCCTGACTCTACG | |
| ZM_11 | GGATTCTCTAGACATGGCTTT | AACGCAGAATTCTCTCCTACT | |
| ZM_12 | ATTGCTGTGCAATCCTTATC | ATCTCTTTCTACCACCACGTT | |
| ZM_13 | GCAGAAGGCAATAAAGTCAT G | GCGTCAGAAGAAAAATACTG | |
| ZM_14 | GGAAGGCGAGTTGATAGATAA | CATGGGATGTTCAAAGAACT | |
| ZM_17 | CTTGCTTCCTCTTTTCTCTCT | ACACTGTACTCAGCGGATTT | |
| ZM_18 | AATACCCTTCAGTATTCAGGTG | CAACAACACAAACACTGCTAC | |
| ZM_20 | GGGATGTTGATAGAGATGTTG | TCTTTCACTCTCACACACACA | |
| ZM_21 | CTCTCTCTCTCTGCTGTTTCA | GCCATACGATCTCAAAATCAC | |
| ZM_22 | ACCACCGATCTACTCACTTTT | CCACTGCACACTACAGTTTTT | |
| ZM_30 | CACTCCACTCATTATCCAAAG | CAAGACACAACTGACACGTAA | |
| ZM_34 | AAGTCCCTTTTCAAGCAATC | GAGAGAGGAAAATGCAGAGAG | |
| ZM_39 | AGAGGCAGAGGAGTTGATAAT | CTTAACTGTAACTCCCTTTTCG | |
| ZM_40 | CGAAAAGGGAGTTACAGTTAAG | CTTCCTCTCCTATCATCCTGT | |
| ZM_44 | GTCTTAAGCCCTCTTAGTTCC | GAAAACCTTCAATGTCAGGA | |
| ZM_45 | GCAAAATCTCTGTTGTCTCAG | GTGTTCCTACCACTCAACACA | |
| ZM_47 | GTTTCCAGGTCTATTCCTTTG | AGGTAGAGCTAATCCTTACCG | |
| Traits | Name of QTL | Markers Type | Marker Code/Number | Mapping Population | Reference |
|---|---|---|---|---|---|
| Production Enhancement | |||||
| Grain yield | Qgn-1, Qgn-6, | SLAF | 9378 | 150 BC1 | [96,122,123] |
| Number of seeds per capsule | Qgn-12 | ||||
| 1000-seed weight | Qtgw-11 | ||||
| Seed coat color | QTL-1, QTL11-1, QTL11-2, QTL13-1 | ||||
| Seed coat color | qSCa-8.2, qSCb-4.1, qSCb-8.1, qSCb-11.1, qSCl-4.1, qSCl-8.1, qSCl-11.1, qSCa-4.1 and qSCa-8.1 | SLAF SNP | 1233 | 107 F2 430 recombinant inbred lines (RILS, F8) | - |
| Seed coat color | SiPPO (SIN_1016759) | SSR | 400 | 500 RILs (F6) | [35] |
| Plant height | Qph-6 and Qph-12 | SNP | 1,800,000 | 705 worldwide accessions | [61] |
| Semi-dwarf plant phenotype | QTL (qPH-3.3), Gene SiGA20ox1 (SIN_1002659) | SNP SSR | 400 | 430 RILS (F8) 500 RILs (F6) | [35,124] |
| Plant height | SiDFL1 (SIN_1014512) andSiILR1 (SIN_1018135) | SNP | 1,800,000 | 705 worldwide accessions | [61] |
| Number of capsules per plant | Qcn-11 | SNP SSR InDels | 1190 22 18 | 224 (RIL), F8:9 | [125] |
| First capsule height | Qfch-4, Qfch-11, and Qfch-12 | ||||
| Capsule axis length | Qcal-5 and Qcal-9 | ||||
| Capsule length | Qcl-3, Qcl-4, Qcl-7, Qcl-8, and Qcl-12 | ||||
| Number of capsules per axil | SiACS (SIN_1006338) | SNP | 1,800,000 | 705 worldwide accessions | [61] |
| Mono flower vs. triple flower | SiFA | SLAF (Marker58311, Marker34507, Marker36337) | 9378 | 150 BC1 | [123] |
| Flowering time | SiDOG1 (SIN_1022538) and SiIAA14 | SNP | - | 705 sesame accessions | [61] |
| Determinate growth habit | Gene SiDt (DS899s00170.023) | NP | 30,193 | 120 F2 | [126] |
| Branching habit | SiBH | SLAF (Marker129539, Marker41538, Marker31462) | 9378 | 150 BC1 | [126] |
| Recessive GMS | Recessive GMS geneSiMs1 | AFLP markers P01MC08, P06MG04, P12EA14 | - | 237 NILs (near-isogenic lines) | [127] |
| Dominant GMS | SBM298 and GB50 | SSR | 1500 | Noval GMS line W1098A (backcrossing and sib-mating); BC2F6 | [128] |
| Stress-Related | |||||
| Waterlogging tolerance | qEZ09ZCL13, qWH09CHL15, qEZ10ZCL07, qWH10ZCL09, qEZ10CHL07, and qWH10CHL09 | SSR (ZM428) closely linked toqWH10CHL09 | 113 | 206 RIL F6 | [122,129] |
| Drought tolerance | TF (transcription factor) families (AP2/ERF and HSF) | - | - | - | [130,131] |
| Tolerance to drought, salinity, oxidative stresses, and charcoal rot | Osmotin-like gene (SindOLP) | - | - | - | [132] |
| Gene for Oil Traits | |||||
| Sesamin production | SiDIR (SIN_1015471), SiPSS (SIN_1025734) | SNP | 1,800,000 | 705 worldwide accessions | [61] |
| Oil content | SIN_1003248, SIN_1013005, SIN_1019167, SIN_1009923 SiPPO (SIN_1016759) SiNST1 (SIN_1005755) | ||||
| Fatty acid composition | SiKASI (SIN_1001803), SiKASII (SIN_1024652), SiACNA (SIN_1005440), SiDGAT2 (SIN_1019256), SiFATA (SIN_1024296), SiFATB (SIN_1022133), SiSAD (SIN_1008977), SiFAD2 (SIN_1009785) | ||||
| Sesamin and sesamolin content | SiNST1 (SIN_1005755) | ||||
| Protein content | SiPPO (SIN_1016759) | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teklu, D.H.; Shimelis, H.; Abady, S. Genetic Improvement in Sesame (Sesamum indicum L.): Progress and Outlook: A Review. Agronomy 2022, 12, 2144. https://doi.org/10.3390/agronomy12092144
Teklu DH, Shimelis H, Abady S. Genetic Improvement in Sesame (Sesamum indicum L.): Progress and Outlook: A Review. Agronomy. 2022; 12(9):2144. https://doi.org/10.3390/agronomy12092144
Chicago/Turabian StyleTeklu, Desawi Hdru, Hussein Shimelis, and Seltene Abady. 2022. "Genetic Improvement in Sesame (Sesamum indicum L.): Progress and Outlook: A Review" Agronomy 12, no. 9: 2144. https://doi.org/10.3390/agronomy12092144
APA StyleTeklu, D. H., Shimelis, H., & Abady, S. (2022). Genetic Improvement in Sesame (Sesamum indicum L.): Progress and Outlook: A Review. Agronomy, 12(9), 2144. https://doi.org/10.3390/agronomy12092144







